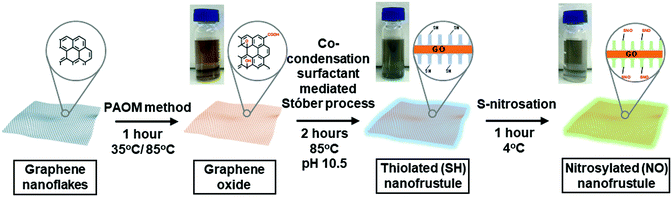
Diatom-inspired 2D nitric oxide releasing anti-infective porous nanofrustules
Hong Kit
Lim†
a,
Shao Jie
Tan†
a,
Zhuoran
Wu
a,
Boon Chong
Ong
 a,
Kwan Wee
Tan
a,
Kwan Wee
Tan
 a,
Zhili
Dong
a,
Zhili
Dong
 a and
Chor Yong
Tay
a and
Chor Yong
Tay
 *abc
*abc
aSchool of Materials Science and Engineering, Nanyang Technological University, 50 Nanyang Avenue, Singapore 639798, Singapore. E-mail: cytay@ntu.edu.sg
bSchool of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore 637551, Singapore
cEnvironmental Chemistry and Materials Centre, Nanyang Environment & Water Research Institute, 1 Cleantech Loop, CleanTech One, Singapore 637141, Singapore
First published on 7th May 2021
Abstract
Two-dimensional (2D) nanomaterials (NM) have emerged as promising platforms for antibacterial applications. However, the inherent “flatness” of 2D NM often limits the loading of antimicrobial components needed for synergistic bactericidal actions. Here, inspired by the highly ornamented siliceous frustules of diatoms, we prepared 2D ultrathin (<20 nm) and rigid “nanofrustule” plates via the out-of-plane growth of cetyltrimethylammonium bromide (CTAB) directed silica mesostructures on the surfaces of 2D graphene oxide nanosheets. The nanofrustules were characterized by the presence of mesoporous channels with a pore size of 3 nm and a high specific surface area of 674 m2 g−1. S-nitrosothiol-modification on the silica surfaces enables the development of a novel anti-infective nitric oxide (NO) releasing NO-nanofrustule system. The cage-like mesoporous silica architecture enabled a controlled and sustainable release of NO from the NO-nanofrustules under physiological conditions. The NO-nanofrustules displayed broad antibacterial effects against Staphylococcus aureus and Escherichia coli with a minimum inhibitory concentration of 250 μg ml−1. Mechanistic studies revealed that the antibacterial property of NO-nanofrustules was attained via a unique “capture-and-release” mode-of-action. The first step entailed the capture of the bacteria by the NO-nanofrustules to form micro-aggregates. This was followed by the release of high levels of NO to the captured bacteria to elicit a potent anti-infective effect. In combination with the lack of cytotoxicity in human dermal cells, the 2D hybrid NO-nanofrustules may be utilized to combat wound infections in clinical settings.
Introduction
Bacterial pathogenicity is a relentless threat to the global public health. Due to the rapid climate change, globalization and urbanization, there is a significant rise in the number of deaths due to pathogens and infections with major public health repercussions in the past decades.1 Based on current trend, infections are projected to kill 10 million people by 2050.2 Currently, the use of a single or combinatorial prescription of antibiotics remains the weapon of choice against bacterial infections. Unfortunately, many years of antibiotics misuse have led to the emergence of drug-resistant “superbugs” which has undermined existing anti-infective treatments.3 Therefore, the development of novel antibiotic-free strategies to combat pathogenic bacteria is urgently needed.In the last two decades, numerous classes of nanomaterials such as 0D nanoparticles (NPs),4 1D nanorods5 and more recently, 2D planar materials6,7 with intrinsic antimicrobial activities have been explored extensively to combat bacterial infections. Compared to antibiotics, the use of nanomaterials is less likely to elicit antimicrobial resistance due to their multiple simultaneous modi operandi.8 Owing to the ease of synthesis, tunable size, tailorable chemistries, 2D nanomaterials such as graphene oxide (GO),9 reduced graphene oxide (rGO),10 transition metal dichalcogenides11 (TMD) and MXenes12 have emerged as promising novel anti-infective candidates. 2D nanomaterials were reported to elicit potent antimicrobial effects through physicochemical means. For instance, the high surface area and reactivity of 2D nanomaterials can exert chemical pressure via overproduction of intracellular reactive oxygen species (ROS) through electron transfer13 or material surface imperfections,14 as well as physical disruption of the bacteria cell membrane.9
However, 2D antimicrobial nanomaterials are not omnipotent; the clinical adoption of 2D nanomaterials is currently curtailed due to their toxic effects in mammalian cells when used at bactericidal concentrations.15 Furthermore, if we solely capitalize on the NP properties such as shape, surface charge and size for antibacterial activity, various optimizations are required to facilitate the cell membrane penetration16 and/or uptake.17 Bacterial uptake of NPs can be difficult especially when dealing with a biofilm.18 At the same time, most of the 2D nanomaterials tend to self-aggregate in biological milieu due to the “plate-plate DLVO interaction”,19 thereby further reducing their antibacterial efficacy as a result of reduced bacteria uptake20 and may even confer resistance.21 Therefore, many studies in this field strive to incorporate other active antimicrobial components, such as peptides,22 drugs23,24 or small molecules25 to the 2D nanomaterials, aiming for synergistic bacteria-killing effects.
Nitric oxide (NO) is a diatomic, free-radical gas that is naturally produced in the body and plays a major role in innate immunity. In response to an infection, NO is produced in large quantities by macrophages via the NO synthase isoforms (iNOS/NOS2) that spontaneously react with O− containing superoxides to generate a variety of antimicrobial molecules in the form of reactive nitrogen species such as such as dinitrogen trioxide (N2O3), nitroxyl (HNO) and peroxynitrite (ONOO−).26 These powerful oxidants can induce irreparable damage to bacterial proteins through nitrosation of reactive thiols and amines, as well as DNA materials.27 NO displays non-specific broad-spectrum antimicrobial activity against a wide range of Gram-positive and Gram-negative bacteria such as Pseudomonas aeruginosa (P. aeruginosa),28Staphylococcus aureus (S. aureus),29Escherichia coli (E. coli),30Acinetobacter baumannii (A. baumannii),31Staphylococcus epidermidis (S. epidermidis),32 and even the Methicillin-resistant Staphylococcus aureus (MRSA).33 NO passes through most physiological barriers easily at the targeted site for effective antimicrobial action.34 As such, the choice of a planar structure to serve as a “mothership” for NO delivery offers a distinctive advantage as this type of structure enables greater interaction with bacteria before releasing the high NO payload.
In this study, we report the development of a novel NO releasing 2D hierarchical porous nanocomposite that is inspired by the bio-silicification process and the ultra-structured “anatomy” of the diatom siliceous cell shells, also called frustules. The process of bio-silicification entails the precise integration of silica materials into an organic matrix to form nanoscale composites with exquisite morphologies.35 Herein, GO was used as the organic template for the growth of surfactant-directed silica to create a mesoporous heterostructure 2D nanomaterial that will enable us to maximize the loading capacity of the pharmacological active S-nitrosothiol (SNO) moiety as an NO source. Because the resultant silica architecture emulates the highly ornamented silica frustules of diatoms, we term this class of porous 2D nanomaterials as 2D “nanofrustule” plates.36 The physiochemical properties such as nanoscale surface topography, porosity and NO loading capacity were systematically characterized. As an antimicrobial agent, the design and working principle of nitrosylated nanofrustule (NO-nanofrustule) are premised on the “capture and release” concept – it is envisioned that once the NO-nanofrustules have successfully docked onto the surface of the bacterial wall, release and diffusion of cell wall-permeable NO into the target cells can occur to exert a potent bactericidal effect. In contrast to the GO based antimicrobial agents, the presence of the mesoporous silica bilayer confers rigidity and structural stability to the nanofrustules, ensuring maximal presentation of the material surface to seek out and capture the bacteria.37 In addition, unlike conventional nanomedicine strategies,38 the bacteria-killing effect of nanofrustules does not require active nanomaterials uptake by the microorganisms. As a proof-of-concept, the anti-microbial activity of NO-nanofrustules was evaluated in S. aureus and E. coli. Furthermore, the biocompatibility of NO-nanofrustules was also validated with keratinocytes and dermal fibroblasts of human origin.
Results and discussion
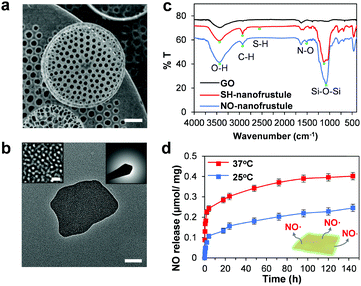
Fig. 1 shows the synthesis scheme of the nanofrustules. Graphene nanopowder with an average planar size of 500 nm to 2 μm range was chosen as the base template for oxidation, whereby the previously developed Preformed Acidic Oxidizing Medium (PAOM) method was used to produce highly oxidized GO sheets.39 Afterwards, the GO sheets were subjected to the modified Stöber method involving co-condensation of two different silica sources, i.e., TEOS and MPTMS, to form the thiol-functionalized (SH) nanofrustules with the mesoporous silica layers covering the top and bottom faces of the GO sheet. The final ratio of the two silica sources was optimized in accordance to an earlier study.40 Finally, the final NO-loaded nanofrustules were formed via S-nitrosation which involves the formation of the S–NO bond from the thiol group. Both SH- and NO-functionalized nanofrustules displayed good dispersibility in water, forming a uniform suspension after 1–2 minutes of ultrasonication.Fig. 2a shows the representative SEM image of Coscinodiscus sp., a genus of centric diatom that is characterized by a distinct honeycomb-shaped array of pores covering the valve surface that is believed to maximize contact with the extracellular environment for efficient nutrient and gas exchange.41 In the case of the synthetic NO-nanofrustule, similar periodic nanoscale 2D pore arrays were grown on the surfaces of the GO template. The as-synthesized CTAB-directed mesoporous silica structure had an average pore size of around 3 nm as shown in Fig. 2b. This contrasts with the TEM image of GO nanosheets which depicts a smooth and non-textured surface (see Fig. S1 in the ESI†). The mesoporous characteristics of the SH-nanofrustules were confirmed in nitrogen sorption measurements, displaying a Type IV isotherm with a hysteresis loop (see Fig. S2 in the ESI†). BET specific surface area of nanofrustules obtained from the nitrogen sorption analysis was 674 m2 g−1, which is significantly higher compared to dried graphite oxide (20–40 m2 g−1)42 and mesoporous silica nanoparticles (18–630 m2 g−1).43 Based on the TEM images of the synthesized products, the nanofrustules not only appeared to be significantly thicker than the GO nanosheets after silicification, but also smaller in terms of the average planar size (see Fig. S1 in the ESI†). Worm-like mesopores were also observed on the surface of the nanofrustules, which is in agreement with the mesoporous structure obtained using a similar synthesis method.44 The pore size of the mesopores was around 3 nm, when measured using BJH analysis (see Fig. S3 in the ESI†).
Shown in Fig. 2c are the FITR spectra of the GO, as well as thiolated (SH) and nitrosylated (NO) nanofrustules. After conjugation of silica and thiol functional groups, the characteristic peaks at approximately 451 cm−1 (Si–O–Si asymmetric stretch), 564 cm−1 (C–S stretch), 694 cm−1 (C–Si stretch), 1128 cm−1 (Si–O–Si asymmetric stretch), 1450 cm−1 (C–H bending in methyl group), 2560 cm−1 (S–H stretch) and 2930 cm−1 (C–H stretch in methyl group) appeared in the nanofrustules samples while they were absent in GO. This indicates that the thiol-functionalized silica was successfully conjugated onto the GO template. Successful thiol conjugation in the nanofrustules was further validated by the quantitative Ellman assay, where we found that the free thiol group concentration in the SH nanofrustules was 0.41 ± 0.08 μmol mg−1. The reduction in wavenumber for the peaks at 1058 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch) and 1384 cm−1 (edge C–C vibration) after conjugation indicates that the C
O stretch) and 1384 cm−1 (edge C–C vibration) after conjugation indicates that the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and edge C bonds in the graphene sp2 structure became weaker. This may be due to the added silica layer covering both sides of the GO nanosheet, therefore reducing the intensity of π–π stacking between two graphene layers of the nanofrustules. In addition, the characteristic peak at approximately 1520 cm−1 (N–O stretch) for NO-nanofrustule samples confirmed the presence of the nitroso functional group.
O and edge C bonds in the graphene sp2 structure became weaker. This may be due to the added silica layer covering both sides of the GO nanosheet, therefore reducing the intensity of π–π stacking between two graphene layers of the nanofrustules. In addition, the characteristic peak at approximately 1520 cm−1 (N–O stretch) for NO-nanofrustule samples confirmed the presence of the nitroso functional group.
Next, to elucidate the NO releasing capability of the NO-nanofrustules, the amount of NO liberated in phosphate buffer solution (PBS) was measured using the Griess reagent method. Fig. 2d shows the time-dependent release of NO at 25 °C and 37 °C. In both experimental groups, we observed an initial burst release of NO in the first 2 hours, followed by a near zero-order sustained release over a period of 4 days. The half-life of NO release from NO-nanofrustules was approximately 1.5 hours. The initial burst NO release may be caused by the spontaneous nature of the S–NO bond cleavage, accompanied by an increase in the local temperature as the suspension equilibrates to 37 °C, which is consistent with the literature.40 The prolonged NO release may be attributed to the “cage effect” of the mesoporous silica structure of NO-nanofrustules, whereby after the homolytic cleavage of the S–NO bond, the resultant thiyl radical and NO radical tend to recombine in the tightly confined spaces of the mesopores.45 The release profile could be modelled using the Korsmeyer–Peppas model for drug release,46 whereby a best-fit straight line could be obtained by plotting log of cumulative percentage of NO release vs. log time. The gradient of the best fit line is m = 0.2891 with a correlation coefficient of 0.9797 (see Fig. S3 in the ESI†). In this model, a value of m equal to or smaller than 0.5 describes a Fickian diffusion release mechanism.47 However, the cumulative amount of NO released at the end of 4 days in 37 °C (0.41 μmol mg−1) was significantly higher than the 25 °C (0.25 μmol mg−1) experimental set, suggesting that the NO-nanofrustule has moderate thermal stability and moderate S–NO bond energy.
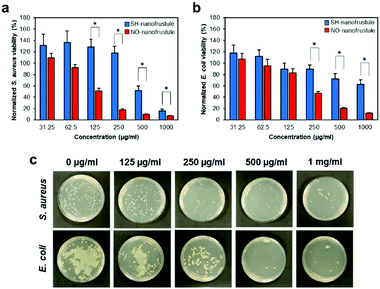
Taking into consideration the half-life of the NO release i.e., 1.5 hours, we conducted the antibacterial broth dilution assay by exposing it to 0.0625 to 1 mg ml−1, in 2-fold increments, of NO-nanofrustules for 2 hours. The corresponding results from the assay are shown in Fig. 3a and b. To decouple the antibacterial contributions from NO from the effects of the 2D nanostructure, SH-nanofrustules were used as an internal control. As expected, both 2D nanomaterials displayed a dose-dependent killing profile against both types of the bacteria. This observation is consistent with numerous reports on the innate anti-microbial properties of 2D nanomaterials. However, compared to SH-nanofrustules, NO-nanofrustules displayed a significant enhancement in the antibacterial efficacy, with a half minimum inhibitory concentration (MIC50) of 250 μg ml−1 in both the S. aureus and E. coli groups. In contrast, the bacteria killing efficacy of SH-nanofrustules was clearly lower as indicated by a MIC50 of 1 mg ml−1. Furthermore, the minimum bactericidal concentration (MBC) of NO-nanofrustules, whereby more than 99% of bacteria were killed, was determined to be 4 mg ml−1 (data not shown). To further demonstrate the antibacterial potential of NO-nanofrustules, a diluted fraction (1/10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) of the surviving bacteria following the 2 hours treatment of various concentrations of the bacteria were plated on agar plates for 24 hours. Fig. 3c shows that the number of viable bacteria colonies of both S. aureus and E. coli groups decreased in a dose-dependent manner, until 1 mg ml−1, whereby agar regrowth of the bacteria was minimum. Therefore, the results show that the NO-nanofrustules have potent non-specific antibacterial activity, in comparison to their predecessor SH-nanofrustules with mild antibacterial activity.
000) of the surviving bacteria following the 2 hours treatment of various concentrations of the bacteria were plated on agar plates for 24 hours. Fig. 3c shows that the number of viable bacteria colonies of both S. aureus and E. coli groups decreased in a dose-dependent manner, until 1 mg ml−1, whereby agar regrowth of the bacteria was minimum. Therefore, the results show that the NO-nanofrustules have potent non-specific antibacterial activity, in comparison to their predecessor SH-nanofrustules with mild antibacterial activity.
Comparing the antibacterial activity of NO-nanofrustules with other similar published materials in retrospect, the best-performing NO-releasing silica nanorods by Yuan's group required at least 70 μM of total NO dose to kill S. aureus,48 while the NO-nanofrustules required less than 50 μM of total released NO to kill S. aureus. Similarly, Grommersch's group who used real diatom frustules bio-templated with S-nitrosothiols achieved good antibacterial efficacy at relatively high concentrations, i.e., in the order of 1–10 mg ml−1.49
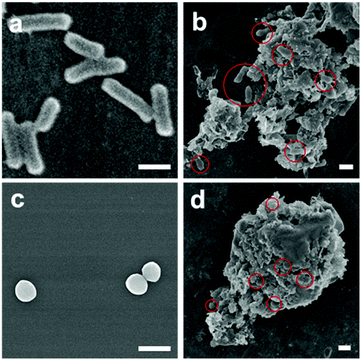
The high antibacterial efficacy of NO-nanofrustules sets the stage for further investigation in the mechanism of bacteria killing by this material. We first hypothesize that due to the high surface area of NO-nanofrustules, it is an excellent platform to trap or interact with the bacteria in the proximity until the localized NO flux is large enough to kill them. FESEM images of the bacteria colonies treated with the NO-nanofrustules confirmed this hypothesis. Fig. 4a and c show the morphology of normal E. coli and S. aureus under SEM, respectively, prior to treatment. Fig. 4b and d show that after treatment with 250 μg ml−1 of NO-nanofrustules, they form micrometer-scale nanomaterial-bacteria aggregates. As a result, almost all the bacteria were observed to interact extensively with the nanomaterials, in such a way that the local NO flux can quickly increase to a lethal level. In addition, some indistinguishable features, other than the flake-like nanofrustules or distorted bacteria, were observed, possibly consisting of the extracellular polymeric matrix (EPS) secreted by the bacteria as a protective attempt against stress.50
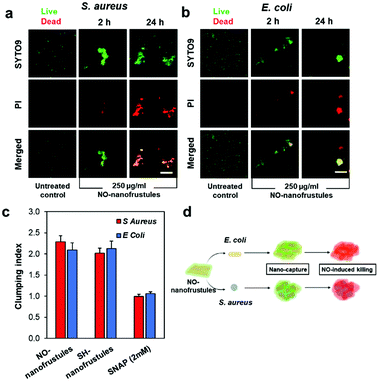
To visualize how the bacteria were killed by NO-nanofrustules, we next conducted a BacLight™ bacterial viability fluorescence assay on the mixture of NO-nanofrustules and bacteria. In Fig. 5a and b, the green and red spots represent the state of bacteria as alive and dead, respectively. The clump appeared as an overlap of slightly brighter green or red spots, representing the bacteria and the aggregated layered mass of the NO-nanofrustules. This observation suggests that most of the bacteria trapped within the clumps were dead. Compared to the images at 2 hours post-treatment, the images at 24 hours post-treatment show that the bacteria were killed in a much greater extent in the NO-nanofrustules treated samples. In contrast, the untreated control displayed mainly green (live) individual free-floating bacteria.
Based on the analysis of the fluorescence BacLight™ assay images, the extent of aggregation of the observed aggregated clumps can be quantified by defining a term called clumping index (CI). The CI is defined as the fraction of observable fluorescent area with a maximum Feret diameter greater than 0.015. As shown in Fig. 5c, the bacterial samples treated with both SH-nanofrustules and NO-nanofrustules showed similar CIs, both values of which were higher than those of the untreated and samples treated with 2 mM of S-nitroso-n-acetylpenicillamine (SNAP), a small molecular donor of NO and a widely recognized antimicrobial.51 No significant difference was found between S. aureus and E. coli in terms of CI, indicating that the formation of aggregated clumps is non-specific to the Gram type or the membrane structure of the bacteria.
Taken together, our findings suggest a unique two-step mechanism of action of NO-nanofrustules. The first step, illustrated in Fig. 5d, involves the tendency of bacteria to adsorb to the nanofrustule surface, together with the size-dependent “canvas effect” which helped to capture and wrap surrounding bacteria forming clumps. This is then followed by subsequent killing of the trapped bacteria due to the high NO flux to elicit a potent bactericidal effect. This clump-forming aggregation process is most likely to be thermodynamically and chemically driven. The bacteria were able to adsorb spontaneously to the nanofrustules due to the porous architecture which provides sufficient interactive anchorage points at the bacteria–nanomaterial interface, which resembles the initial step of biofilm formation. However, in our case, instead of being able to propagate healthily on the surface, the adsorption process is clearly an act of suicide for the bacteria due to the high NO flux on the nanofrustule surface. Our approach is markedly different from using conventional antibacterial NO-releasing nanoparticles, which rely on the bacteria as the anchorage point and subsequent bacterial uptake.52 Notably, other types of 2D nanomaterials such as GO,53 MoS26 and MXenes12 with a similar lateral size range also demonstrate the specialty of wrapping and capturing bacteria. Further evidence also points to a van der Waals force dominated interaction between amorphous colloidal silica nanoparticles and the dioleoyl phosphatidylcholine (a component of bio-membranes) monolayer, independent of the particle size, pH, and the ionic strength of the solution,54 hence ruling out the possibility of electrostatic nature of the interaction. Furthermore, a silica substrate with a nanostructured rough surface or porous surface has higher adhesion force with bacteria compared to the non-porous surface, as quantified by surface morphometry.55,56 As for the observed shape distortion in both types of bacteria, because GO sheets with lateral sizes of more than 1 μm are found to wrap and trap bacteria, causing bacterial cells to be intertwined together alongside a perturbation to the bacterial membrane potential,57 the nanofrustules with a flake-like structure may have inherited this specialty from their GO predecessor. Another possible explanation for this aggregation process is the previously reported “phase-to-size transformation” as seen in thermal responsive polymer brush-conjugated Zn-doped carbon nanosheets, involving a thermal-driven hydrophilic-to-hydrophobic switching of the nanosheet surface. This transformation was found to improve trapping and wrapping-induced local bacteria eradication. However, the transformation of our study is not as drastic because the attempt to alter the hydrophilicity state using external electromagnetic radiation such as NIR has not been employed in this study.
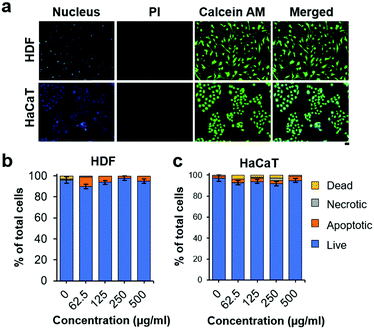
Biocompatibility must be established for the exploitation of NO-nanofrustules to combat infections. We first examined the viability of NO-nanofrustules treated human dermal fibroblasts (HDF) and human keratinocytes (HaCaT) using the Calcein AM/PI Live/Dead staining assay. In Fig. 6a, the assay highlighted that when exposed to NO-nanofrustules at the MIC50 (250 μg ml−1) for 24 hours, majority of the cells remained viable (green), while the occurrence of PI-stained cells (red) was negligible. Furthermore, the nanomaterials treated cells retained their characteristic cellular morphologies of epithelial cells (HaCaT) and fibroblasts (HDF), suggesting that those cells remained healthy.
Consistently, for both cell types, the annexin V/PI cell viability assay results in Fig. 6b show that more than 90% of the cells were viable, with less than 5% undergoing apoptosis, when treated with up to 500 μg ml−1 of NO-nanofrustules for 2 hours, which is within the time scale of antibacterial studies. The observations are in line with previous findings in the literature, as a similar dose of NO-releasing particles can be detrimental to bacteria but not mammalian cells due to sheer differences in size, uptake mechanism and membrane structures.52
The results led us to conclude that NO-nanofrustules are cytocompatible with human dermal cells and shows great potential to be used for skin-related anti-infective therapies such as treatment of lesions in wounds and eczema. It is a proven strategy to use such nanomaterials under inflammatory conditions, whereby the nanomaterials can be absorbed on the surface of pathogenic microbes, not only disrupting any imminent biofilm formation but also blocking the active membrane fragments and adhesion factors, finally leading to their clearance from the wound.58
Conclusions
In summary, we developed novel diatom-inspired NO-releasing nanofrustules with high efficacy in eradicating Gram-positive S. aureus and Gram-negative E. coli bacteria. Besides showing the non-specific broad-spectrum lethality of the material, we also elucidated the mechanism of bacteria killing which leverages on the entropy driven aggregation behavior of the unique 2D porous architecture. The aggregation not only helps to capture and trap the bacteria in the process, but also creates a favorable environment for NO diffusive action. Importantly, NO-nanofrustules were observed to elicit negligible cytotoxicity within our experimental conditions, suggesting that the platform is biocompatible for future clinical exploitation. In the broader context, our nano-enabled “capture-and-release” bactericidal strategy can be extended to design a range of multifunctional 2D nanomaterials to combat wound infections.Experimental
Materials
The following materials were used without any further modifications, unless stated otherwise. Sulfuric acid (H2SO4, 98%), phosphorus pentoxide (P2O5), potassium permanganate (KMnO4), sodium nitrate (NaNO3), hydrogen peroxide (30%, H2O2), cetyltetraammonium bromide (CTAB), tetraethyl orthosilicate (TEOS), 3-mercaptopropyl trimethoxysilane (MPTMS), sodium nitrite (NaNO2), sodium hydroxide (NaOH) and S-nitroso-n-acetylpenicillamine (SNAP) were purchased from Sigma-Aldrich. Pentetic acid (DTPA) was purchased from Ark Pharm. Graphene nanopowder (0541DX) was purchased from SkySpring Nanomaterials. Anhydrous ethanol (99.9%, EtOH) and anhydrous methanol (99.9% MeOH) were purchased from Aik Moh Pte Ltd.Synthesis procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) minutes before cooling to room temperature. 5 ml of H2O2 was added dropwise to the flask and a bright yellow coloration was observed indicating the presence of highly oxidized GO. To remove metal impurities, the flask's contents were washed thrice with 1 M HCl and twice with DI water. The resultant GO dispersion was then dialyzed in DI water using a cellulose membrane dialysis tube (10 kDa, Thermo Scientific) for at least 72 hours, changing the dialysate at least once every 12 hours. The neutralized, dialyzed solution was then centrifuged at 8500 rpm for 5 minutes to increase the lateral size homogeneity of GO. Part of the dialysis product was then lyophilized for 24 hours to obtain solid GO nanoflakes.
minutes before cooling to room temperature. 5 ml of H2O2 was added dropwise to the flask and a bright yellow coloration was observed indicating the presence of highly oxidized GO. To remove metal impurities, the flask's contents were washed thrice with 1 M HCl and twice with DI water. The resultant GO dispersion was then dialyzed in DI water using a cellulose membrane dialysis tube (10 kDa, Thermo Scientific) for at least 72 hours, changing the dialysate at least once every 12 hours. The neutralized, dialyzed solution was then centrifuged at 8500 rpm for 5 minutes to increase the lateral size homogeneity of GO. Part of the dialysis product was then lyophilized for 24 hours to obtain solid GO nanoflakes.
Characterization methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 and pressed to form disks.
20 and pressed to form disks.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× with fresh LB medium and plated on agar as a part of the agar dilution assay.
000× with fresh LB medium and plated on agar as a part of the agar dilution assay.
For the broth microdilution assay, 10 μl of the PrestoBlue reagent (Invitrogen) was added to each well and incubated for 1 hour. Fluorescence readings were taken with a spectrophotometer at an excitation wavelength of 560 nm and an emission wavelength of 590 nm; the bacterial cell viability was calculated as a percentage of fluorescence intensity I of the sample compared to that of the negative control in eqn (1).
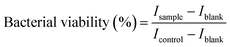 | (1) |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge support from the Singapore's Ministry of Education (MOE) Academic Research Fund Tier 1 (RG38/20 2020-T1-001-152) and the Nanyang Technological University (NTU)-Harvard School of Public Health (HSPH) Initiative for Sustainable Nanotechnology Program (NTU-HSPH 18002). The authors would also like to acknowledge the Facility for Analysis, Characterization, Testing and Simulation (FACTS), Nanyang Technological University, Singapore, for the use of their electron microscopy facilities.References
- S. M. Diene and J. M. Rolain, Clin. Microbiol. Infect., 2014, 20, 831–838 Search PubMed.
- L. S. J. Roope, R. D. Smith, K. B. Pouwels, J. Buchanan, L. Abel, P. Eibich, C. C. Butler, P. S. Tan, A. S. Walker, J. V. Robotham and S. Wordsworth, Science, 2019, 364 Search PubMed.
- D. Ferber, Science, 2000, 288, 792–794 Search PubMed.
- G. Zhu, Z. Sun, P. Hui, W. Chen and X. Jiang, ACS Biomater. Sci. Eng., 2020, 7, 1827–1835 Search PubMed.
- Y. Zhu, M. Ramasamy and D. K. Yi, ACS Appl. Mater. Interfaces, 2014, 6, 15078–15085 Search PubMed.
- A. C. Ferrari, F. Bonaccorso, V. Fal'ko, K. S. Novoselov, S. Roche, P. Bøggild, S. Borini, F. H. L. Koppens, V. Palermo and N. Pugno, et al. , Nanoscale, 2015, 7, 4598–4810 Search PubMed.
- G. Sai-Anand, A. Sivanesan, M. R. Benzigar, G. Singh, A.-I. Gopalan, A. V. Baskar, H. Ilbeygi, K. Ramadass, V. Kambala and A. Vinu, Bull. Chem. Soc. Jpn., 2019, 92, 216–244 Search PubMed.
- L. Wang, C. Hu and L. Shao, Int. J. Nanomed., 2017, 12, 1227–1249 Search PubMed.
- S. Liu, T. H. Zeng, M. Hofmann, E. Burcombe, J. Wei, R. Jiang, J. Kong and Y. Chen, ACS Nano, 2011, 5, 6971–6980 Search PubMed.
- T. Sreeprasad, M. S. Maliyekkal, K. Deepti, K. Chaudhari, P. L. Xavier and T. Pradeep, ACS Appl. Mater. Interfaces, 2011, 3, 2643–2654 Search PubMed.
- T. I. Kim, B. Kwon, J. Yoon, I. J. Park, G. S. Bang, Y. Park, Y. S. Seo and S. Y. Choi, ACS Appl. Mater. Interfaces, 2017, 9, 7908–7917 Search PubMed.
- R. P. Pandey, P. A. Rasheed, T. Gomez, K. Rasool, J. Ponraj, K. Prenger, M. Naguib and K. A. Mahmoud, ACS Appl. Nano Mater., 2020, 3, 11372–11382 Search PubMed.
- H. Zhao, C. Zhang, Y. Wang, W. Chen and P. J. J. Alvarez, Environ. Sci. Technol., 2018, 52, 12783–12791 Search PubMed.
- F. Perreault, A. F. de Faria, S. Nejati and M. Elimelech, ACS Nano, 2015, 9, 7226–7236 Search PubMed.
- W. Lu, K. Yao, J. Wang and J. Yuan, J. Colloid Interface Sci., 2015, 437, 35–41 Search PubMed.
- K. H. Tan, S. Sattari, S. Beyranvand, A. Faghani, K. Ludwig, K. Schwibbert, C. Böttcher, R. Haag and M. Adeli, Langmuir, 2019, 35, 4736–4746 Search PubMed.
- B. Ahmed, B. Solanki, A. Zaidi, M. S. Khan and J. Musarrat, Toxicol. Res., 2019, 8, 246–261 Search PubMed.
- S. Fulaz, S. Vitale, L. Quinn and E. Casey, Trends Microbiol., 2019, 27, 915–926 Search PubMed.
- T. M. Mohona, A. Gupta, A. Masud, S.-C. Chien, L.-C. Lin, P. C. Nalam and N. Aich, Environ. Sci. Technol., 2019, 53, 4161–4172 Search PubMed.
- S. Su, J. Wang, E. Vargas, J. Wei, R. Martínez-Zaguilán, S. R. Sennoune, M. L. Pantoya, S. Wang, J. Chaudhuri and J. Qiu, ACS Biomater. Sci. Eng., 2016, 2, 1357–1366 Search PubMed.
- A. Panáček, L. Kvítek, M. Smékalová, R. Večeřová, M. Kolář, M. Röderová, F. Dyčka, M. Šebela, R. Prucek, O. Tomanec and R. Zbořil, Nat. Nanotechnol., 2018, 13, 65–71 Search PubMed.
- E. de Alteriis, V. Maselli, A. Falanga, S. Galdiero, F. M. Di Lella, R. Gesuele, M. Guida and E. Galdiero, Infect. Drug Resist., 2018, 11, 915 Search PubMed.
- J. Xue, Y. Niu, M. Gong, R. Shi, D. Chen, L. Zhang and Y. Lvov, ACS Nano, 2015, 9, 1600–1612 Search PubMed.
- F. Han, S. Lv, Z. Li, L. Jin, B. Fan, J. Zhang, R. Zhang, X. Zhang, L. Han and J. Li, NPG Asia Mater., 2020, 12, 1–11 Search PubMed.
- S. R. Ali, S. Pandit and M. De, ACS Appl. Bio Mater., 2018, 1, 967–974 Search PubMed.
- K. T. Nguyen, Z. Wu, T. Huang and C. Y. Tay, Curr. Med. Chem., 2018, 25, 1420–1432 Search PubMed.
- F. C. Fang, J. Clin. Invest., 1997, 99, 2818–2825 Search PubMed.
- K. E. A. Darling and T. J. Evans, Infect. Immun., 2003, 71, 2341–2349 Search PubMed.
- V. F. Cardozo, C. A. C. Lancheros, A. M. Narciso, E. C. S. Valereto, R. K. T. Kobayashi, A. B. Seabra and G. Nakazato, Int. J. Pharm., 2014, 473, 20–29 Search PubMed.
- G. Narin, Ç. B. Albayrak and S. Ülkü, Appl. Clay Sci., 2010, 50, 560–568 Search PubMed.
- M. R. Mihu, U. Sandkovsky, G. Han, J. M. Friedman, J. D. Nosanchuk and L. R. Martinez, Virulence, 2010, 1, 62–67 Search PubMed.
- E. M. Hetrick, J. H. Shin, H. S. Paul and M. H. Schoenfisch, Biomaterials, 2009, 30, 2782–2789 Search PubMed.
- L. R. Martinez, G. Han, M. Chacko, M. R. Mihu, M. Jacobson, P. Gialanella, A. J. Friedman, J. D. Nosanchuk and J. M. Friedman, J. Invest. Dermatol., 2009, 129, 2463–2469 Search PubMed.
- R. B. Weller, J. Invest. Dermatol., 2009, 129, 2335–2337 Search PubMed.
- N. Kröger, Curr. Opin. Chem. Biol., 2007, 11, 662–669 Search PubMed.
- J. T. Chao, M. J. P. Biggs and A. S. Pandit, Expert Opin. Drug Delivery, 2014, 11, 1687–1695 Search PubMed.
- F. Zou, H. Zhou, D. Y. Jeong, J. Kwon, S. U. Eom, T. J. Park, S. W. Hong and J. Lee, ACS Appl. Mater. Interfaces, 2017, 9, 1343–1351 Search PubMed.
- Y. Wang, Y. Yang, Y. Shi, H. Song and C. Yu, Adv. Mater., 2020, 32, 1904106 Search PubMed.
- C. H. Chen, S. Hu, J. F. Shih, C. Y. Yang, Y. W. Luo, R. H. Jhang, C. M. Chiang and Y. Hung, Sci. Rep., 2017, 7, 3908 Search PubMed.
- D. A. Riccio, J. L. Nugent and M. H. Schoenfisch, Chem. Mater., 2011, 23, 1727–1735 Search PubMed.
- M. Mishra, A. P. Arukha, T. Bashir, D. Yadav and G. B. K. S. Prasad, Front. Microbiol., 2017, 8, 1239 Search PubMed.
- L. Li, X. L. Liu, H. Y. Geng, B. Hu, G. W. Song and Z. S. Xu, J. Mater. Chem. A, 2013, 1, 10292–10299 Search PubMed.
- M. Szekeres, J. Tóth and I. Dékány, Langmuir, 2002, 18, 2678–2685 Search PubMed.
- K. T. Nguyen, S. Sreejith, J. Joseph, T. He, P. Borah, E. Y. Guan, S. W. Lye, H. Sun and Y. Zhao, Part. Part. Syst. Charact., 2014, 31, 1060–1066 Search PubMed.
- M. J. Malone-Povolny and M. H. Schoenfisch, ACS Appl. Mater. Interfaces, 2019, 11, 12216–12223 Search PubMed.
- J. O. Kim, J.-K. Noh, R. K. Thapa, N. Hasan, M. Choi, J. H. Kim, J.-H. Lee, S. K. Ku and J.-W. Yoo, Int. J. Biol. Macromol., 2015, 79, 217–225 Search PubMed.
- P. L. Ritger and N. A. Peppas, J. Controlled Release, 1987, 5, 23–36 Search PubMed.
- Y. Lu, D. L. Slomberg, B. Sun and M. H. Schoenfisch, Small, 2013, 9, 2189–2198 Search PubMed.
- B. M. Grommersch, J. Pant, S. P. Hopkins, M. J. Goudie and H. Handa, ACS Appl. Mater. Interfaces, 2018, 10, 2291–2301 Search PubMed.
- T. Trunk, H. S. Khalil and J. C. Leo, AIMS Microbiol., 2018, 4, 140–164 Search PubMed.
- J. Pant, J. Gao, M. J. Goudie, S. P. Hopkins, J. Locklin and H. Handa, Acta Biomater., 2017, 58, 421–431 Search PubMed.
- H. Jeong, J.-H. Park, J. H. Shin, J.-C. Yoo, C. Y. Park and J. Hong, Chem. Mater., 2018, 30, 8528–8537 Search PubMed.
- O. Akhavan and E. Ghaderi, ACS Nano, 2010, 4, 5731–5736 Search PubMed.
- A. Vakurov, R. Brydson and A. Nelson, Langmuir, 2012, 28, 1246–1255 Search PubMed.
- C. Spengler, F. Nolle, J. Mischo, T. Faidt, S. Grandthyll, N. Thewes, M. Koch, F. Müller, M. Bischoff, M. A. Klatt and K. Jacobs, Nanoscale, 2019, 11, 19713–19722 Search PubMed.
- H. Song, Y. Ahmad Nor, M. Yu, Y. Yang, J. Zhang, H. Zhang, C. Xu, N. Mitter and C. Yu, J. Am. Chem. Soc., 2016, 138, 6455–6462 Search PubMed.
- J. Chen, H. Peng, X. Wang, F. Shao, Z. Yuan and H. Han, Nanoscale, 2014, 6, 1879–1889 Search PubMed.
- S. Lavie and G. Stotzky, Appl. Environ. Microbiol., 1986, 51, 65–73 Search PubMed.
- Z.-M. Wang, W. Peng, Y. Takenaka, N. Yoshizawa, K. Kosuge, W. Wang and G. A. Ozin, Adv. Funct. Mater., 2017, 27, 1704066 Search PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |