Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Targeting transdifferentiated hepatic stellate cells and monitoring the hepatic fibrogenic process by means of IGF2R-specific peptides designed in silico†
Florian
Weber
ab,
Tommaso
Casalini
 cd,
Gina
Valentino
ab,
Lorine
Brülisauer
b,
Nico
Andreas
e,
Andreas
Koeberle
cd,
Gina
Valentino
ab,
Lorine
Brülisauer
b,
Nico
Andreas
e,
Andreas
Koeberle
 fg,
Thomas
Kamradt
e,
Alessandro
Contini
fg,
Thomas
Kamradt
e,
Alessandro
Contini
 h and
Paola
Luciani
h and
Paola
Luciani
 *ab
*ab
aDepartment of Chemistry, Biochemistry and Pharmaceutical Sciences, University of Bern, Bern, Switzerland. E-mail: paola.luciani@dcb.unibe.ch
bDepartment of Pharmaceutical Technology, Institute of Pharmacy, Friedrich Schiller University Jena, Jena, Germany
cInstitute of Mechanical Engineering and Material Technology, Department of Innovative Technology, SUPSI, Manno, Switzerland
dInstitute for Chemical and Bioengineering, Department of Chemistry and Applied Biosciences, ETH Zurich, Zurich, Switzerland
eInstitute of Immunology, Jena University Hospital, Jena, Germany
fMichael Popp Institute and Center for Molecular Biosciences (CMBI), University of Innsbruck, Innsbruck, Austria
gChair of Pharmaceutical/Medicinal Chemistry, Institute of Pharmacy, Friedrich Schiller University Jena, Jena, Germany
hDipartimento di Scienze Farmaceutiche-Sezione di Chimica Generale e Organica “A. Marchesini”, Università degli Studi di Milano, Milano, Italy
First published on 15th January 2021
Abstract
The lack of accurate and easily applicable methods for the diagnosis of liver fibrosis, a disease characterized by an accumulation of the extracellular matrix released by activated hepatic stellate cells (HSCs), has been a major limitation for the clinical management of liver diseases. The identification of biomarkers specific to liver microstructure alterations, combined with a non-invasive optical imaging modality, could guide clinicians towards a therapeutic strategy. In this study, structural information of the insulin-like growth factor 2 receptor (IGF2R), an overexpressed protein on activated HSCs, was used for in silico screening of novel IGF2R-specific peptide ligands. Molecular dynamics simulations, followed by computational alanine scanning of the IGF2R/IGF2 complex, led to the identification of a putative peptide sequence containing the most relevant amino acids for the receptor–ligand interaction (IGF2 E12-C21). The Residue Scan tool, implemented in the MOE software, was then used to optimize the binding affinity of this sequence by amino acid mutations. The designed peptides and their associated scrambled sequences were fluorescently labelled and their binding affinity to LX-2 cells (model for activated human HSCs) was tested using flow cytometry and confocal microscopy. In vitro binding was verified for all sequences (KD ≤ 13.2 μM). With respect to the putative binding sequence, most mutations led to an increased affinity. All sequences have shown superior binding compared to their associated scrambled sequences. Using HPLC, all peptides were tested in vitro for their proteolytic resistance and showed a stability of ≥60% intact after 24 h at 37 °C in 50% v/v FBS. In view of their prospective diagnostic application, a comparison of binding affinity was performed in perpetuated and quiescent-like LX-2 cells. Furthermore, the IGF2R expression for different cell phenotypes was analysed by a quantitative mass spectrometric approach. Our peptides showed increased binding to the perpetuated cell state, indicating their good selectivity for the diagnostically relevant phenotype. In summary, the increased binding affinity of our peptides towards perpetuated LX-2 cells, as well as the satisfactory proteolytic stability, proves that the in silico designed sequences offer a new potential strategy for the targeting of hepatic fibrosis.
Introduction
Fibrosis is a progressive dysregulated pathway of the natural wound healing response, defined as an excessive accumulation and deposition of the extracellular matrix (ECM) released by activated myofibroblasts after acute tissue injury of any origin.1 The replacement of healthy functional tissue with collagen-producing cells forming scar tissue and the resulting architectural deformation lead to fibrosis, cirrhosis, and hepatocellular carcinoma in its final and pathological manifestation.2 According to recent epidemiological studies, 10 million European citizens with undiagnosed liver disease are bearing significant liver fibrosis and approximately 1.5 million have silent cirrhosis.3 At present, a definite diagnosis of early or severe fibrosis can only be achieved by means of liver biopsy, an invasive and painful methodology that often has a high risk for complications and misjudgement.4 Hence, to improve prognosis for the patients, it is of paramount importance to develop a more convenient method for comprehensive and early diagnosis of hepatic fibrosis.5 As a first assessment, the ratios of combinatorial markers like aspartate transaminase/alanine aminotransferase (AST/ALT) and the AST-to-platelet ratio index (APRI), or various direct and indirect markers, such as collagens and collagenases, glycoproteins, or cytokines released by inflamed tissue, are only preliminary indicators monitored prior to proceeding with further diagnoses.6 These serum markers are mostly unspecific to liver tissue or liver fibrosis and do not provide any information concerning the grade and stage of the disease.5,6 Another approach to determine fibrosis-related transformations in hepatic tissue is to use clinical imaging techniques such as ultrasound and contrast-enhanced ultrasonography, which enable the alteration of the liver surface and of the intrahepatic microcirculation to be evaluated by assessing changes in the hepatic architecture with a satisfactory specificity (95%) for severe liver fibrosis.6 However, due to inadequate sensitivity and high inter-observer variability, these techniques offer limited potential for the evaluation of early fibrosis progression, staging, and grading.7 FibroScan TM® is a widely used, well-validated, and user-friendly elastographic technique that can be performed at the bedside with moderate reproducibility. As a function of increased liver stiffness, which occurs in hepatic fibrosis, the transfer of shear waves will be attenuated.8 The applicability depends on the experience of the operator and on the physical constitution of the patient, bringing along substantial limitations for obese patients suffering from ascites or a low diaphragm.8 Magnetic resonance imaging has the potential to become a reference technique to determine the stage, grade, and progress of liver fibrosis, but it suffers from a lack of appropriate and selective contrast agents, especially at early stage fibrosis.4Over the last decade, the development of peptide-based contrast agents for optical medical imaging of acute and chronic inflammations and their underlying pathways has become increasingly prominent.9 The peptide contrast agents are produced by linking the peptide sequence directly to a fluorescent probe, to magnetic resonance imaging and photo-acoustically active agent, or onto the surface of nanoscale lipid carrier systems incorporating contrast agents as an additional payload.10,11 Direct binding of these labelled peptides to biomarkers overexpressed on the surface of a disease-specific cell type would result in site-specific labelling and a reduction in background noise.12
In liver fibrosis, hepatic stellate cells (HSCs) are central disease mediators and thus considered optimal targets.13 HSCs undergo a mesenchymal transdifferentiation during fibrosis progression from a quiescent cell state with vitamin A storing capacity to an activated myofibroblast-like phenotype.13,14 Upon activation, it has been observed that the insulin growth factor 2 receptor (IGF2R), also known as the cation-independent mannose-6-phosphate receptor, is selectively overexpressed on the HSC surface.15 IGF2R is a 300 kDa dimeric transmembrane multifunctional protein receptor characterized by a large extracellular domain and a small transmembrane/intracellular domain, responsible for the regulation of the insulin growth factor 2 receptor (IGF2) and the corresponding transport of newly synthesized lysosomal enzymes from the trans-Golgi vesicles to the lysosomes.16–18
Owing to the ability of the receptor to internalize rather rapidly extracellular molecules and its ubiquitous expression on activated HSCs, IGF2R represents an optimal candidate for the development of a functional diagnostic platform for liver fibrosis.17,19 By means of a combinatorial bio-panning strategy on immobilized IGF2R and rat HSCs, a dodecamer peptide (VHWDFRQWWQPS, P431) was identified by Chen and co-workers as the best ligand for IGRF2R and was validated for its specificity to HSCs using LX-2 cells, an immortalized human HSC line.20 In the present study, the structural information of IGF2R derived from the PDB database was used for an in silico screening of novel peptide ligands specific to this receptor. Subsequently, the sequences showing the best in silico binding affinity for IGF2R were chosen to assess their selectivity to target activated HSCs over quiescent-like HSCs in comparison to peptide P431. The selected peptides and their scrambled sequences were labelled with fluorescein isothiocyanate (FITC) (Table 1). Their binding to different IGF2R-expressing human cell lines as well as cell uptake was assessed using flow cytometry and confocal microscopy. Furthermore, the expression of IGF2R was analysed for all tested cell lines by means of a quantitative mass spectrometric approach. Unlabelled sequences were tested in vitro for their proteolytic resistance in view of their prospective diagnostic application.
| Compound | Sequence (N → C) | Exact mass [Da] | [M + zH]z+ | Purity by HPLC [%] |
|---|---|---|---|---|
| a The N-terminus was blocked by acetylation to prevent cyclization reactions, while N-methyl amine was used as a protection group for the C-terminus. b The N-terminus was blocked by acetylation to prevent cyclization reactions. c A reactive primary amine was additionally introduced into the peptides by including a terminal lysine at the C-terminus, which was then used to conjugate FITC. | ||||
| Tc | ELVDTLQFVSGGGK | 1504.68 | 753 (z = 2) | 96.8 |
| T1 | ELVD![[W with combining low line]](https://www.rsc.org/images/entities/b_char_0057_0332.gif) LQFVSGGGK LQFVSGGGK |
1589.79 | 795 (z = 2) | 95.8 |
| T2 | ELVD![[Y with combining low line]](https://www.rsc.org/images/entities/b_char_0059_0332.gif) LQFVSGGGK LQFVSGGGK |
1566.76 | 784 (z = 2) | 97.3 |
| T3 | ELVDTLQFV![[W with combining low line]](https://www.rsc.org/images/entities/b_char_0057_0332.gif) GGGK GGGK |
1602.82 | 802 (z = 2) | 95.3 |
| T4 | ELVD![[F with combining low line]](https://www.rsc.org/images/entities/b_char_0046_0332.gif) LQFVSGGGK LQFVSGGGK |
1550.76 | 776 (z = 2) | 96.6 |
| T5 | ELVDTLQFV![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) GGGK GGGK |
1573.79 | 757 (z = 2) | 96.6 |
| P431 | VHWDFRQWWQPSGGGK | 2013.13 | 1007 (z = 2) | 99.9 |
| SCR431 | RPWSQVHWQDFWGGGK | 2013.13 | 1007 (z = 2) | 95.4 |
| Tc_FITC | ELVDTLQFVSGGGK{FITC} | 1894.06 | 948 (z = 2) | 98.8 |
| T1_FITC | ELVD![[W with combining low line]](https://www.rsc.org/images/entities/char_0057_0332.gif) LQFVSGGGK{FITC} LQFVSGGGK{FITC} |
1979.17 | 990 (z = 2) | 99.2 |
| T2_FITC | ELVD![[Y with combining low line]](https://www.rsc.org/images/entities/b_char_0059_0332.gif) LQFVSGGGK{FITC} LQFVSGGGK{FITC} |
1956.14 | 979 (z = 2) | 99.1 |
| T3_FITC | ELVDTLQFV![[W with combining low line]](https://www.rsc.org/images/entities/b_char_0057_0332.gif) GGGK{FITC} GGGK{FITC} |
1993.20 | 997 (z = 2) | 98.0 |
| T4_FITC | ELVD![[F with combining low line]](https://www.rsc.org/images/entities/b_char_0046_0332.gif) LQFVSGGGK{FITC} LQFVSGGGK{FITC} |
1940.14 | 971 (z = 2) | 94.0 |
| T5_FITC | ELVDTLQFV![[R with combining low line]](https://www.rsc.org/images/entities/b_char_0052_0332.gif) GGGK{FITC} GGGK{FITC} |
1963.17 | 982 (z = 2) | 98.6 |
| P431_FITC | VHWDFRQWWQPSGGGK{FITC} | 2402.51 | 801 (z = 3) | 94.1 |
| SCR_Tc_FITC | LETQVSVDFLGGGK{FITC} | 1894.06 | 948 (z = 2) | 99.4 |
| SCR_T1_FITC | LEWQVSVDFLGGGK{FITC} | 1979.17 | 990 (z = 2) | 96.9 |
| SCR_T2_FITC | LEYQVSVDFLGGGK{FITC} | 1956.14 | 979 (z = 2) | 96.5 |
| SCR_T3_FITC | LETQVWVDFLGGGK{FITC} | 1993.20 | 997 (z = 2) | 96.6 |
| SCR_T4_FITC | LEFQVSVDFLGGGK{FITC} | 1940.14 | 971 (z = 2) | 97.0 |
| SCR_T5_FITC | LETQVRVDFLGGGK{FITC} | 1963.17 | 982 (z = 2) | 97.8 |
| SCR431_FITC | RPWSQVHWQDFWGGGK{FITC} | 2402.51 | 802 (z = 3) | 96.9 |
Experimental details
Materials
Peptide P431 and its scrambled version (SCR431) were purchased from GenScript (Piscataway, NJ 08854, USA); all the other labelled (Tc_FITC, T1_FITC, T2_FITC, T3_FITC, T4_FITC, T5_FITC, SCR_Tc_FITC, SCR_T1_FITC, SCR_T2_FITC, SCR_T3_FITC, SCR_T4_FITC, SCR_T5_FITC) and unlabelled (Tc, T1, T2, T3, T4, T5) peptides were bought from Peptide Synthetics (Farnham, UK). The sequences are listed in Table 1. The immortalized human HSC line LX-2, Dulbecco's Modified Eagle Medium (DMEM, high glucose), penicillin/streptomycin solution (penicillin: 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1, streptomycin: 10
000 U mL−1, streptomycin: 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg mL−1), L-glutamine (200 mM), fetal bovine serum (FBS), non-enzymatic cell dissociation solution, and transforming growth factor β1 (TGF-β1) were obtained from Merck Millipore (Darmstadt, Germany). RPMI 1640 was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Cryopreserved immortalized human hepatocellular carcinoma (HepG2) cells were purchased from DSMZ (Braunschweig, Germany). The human embryonic kidney cell line 293 (HEK293) was obtained from the European Collection of Authenticated Cell Cultures (ECACC, Porton Down, United Kingdom). Retinol (Rol), palmitic acid (PA), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), and dimethyl sulfoxide (DMSO) were bought from Sigma-Aldrich (Schnelldorf, Germany). Cell culture plates and disposable materials were from Greiner Bio One International GmbH (Frickenhausen, Germany). Phenol red-free DMEM was from HyClone (Logan, UT, USA). Dulbecco's phosphate buffered saline (DPBS) without calcium and magnesium, ethylenediaminetetraacetic acid (EDTA), formic acid, fluorescein isothiocyanate (FITC) isomer I, propidium iodide (PI), buffer (boric acid/potassium chloride/sodium hydroxide) pH = 9, methanol (MeOH, HPLC grade), acetonitrile (ACN, HPLC grade), trans-ferulic acid, and trifluoracetic acid (TFA) were obtained from Carl Roth (Karlsruhe, Germany). A 70 μm mesh was bought from Hartenstein (Würzburg, Germany). A BD Falcon conical tube was purchased from BD Bioscience (Franklin Lakes, USA). Purified and deionized water was prepared with Milli-Q + Q-POD (Merck Millipore, Darmstadt, Germany).
000 μg mL−1), L-glutamine (200 mM), fetal bovine serum (FBS), non-enzymatic cell dissociation solution, and transforming growth factor β1 (TGF-β1) were obtained from Merck Millipore (Darmstadt, Germany). RPMI 1640 was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Cryopreserved immortalized human hepatocellular carcinoma (HepG2) cells were purchased from DSMZ (Braunschweig, Germany). The human embryonic kidney cell line 293 (HEK293) was obtained from the European Collection of Authenticated Cell Cultures (ECACC, Porton Down, United Kingdom). Retinol (Rol), palmitic acid (PA), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), and dimethyl sulfoxide (DMSO) were bought from Sigma-Aldrich (Schnelldorf, Germany). Cell culture plates and disposable materials were from Greiner Bio One International GmbH (Frickenhausen, Germany). Phenol red-free DMEM was from HyClone (Logan, UT, USA). Dulbecco's phosphate buffered saline (DPBS) without calcium and magnesium, ethylenediaminetetraacetic acid (EDTA), formic acid, fluorescein isothiocyanate (FITC) isomer I, propidium iodide (PI), buffer (boric acid/potassium chloride/sodium hydroxide) pH = 9, methanol (MeOH, HPLC grade), acetonitrile (ACN, HPLC grade), trans-ferulic acid, and trifluoracetic acid (TFA) were obtained from Carl Roth (Karlsruhe, Germany). A 70 μm mesh was bought from Hartenstein (Würzburg, Germany). A BD Falcon conical tube was purchased from BD Bioscience (Franklin Lakes, USA). Purified and deionized water was prepared with Milli-Q + Q-POD (Merck Millipore, Darmstadt, Germany).
Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 explicit TIP3P22 water molecules and 14 Na+ ions were added to assure electroneutrality; ion parameters optimized for the TIP3P water model were taken from Joung and Cheatham.23,24
000 explicit TIP3P22 water molecules and 14 Na+ ions were added to assure electroneutrality; ion parameters optimized for the TIP3P water model were taken from Joung and Cheatham.23,24
Molecular dynamics (MD) simulations were carried out as follows: first, energy minimization was performed applying a harmonic restraint on the solute (harmonic constant equal to 500 kcal mol−1 Å−2) in order to remove bad solute/solvent and solvent/solvent contacts due to the random placement of explicit water molecules; energy minimization was subsequently repeated on the overall system, removing the restraint. The temperature was raised to 310 K by means of 20 ps in an NVT ensemble (i.e., at a constant number of particles N, volume V, and temperature T) and system density was equilibrated through 1 ns in NPT ensemble (i.e., at a constant number of particles N, pressure P, and temperature T) at 310 K and 1 atm. Finally, 100 ns of MD simulations were carried out in NPT ensemble at 310 K and 1 atm. Temperature and pressure were kept at desired values using Langevin dynamics (with a collision frequency equal to 1 ps−1) and a Berendsen barostat with isotropic position scaling, respectively.
Simulations were performed adopting periodic boundary conditions; a cut-off value equal to 10 Å was chosen for non-bonded interactions. The SHAKE25 algorithm was employed to restrain all covalent bonds involving hydrogen atoms: this allowed adopting a time step equal to 2 fs to propagate dynamics by integrating Newton's equation of motion using the Leap-Frog algorithm. Molecular coordinates were collected every 20 ps. The attainment of an equilibrated complex structure was verified by evaluating root mean square displacement (RMSD) and interaction energy over time by means of Molecular Mechanics Generalized Born Surface Area (MMGBSA, vide infra). Plots are presented in the ESI† (Fig. S1 and S2).
| ΔEint = ΔEcomplex − Ereceptor − Eligand | (1) |
| E = Egas + ΔGsolv | (2) |
| ΔGsolv = ΔGpolar + ΔGnp | (3) |
| ΔGnp = a·SASA | (4) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The reaction mixture was stirred at room temperature overnight in the dark and purified by preparative high performance liquid chromatography (HPLC, vide infra). The purity was confirmed by analytical HPLC (vide infra) and the compounds were characterized by mass spectrometry (LC-MS, vide infra).
1. The reaction mixture was stirred at room temperature overnight in the dark and purified by preparative high performance liquid chromatography (HPLC, vide infra). The purity was confirmed by analytical HPLC (vide infra) and the compounds were characterized by mass spectrometry (LC-MS, vide infra).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 v/v) + 0.07% formic acid were analyzed using a QTRAP 5500 mass spectrometer (Sciex, Darmstadt, Germany) equipped with an electrospray ionization source and operating in the positive ion mode. For full scans (m/z 600–900), the ion spray voltage was set to 3000 V, the sheath gas pressure to 16 psi, and the declustering potential to 50 V. In contrast to the settings described above, for product ion scans (m/z 50–1200), a sheath gas pressure of 13 psi and a declustering potential of 150 V were used, and the collision energy was ramped from 10 to 60 eV in 2 eV steps. Mass spectra were processed using Analyst 1.6 (Sciex, Darmstadt, Germany).
5 v/v) + 0.07% formic acid were analyzed using a QTRAP 5500 mass spectrometer (Sciex, Darmstadt, Germany) equipped with an electrospray ionization source and operating in the positive ion mode. For full scans (m/z 600–900), the ion spray voltage was set to 3000 V, the sheath gas pressure to 16 psi, and the declustering potential to 50 V. In contrast to the settings described above, for product ion scans (m/z 50–1200), a sheath gas pressure of 13 psi and a declustering potential of 150 V were used, and the collision energy was ramped from 10 to 60 eV in 2 eV steps. Mass spectra were processed using Analyst 1.6 (Sciex, Darmstadt, Germany).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g in a MiniSpin centrifuge (Eppendorf, Hamburg, Germany). The supernatant was collected. The analysis was performed using an Agilent 1260 Infinity II HPLC. The injection volume of the purified peptides was 20 μL and the temperature of the autosampler was adjusted to room temperature (RT). For the analysis, a previously described method was applied (peptide purification). Peptide T4 was analysed differently due to an overlap with an FBS-derived peak in the chromatogram. Briefly, the analysis was started with 40% v/v of mobile phase A and 60% v/v of mobile phase B, which were kept constant for 25 min, followed by a rapid increase to 95% v/v of mobile phase A. After 28 min, the mobile phase contained 25% v/v of mobile phase A. These parameters were kept constant until the end of the run at 35 min. In order to determine the amount of intact peptide, the following equations were used to calculate the percentages at certain time points:
000 × g in a MiniSpin centrifuge (Eppendorf, Hamburg, Germany). The supernatant was collected. The analysis was performed using an Agilent 1260 Infinity II HPLC. The injection volume of the purified peptides was 20 μL and the temperature of the autosampler was adjusted to room temperature (RT). For the analysis, a previously described method was applied (peptide purification). Peptide T4 was analysed differently due to an overlap with an FBS-derived peak in the chromatogram. Briefly, the analysis was started with 40% v/v of mobile phase A and 60% v/v of mobile phase B, which were kept constant for 25 min, followed by a rapid increase to 95% v/v of mobile phase A. After 28 min, the mobile phase contained 25% v/v of mobile phase A. These parameters were kept constant until the end of the run at 35 min. In order to determine the amount of intact peptide, the following equations were used to calculate the percentages at certain time points:| Ct = AUC220nm sample/AUC220nm internal standard | (5) |
| Intact peptide (%) = (Ct/C0) × 100 | (6) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm) of the sample divided by the AUC220
nm) of the sample divided by the AUC220![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm of the internal standard observed at different time points and C0 represents the ratio of the AUC220
nm of the internal standard observed at different time points and C0 represents the ratio of the AUC220![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm of the sample divided by the AUC220
nm of the sample divided by the AUC220![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm of the internal standard at time zero.
nm of the internal standard at time zero.
| Cell metabolic activity (%) = (ODsample/ODcontrol) × 100 | (7) |
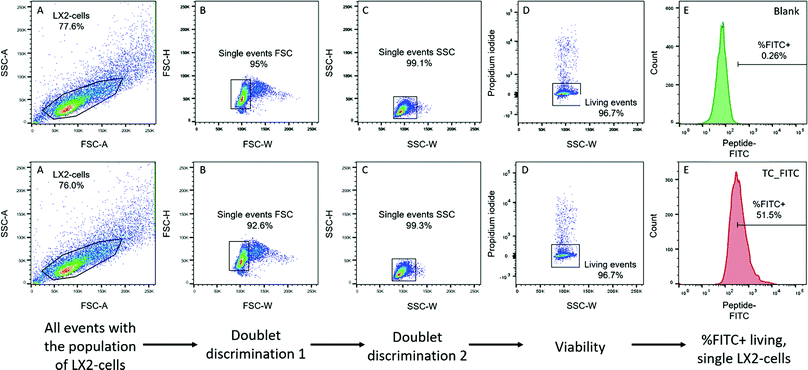
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events in total per sample were recorded. A 488 nm laser was used for fluorescence excitation. The FITC channel (530/30 nm band-pass) was used to acquire FITC emission on the peptides and the PerCP channel (695/40 nm band-pass) was used to detect PI emission. The acquisition software was BD FACS Diva 8.0.1 (BD Biosciences, Franklin Lakes, USA). The data were analysed using FlowJo V10.0 software (BD Biosciences, Franklin Lakes, USA). The fluorescence intensity of the peptides bound to the cell surface and/or internalized by the cells was assessed by means of a gating strategy, as displayed in Fig. 1, in order to ensure that unwanted events and artefacts were minimized. Cell-specific signals were used for doublet discrimination. FSC-W/FSC-H was used to detect double positive signals.
000 events in total per sample were recorded. A 488 nm laser was used for fluorescence excitation. The FITC channel (530/30 nm band-pass) was used to acquire FITC emission on the peptides and the PerCP channel (695/40 nm band-pass) was used to detect PI emission. The acquisition software was BD FACS Diva 8.0.1 (BD Biosciences, Franklin Lakes, USA). The data were analysed using FlowJo V10.0 software (BD Biosciences, Franklin Lakes, USA). The fluorescence intensity of the peptides bound to the cell surface and/or internalized by the cells was assessed by means of a gating strategy, as displayed in Fig. 1, in order to ensure that unwanted events and artefacts were minimized. Cell-specific signals were used for doublet discrimination. FSC-W/FSC-H was used to detect double positive signals.
It is well documented that cells with compromised membrane integrity, such as apoptotic cells, are prone to uncontrolled and unspecific uptake of proteins and peptides, with a resultant higher fluorescence emission due to the increased uptake.31 Consequently, an exclusion of dead cells from the final population was performed using propidium iodide. After excluding all unwanted events that could be responsible for artefacts, the percentage of FITC-positive labelled cells was assessed by gating all cells exceeding a certain value. To determine the threshold, unstained cells were used, and the threshold was set at 0.25% positive events in the control. In every set of experiments, the gating strategy and the compensation were separately adjusted optimizing the signal for each cell type to exclude cell type-specific differences in auto-fluorescence and complexity that could lead to shifts in the detected signal.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 w/v) in hypotonic buffer (50 mM mannitol and 5 mM HEPES, pH 7.4, supplemented with protease inhibitor) and left in an ultrasound bath for 1 min (in ice). Subsequently, 1 M calcium chloride solution was added to a final concentration of 10 mM and the suspension was vortexed for 10 min at RT. After centrifugation for 15 min at 3000 × g (4 °C), the supernatant was transferred into a new tube and ultra-centrifuged at 48
30 w/v) in hypotonic buffer (50 mM mannitol and 5 mM HEPES, pH 7.4, supplemented with protease inhibitor) and left in an ultrasound bath for 1 min (in ice). Subsequently, 1 M calcium chloride solution was added to a final concentration of 10 mM and the suspension was vortexed for 10 min at RT. After centrifugation for 15 min at 3000 × g (4 °C), the supernatant was transferred into a new tube and ultra-centrifuged at 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g (4 °C, 30 min). The pellet was suspended in 8 M urea/100 mM Tris (pH = 8), and a BCA assay was performed to quantify the absolute amount of proteins. Subsequently, 10 μg of protein was reduced, alkylated, and digested by Lys C for 2 h and then by trypsin overnight. The digests were analyzed by liquid chromatography (LC)-MS/MS (PROXEON coupled to a Q Exactive HF mass spectrometer, Thermo Fisher Scientific, Männedorf, Switzerland) with one injection of 5 μL digests. Peptides were trapped on a μPrecolumn PepMap100 C18 (5 μm, 100 Å, 300 μm × 5 mm, Thermo Fisher Scientific, Männedorf, Switzerland) and separated by backflush on a C18 column (5 μm, 100 Å, 75 μm × 15 cm) by applying a 60 minute gradient of 5–40% v/v ACN (+0.1% v/v formic acid) in ultrapure water (+0.1% v/v formic acid) at a flow rate of 350 nL min−1. A parallel reaction monitoring approach was used with the inclusion list of MPRI human peptides, the resolution being set at 30
000 × g (4 °C, 30 min). The pellet was suspended in 8 M urea/100 mM Tris (pH = 8), and a BCA assay was performed to quantify the absolute amount of proteins. Subsequently, 10 μg of protein was reduced, alkylated, and digested by Lys C for 2 h and then by trypsin overnight. The digests were analyzed by liquid chromatography (LC)-MS/MS (PROXEON coupled to a Q Exactive HF mass spectrometer, Thermo Fisher Scientific, Männedorf, Switzerland) with one injection of 5 μL digests. Peptides were trapped on a μPrecolumn PepMap100 C18 (5 μm, 100 Å, 300 μm × 5 mm, Thermo Fisher Scientific, Männedorf, Switzerland) and separated by backflush on a C18 column (5 μm, 100 Å, 75 μm × 15 cm) by applying a 60 minute gradient of 5–40% v/v ACN (+0.1% v/v formic acid) in ultrapure water (+0.1% v/v formic acid) at a flow rate of 350 nL min−1. A parallel reaction monitoring approach was used with the inclusion list of MPRI human peptides, the resolution being set at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, with an automatic gain control target of 2E05, a maximum ion injection time of 130 ms, HCD collision energy to 27, a fill ratio of 1%, charge exclusion of unassigned and 1+ ions, and peptide match preferred, respectively. The results were analyzed using Skyline v20.1.0.155 software (MacCoss Lab Software, University of Washington, Seattle, USA) at the MS2 fragment level.
000, with an automatic gain control target of 2E05, a maximum ion injection time of 130 ms, HCD collision energy to 27, a fill ratio of 1%, charge exclusion of unassigned and 1+ ions, and peptide match preferred, respectively. The results were analyzed using Skyline v20.1.0.155 software (MacCoss Lab Software, University of Washington, Seattle, USA) at the MS2 fragment level.
Results and discussion
Molecular dynamics simulations of the IGF2/IGF2R complex
A 100 ns MD simulation was performed in order to obtain a reasonably equilibrated structure of the complex; this was successfully verified by computing the RMSD and complex energetics as a function of simulation time (Fig. S1 and S2, ESI†). Interaction energy was obtained through the MMGBSA approach considering the last 20 ns of the molecular trajectory; this led to a ΔEint value equal to −48.54 ± 7.58 kcal mol−1. Specific contributions are listed in Table 2 where ΔEelec and ΔEvdW are the contributions to the binding due to electrostatic and van der Waals interactions, respectively, and ΔΔGsolv is the change in solvation free energy after the attainment of the non-covalent complex.| Contributions | Specific energy [kcal mol−1] |
|---|---|
| ΔEelec [kcal mol−1] | −93.67 ± 57.76 |
| ΔEvdW [kcal mol−1] | −74.21 ± 7.68 |
| ΔΔGsolv [kcal mol−1] | 119.34 ± 53.62 |
| ΔEint [kcal mol−1] | −48.54 ± 7.58 |
Trajectory post-processing revealed that both electrostatic (which include salt bridges) and van der Waals (that also account for hydrophobic effects) interactions contribute to the formation of the complex. The positive value of ΔΔGsolv indicates that the complex attainment slightly penalizes the overall solubility; this is due to the charged moieties that are no more exposed to the solvent when the complex is formed. Per-residue decomposition allowed highlighting the relevant contribution of the fragment E21–S21 of IGF2, which is almost entirely included in the binding pocket; detailed outcomes are reported in Table S1 (ESI†).
Alanine scanning was subsequently performed with the selected amino acids from the previous analysis, in order to underline the impact of their mutation. The results are summarized in Table 3.
| Mutated amino acid | ΔΔEint [kcal mol−1] |
|---|---|
| E6 | −10.50 ± 3.64 |
| E12 | −9.09 ± 2.23 |
| D15 | −1.91 ± 1.92 |
| Q18 | −1.07 ± 2.34 |
| F19 | −6.43 ± 1.08 |
| V20 | −0.82 ± 0.39 |
| D23 | −2.80 ± 3.10 |
As discussed in detail below (vide infra), alanine scanning highlighted two hot spots for the binding: E12 (that leads to a salt bridge with R1571) and F19, whose contribution is related to hydrophobic effects. Although the analysis also identified E6 as relevant for the binding, the E12–S21 segment is better enclosed in the binding pocket and thus constitutes an ideal starting point for the in silico design of new peptides.
In silico design of peptides
Fig. 2 presents the WT peptide (sequence E12–S21) bound to IGF2R. According to CAS analysis, two principal hot-spots mediate the interaction: E12, which makes a salt-bridge with R1571, and F19, whose benzyl group makes π–π interactions with F1567 and hydrophobic contacts with the pocket formed by Y1542, F1567, Y1606, L1626, L1629 and L1636. Additionally, hydrogen bonds are observed between the D15 sidechain carboxy group and the Y1542 hydroxy group, and between the Q18 sidechain carbonyl and the S1543 hydroxy group. Starting from the WT peptide a set of 68-point mutations, aimed to improve binding affinity (ΔGbind) and complex thermodynamic stability (ΔGtot), were evaluated according to Table 4.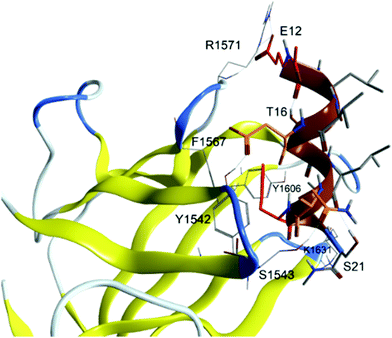 | ||
| Fig. 2 Representation of the WT peptide in complex with the IGF2 receptor. Hot-spot and warm-spot residues, identified by CAS analysis, are coloured in red and orange, respectively. | ||
| IGF2 residue | Mutation |
|---|---|
| L13 | ANQILSTV |
| V14 | ANQILSTV |
| D15 | NDQE |
| T16 | HILFPSTWYV |
| L17 | ANQILSTV |
| Q18 | RNDQEHKSTY |
| F19 | FWY |
| V20 | ILFTWV |
| S21 | RNDQEHKSTWY |
The results of the RS analysis, summarized in Table 5, show that the T16W mutation positively affects both the binding affinity (ΔGbind) and the thermodynamic stability (ΔGtot) by −7.9 and −0.1 kcal mol−1, respectively. A T16F mutation is also beneficial for both ΔGbind (−3.9 kcal mol−1) and ΔGtot (−0.2 kcal mol−1), while the T16Y slightly deteriorates the predicted total stability (ΔGtot = 0.1 kcal mol−1), but not the binding affinity (ΔGbind = −5.9 kcal mol−1).
| Mutation | ΔGbind [kcal mol−1] | ΔGtot [kcal mol−1] | Final sequence |
|---|---|---|---|
| T16W | −7.9 | −0.1 | ELVDWLQFVS |
| T16Y | −5.9 | 0.1 | ELVDYLQFVS |
| S21W | −10.8 | 0.4 | ELVDTLQFVW |
| T16F | −3.9 | −0.2 | ELVDFLQFVS |
| S21R | −6.5 | 0.4 | ELVDTLQFVR |
Mutations at S21 (S21W and S21R) also provide improved affinity, although at the expense of overall stability (ΔGtot = 0.4 kcal mol−1). In particular, the highest decrease in binding affinity (coupled with the highest loss in thermodynamic stability, within the selected peptides) is observed for the S21W mutation (ΔGbind = −10.8 kcal mol−1). All the peptides described herein were selected for experimental evaluation.
Production and characterization of FITC-labelled, in silico identified IGF2R peptide ligands
The previously described dodecamer (P431) identified by Chen et al. has been selected as a positive reference for the binding affinity to the IGF2R in the screening of the newly in silico identified IGF2R ligands.20 Randomly generated scrambled sequences of all peptides were implemented into the binding assay for a comprehensive evaluation and to identify unspecific binding to the cell membrane.33 All peptides were labelled with FITC (Table 1), and in order to minimize the intramolecular hydrophobic interactions between the fluorophore and the amino acids of the peptides essential for IGF2R binding, a 3-glycine spacer was included in the sequence at the C-terminus.34 A reactive primary amine was introduced in the peptides by including a terminal lysine at the C-terminus, which was then used to attach FITC. The N-terminus was blocked by acetylation to prevent cyclization reactions, while N-methyl amine was used as a protection group for the C-terminus. Amino acid sequences and masses of all peptide ligands are reported in Table 1. A purity of ≥94% was confirmed for all ligands.Serum stability
When aiming at identifying novel targeting ligands to functionalize nanoparticles or biopolymers for diagnostic or theragnostic purposes, testing the serum stability of the targeting sequence is one of the pivotal parameters to investigate. To simulate the biological environment, non-FITC-labelled peptides were incubated in 50% v/v FBS in PBS and the amount of intact peptide was analyzed over time using analytical HPLC. A minimum proteolytic stability of 80% intact after 8 h and 60% intact after 24 h was set as the satisfactory threshold in this experiment.35,36In contrast to the previously described P431 for which only traces (approx. 3%) could be detected after 8 h of incubation in the serum-containing medium, the six in silico designed peptides (Tc, T1–T5) showed almost no proteolytic degradation after 8 h (intact peptide >85%; Fig. 3). After 24 h, P431 was completely degraded, while ≥60% remained intact for most of the other candidates (except T3: 58.2 ± 2.7%), indicating their potential as targeting ligands for in vivo investigations.
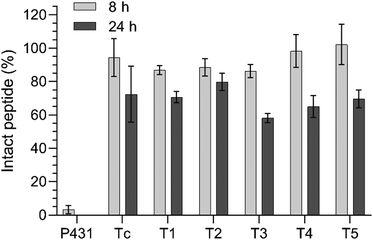 | ||
| Fig. 3 Proteolytic stability of the different peptides (25 μM) over 8 and 24 h in the presence of 50% v/v FBS in PBS at 37 °C. | ||
Cellular metabolism assay
The in silico derived sequences were investigated for their role as targeting ligands for HSCs. Optimally, a targeting ligand for optical imaging should show a low intrinsic activity (competitive agonism) and should not induce apoptosis or alterations in metabolic activity.37 Therefore, the mitochondrial dehydrogenase activity was analyzed as a measure of metabolic activity after incubation with the peptide (5 and 10 μM) over a period of 8 h to reflect an expected timeframe of the receptor–ligand interaction in vivo. Untreated cells were used as the 100% activity control. All other results were calculated as the percentage of metabolic activity in relation to untreated cells. In this experiment, values ≥70% were considered as indicating unchanged or slightly changed metabolic activity (Fig. 4). No remarkable effects on metabolic activity could be detected for the concentrations of 5 and 10 μM for all sequences. The designed peptides are suitable targeting ligands that do not induce substantial cytotoxicity.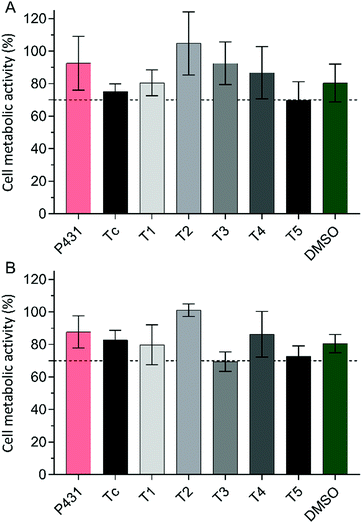 | ||
| Fig. 4 Cell metabolic activity in % after an 8 h incubation of LX-2 cells with purified peptides at a concentration of 5 μM (A) or 10 μM (B) in complete growth medium at 37 °C. | ||
Cellular uptake by the hepatic stellate cell line LX-2
In order to evaluate the potential of our peptide sequences as targeting ligands to be anchored to the surface of diagnostic liposomes, nanoparticles, or gadolinium complexes, it was of crucial importance to evaluate their binding efficiency to different cell lines. Accordingly, naïve LX-2 cells were used as a model of activated HSCs that highly express IGF2R (vide infra). In healthy liver tissue, HSCs are surrounded by hepatocytes forming liver sinusoids. Hence, HepG2 cells, a model for polarized human hepatocytes, were used to demonstrate superior peptide binding on LX-2 cells compared to other liver tissue. HEK293, immortalized human embryonic kidney cells, were used as a negative control. Cells were incubated for 1 h with the fluorescent peptides (10 μM) or their scrambled sequences (10 μM) and further analysed via flow cytometry. Percentages of FITC positive labelled cells (%FITC+) were obtained for LX-2 cells (Fig. 5A), HepG2 and HEK293 (data shown in the ESI,† Fig. S4B and C, respectively). Furthermore, the binding was performed by three different observers in two different labs (Fig. S5, ESI†). In order to display the superiority of the binding peptides to their scrambled sequences on LX-2 cells (Fig. 5B) and their intercellular selectivity compared to HepG2 (Fig. 5C) or HEK293 (Fig. 5D), the flow cytometry data were plotted as the ratio RSCR between the percentage of sequence-specific %FITC+ LX-2 cells and the percentage of sequence-specific %FITC+ of the scrambled peptides, HepG2 or HEK293. Furthermore, to compare the strength of the bimolecular interaction with LX-2-cells expressing IGF2R, the equilibrium dissociation constant (KD) was determined for all sequences. Bmax [μM] is defined as the maximum specific binding extrapolated to higher concentrations (>Cmax) than those tested in the experiment. To determine KD, the %FITC+ living LX-2 cells were tested after incubation with different concentrations, i.e. 1, 5, 10, 25, and 50 μM, of the peptide. The results were calculated by means of nonlinear regression of the specific binding with Hill slope calculation as displayed in Fig. 6. The obtained KD, Bmax, coefficient of determination (R2), and standard error of regression (Sx·y) values are shown in Table 6.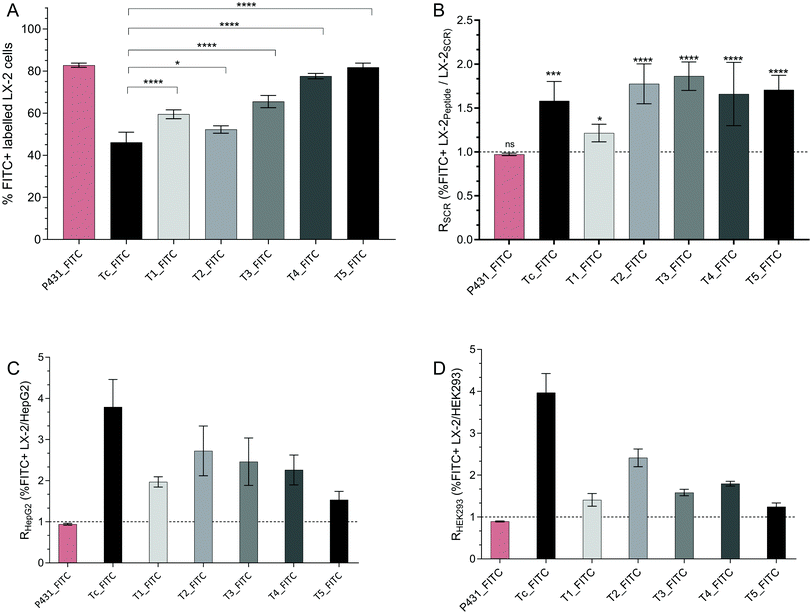 | ||
| Fig. 5 (A) Flow cytometry data expressing %FITC+ LX-2 cells after incubation with targeting peptides in comparison with the putative binding sequence Tc. P431 was used as the positive control. Multiple comparison tests (Holm–Sidak) between all peptides are displayed in Table S2 (ESI†). (B) Ratio of %FITC+ LX-2 incubated with binding peptide/%FITC+ LX-2 incubated with the scrambled peptide (RSCR); significance test has been performed relative to P431-FITC. (C) Ratio of %FITC+ LX-2/%FITC+ HepG2 (RHepG2). (D) Ratio of %FITC+ LX-2/%FITC+ HEK293 (RHEK293). The concentration of the peptides was 10 μM (1 h at 37 °C). | ||
| Peptide | Tc | T1 | T2 | T3 | T4 | T5 | P431 |
|---|---|---|---|---|---|---|---|
| B max [μM] | 107.5 | 105.9 | 108.4 | 99.15 | 104.6 | 100.8 | 101.2 |
| K D [μM] | 13.03 | 8.71 | 13.15 | 8.44 | 6.47 | 5.01 | 6.26 |
| R 2 | 0.98 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 | 0.99 |
| Sy·x | 3.96 | 1.77 | 2.65 | 1.49 | 2.11 | 2.09 | 3.87 |
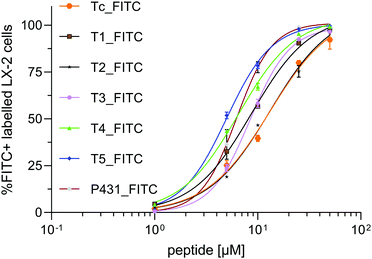
In contrast to cells treated with vehicle control (DMSO), all cells incubated with FITC-modified peptides generated a detectable fluorescence signal higher than the auto-fluorescence of untreated cells. The sequence Tc_FITC, a decamer representing the putative natural peptide sequence containing the most relevant amino acids for the interaction of IGF2 with IGF2R, was used as the reference peptide. Favourably, by replacing one amino acid in the sequence Tc, the binding affinity towards the target should be increased without impairing the natural binding to IGF2R. The selectivity toward LX-2 cells compared to HepG2 or HEK293 is expressed as RHepG2 or RHEK293 and should be > 1 (Fig. 5C and D, respectively).
As shown in Fig. 5 and 6, the reference ligand Tc_FITC showed the lowest binding affinity with KD = 13.03 μM on LX-2 cells (46.2% positively labelled cells (%FITC+)) while demonstrating the highest selectivity toward LX-2 cells relative to HepG2 and HEK293 (RHepG2 = 3.79, RHEK293 = 3.97). The highest peptide uptake was observed after treatment with the previously described sequence P431_FITC (%FITC+ = 82.8%). Interestingly, also its scrambled sequence SCR431_FITC, a negative control not included in the original publication but generated in this study to validate the binding properties of this allegedly IGF2R-specific peptide, bound in a similar fashion to LX-2 cells (%FITC+ = 85.2%, RSCR = 0.97, Fig. 5B), suggesting ultimately the lack of specificity of P431 for these cells. The binding affinity of P431_FITC taken from the original publication (KD = 6.19 μM) could be confirmed (KD = 6.26 μM) in this set of experiments, proving the validity of our testing method.20 However, for both sequences P431_FITC and its scrambled version SCR431_FITC, no specific selectivity for HSCs could be detected as the RHepG2 (0.94 and 0.92, respectively) and RHEK293 (0.89 and 0.91, respectively) values prove (Fig. 5C and D). Contrariwise, the in silico generated target-specific peptides were characterized by a 1.2- to 1.9-fold increase in binding compared to their corresponding scrambled sequences (RSCR, Fig. 5B) which was demonstrated as significantly effective in vivo.38 Thus, the increased binding of our peptides compared to their equivalent scrambled sequence indicates their potential as IGF2R-selective targeting ligands.
For the target-specific peptides tested on LX-2 cells, all sequences showed a significantly increased binding affinity compared to the reference Tc_FITC. The highest %FITC+ values were observed for T4_FITC (KD = 6.47 μM, %FITC+ = 77.6%) and T5_FITC (KD = 5.00 μM, %FITC+ = 81.8%), with ca. 2-fold and 2.6-fold higher signals, respectively, than Tc_FITC (Fig. 6). However, this significant (p < 0.0001) increase in binding affinity for T4 and T5 is accompanied by a decrease in intercellular selectivity in comparison to Tc (>2-fold decreased) (Fig. 5C and D). The sequences T1_FITC (KD = 8.71 μM, %FITC+ = 59.5%, RHepG2 = 1.97, RHEK293 = 2.41), T2_FITC (KD = 13.15 μM, %FITC+ = 52.3%, RHepG2 = 2.72, RHEK293 = 2.41), and T3_FITC (KD = 8.44 μM, %FITC+ = 65.5%, RHepG2 = 2.46, RHEK293 = 1.58) also showed an increased (T1_FITC: 1.5-fold and T3_FITC: 1.5-fold) binding affinity and a decreased selectivity compared to Tc_FITC. However, in comparison to T4 and T5, the binding of T1, T2, and T3 is significantly lower (p ≤ 0.01). T2_FITC showed the best cell selectivity of all tested mutated sequences (Fig. 5C and D) but it was significantly decreased compared to Tc_FITC.
The sequence T1 was generated by mutating the hydrophilic amino acid threonine (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = −0.26) at position 5 replacing its hydroxyl side group with an aromatic indole-functionalized amino acid such as tryptophan (log
P = −0.26) at position 5 replacing its hydroxyl side group with an aromatic indole-functionalized amino acid such as tryptophan (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 1.46) in the middle of the sequence. A similar mutation was performed for peptide T3. At position 10 in sequence T3, the hydrophilic serine (log
P = 1.46) in the middle of the sequence. A similar mutation was performed for peptide T3. At position 10 in sequence T3, the hydrophilic serine (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = −0.45) was replaced with a tryptophan. The resulting increase in binding and decrease in selectivity were nearly identical to those of T1. Replacing the tyrosine at position 5 with a threonine (log
P = −0.45) was replaced with a tryptophan. The resulting increase in binding and decrease in selectivity were nearly identical to those of T1. Replacing the tyrosine at position 5 with a threonine (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 0.55) in sequence T2 (KD = 13.15 μM) did not substantially improve the binding affinity. However, T2 (RHepG2 = 2.73, RHEK293 = 2.41) is characterized by a noteworthy intercellular selectivity compared to the other mutated sequences.
P = 0.55) in sequence T2 (KD = 13.15 μM) did not substantially improve the binding affinity. However, T2 (RHepG2 = 2.73, RHEK293 = 2.41) is characterized by a noteworthy intercellular selectivity compared to the other mutated sequences.
The hydroxyphenyl side group of tyrosine is more lipophilic than the hydroxyl group in threonine but may also form hydrogen bonds which ultimately leads to only a small change in molecular charge.39 It can be hypothesized that the replacement of the non-aromatic hydrogen bond-forming amino acid with a lipophilic hydroxyl group-free amino acid at position 5 or 10 of the sequence led to an increased IGF2R affinity but to decreased selectivity, underlying the primary role of a hydrogen bond at position 5 to achieve pronounced selective binding. Replacing threonine with phenylalanine (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 1.16) at position 5 in sequence T4 resulted in a considerable 68% increase of the uptake. Consistent with the results obtained for peptides T1 and T3, a mutation towards a more lipophilic amino acid without being able to form hydrogen bonds resulted in higher binding and lower selectivity of the peptide. Besides the loss of hydrogen bonds, a decreased steric interaction associated with minor changes in the 3D structure compared to sequence T1 could be a pivotal parameter for the observed differences. An exception to the previously described hypothesis is represented by peptide T5, in which the amino acid serine at the extremity of the sequence was replaced by arginine (log
P = 1.16) at position 5 in sequence T4 resulted in a considerable 68% increase of the uptake. Consistent with the results obtained for peptides T1 and T3, a mutation towards a more lipophilic amino acid without being able to form hydrogen bonds resulted in higher binding and lower selectivity of the peptide. Besides the loss of hydrogen bonds, a decreased steric interaction associated with minor changes in the 3D structure compared to sequence T1 could be a pivotal parameter for the observed differences. An exception to the previously described hypothesis is represented by peptide T5, in which the amino acid serine at the extremity of the sequence was replaced by arginine (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = −0.79). In this case, however, the mutation introduces a positive charge of the side chain, resulting in an overall neutral charge of the sequence at physiological pH, likely responsible for unspecific interactions with the cell membrane, as confirmed by the lower selectivity of peptide T5 (RHepG2 = 1.53, RHEK293 = 1.24).40 This could also be an explanation for the high uptake of FITC_P431 and its scrambled version in comparison to the other sequences. Both sequences (P431 and SCR431) consist of the amino acid arginine and histidine, ultimately resulting in a positive net charge at physiological pH leading to RSCR = 1 due to charge-related interactions between the cell membrane and the peptide.37
P = −0.79). In this case, however, the mutation introduces a positive charge of the side chain, resulting in an overall neutral charge of the sequence at physiological pH, likely responsible for unspecific interactions with the cell membrane, as confirmed by the lower selectivity of peptide T5 (RHepG2 = 1.53, RHEK293 = 1.24).40 This could also be an explanation for the high uptake of FITC_P431 and its scrambled version in comparison to the other sequences. Both sequences (P431 and SCR431) consist of the amino acid arginine and histidine, ultimately resulting in a positive net charge at physiological pH leading to RSCR = 1 due to charge-related interactions between the cell membrane and the peptide.37
A negative net charge and a mutation towards a more lipophilic amino acid not able to form hydrogen bonds at position 5 or 10 of the sequence resulted overall in the best binding with respect to activated HSCs. However, these mutations negatively impacted cell selectivity. To generate a peptide with improved binding and selectivity, additional mutations need to be performed on the tested sequences.
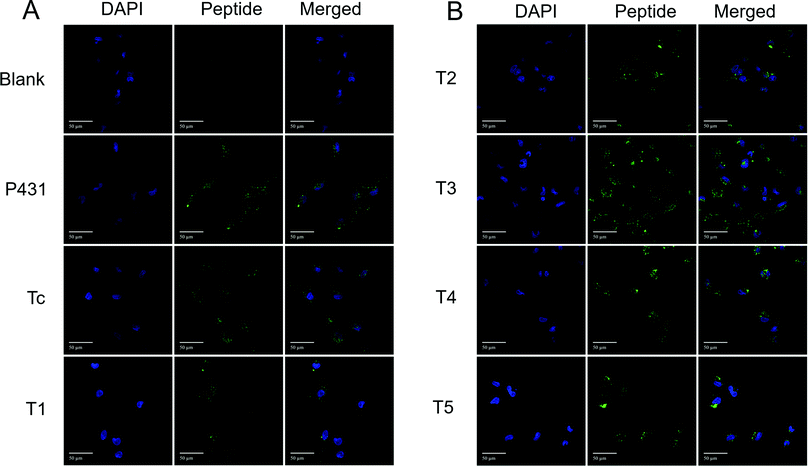
To further confirm the binding affinity observed with FACS and to analyse the cellular location of the FITC-labelled peptides after incubation with cells, confocal microscopy imaging was carried out. LX-2 cells were incubated with 10 μM of FITC-labelled peptides or their corresponding scrambled sequences for 1 h. To determine the absolute number of observed cells, the nucleus was stained with DAPI. In order to quantify the fluorescence signal of FITC, the corrected total cell fluorescence (CTCF) was calculated. As displayed in Fig. 7A and B, the majority of the FITC+ signal after 1 h is mainly located in the cellular lumen. Representative confocal microscopy images of LX-2 cells after the incubation with the scrambled sequences are available in the ESI† (Fig. S6A and B). This result confirms the ability of the receptor to rapidly internalize the specific ligand. Furthermore, the analysis of the CTCF (Fig. S7, ESI†) confirmed the results obtained by flow cytometry. The obtained pattern of the fluorescence intensity for all peptides (Fig. S7, ESI†) is comparable to the flow cytometry data displaying the %FITC+ events (Fig. 5A). The ability of being rapidly internalized makes our peptides promising candidates to decorate theragnostic carriers designed for early detection of alterations in HSCs and treatment of hepatic fibrosis.
Cellular uptake in LX-2 cells in different states of activation
The progression and resolution of hepatic fibrosis is characterized as a complex dynamic process of interactions between parenchymal and non-parenchymal liver cells. HSCs, represented as a quiescent-like phenotype in a non-fibrotic liver, undergo a (reversible) transition towards an activated proliferative myofibroblast-like phenotype after being exposed to chronic apoptosis of hepatocytes associated with immune cell infiltration and the resulting release of pro-inflammatory and pro-fibrogenic cytokines.41 Furthermore, activated HSCs are characterized by attenuated proliferation and aberrant production of proteins, such as collagen type I and type IV, desmin, vimentin, and α-smooth muscle actin that form the major components of the ECM.42Particularly relevant for the present investigation are the different expression levels of IGF2R as a function of the activation state, with a higher amount of IGF2R expressed on transdifferentiated HSCs in comparison with the ones in a quiescent state.43 LX-2 cells retain crucial features of activated HSCs, but it is possible to perpetuate their activation through supplementary treatment with TGF-β1.30 On the contrary, by exposing LX-2 cells to a mixture of retinol and palmitic acid (Rol + PA), their quiescent-like phenotype can be promoted.44 In line with the work published by Chen et al.,20 we also shared the final goal to identify a ligand specific for activated HSCs. Aiming at following the progression and/or regression of hepatic fibrosis, it was important to prove that the uptake of the identified peptide sequences would be higher in naive and perpetuated LX-2 cells in comparison to quiescent-like LX-2 cells. The results of the ratio Rfibrogenic (%FITC+ perpetuated LX-2/%FITC+ quiescent LX-2) are displayed in Fig. 8.
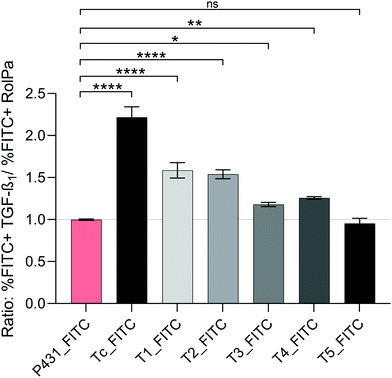 | ||
| Fig. 8 Ratio Rfibrogenic (%FITC+perpetuated LX-2/%FITC+quiescent LX-2) after incubation of perpetuated and quiescent-like LX-2 cells with FITC-labelled peptides at a concentration of 10 μM for 1 h at 37 °C. LX-2 cells were transdifferentiated into the perpetuated state with 10 ng mL−1 TGF-β1 or into the quiescent-like state with Rol (10 μM) and PA (300 μM) for 24 h at 37 °C prior to incubation with peptides. Rfibrogenic > 1 indicates selectivity for the perpetuated phenotype over quiescent-like LX-2 cells. In Table S3 (ESI†), the results of the multiple comparison tests (Holm–Sidak) between all peptides are displayed. | ||
A Rfibrogenic value > 1 is defined as increased selectivity towards the activated state. Rfibrogenic ≤ 1 characterizes no (=1) or reciprocal selectivity (<1) which is not desired for a future approach regarding the diagnosis of hepatic fibrosis. The treatment-specific %FITC+ for the quiescent-like and perpetuated LX-2-phenotypes are displayed in the ESI† in Fig. S8. Upon treatment of naïve LX-2 with TGF-β1, an increase of peptide uptake was observed for all the tested in silico derived peptides with respect to untreated LX-2 cells (Fig. S8 and Fig. 5A). As expected, after the treatment of naïve LX-2 with Rol + PA, a reduced binding could be detected for all in silico sequences, except T5 and P431. For P431, we obtained a Rfibrogenic value of 0.99, indicating the same affinity for all states of activation and thus a lack of selectivity. Tc (Rfibrogenic = 2.21), T1 (Rfibrogenic = 1.58), and T2 (Rfibrogenic = 1.53) showed a significantly decreased uptake after cells were treated with Rol + PA and an increased uptake after the treatment with TGF-β1 (p ≤ 0.0001), making them the most promising sequences for a phenotype-specific uptake into HSCs.
Sequence T4 (Rfibrogenic = 1.25) was only characterized by a moderate difference towards the quiescent-like cells after perpetuation. T3 (Rfibrogenic = 1.18) showed a significant but minor selective uptake to the perpetuated state.
For T5 (Rfibrogenic = 0.95), no significant difference in uptake in naïve, perpetuated, or quiescent-like LX-2 was detected. This supports the hypothesis that T5 and P431 mainly or fully have unspecific interactions with the cellular membrane due to the presence of multiple arginines in the sequence, which elevate the positive charge of the molecule at physiological pH. Although some of our mutated peptides show selectivity towards the activated LX-2 cells, when compared to the putative binding ligand Tc, the implemented amino acid mutations seem to negatively impact (slightly for T1 and T2, and moderately for T3 and T4) selectivity.
Detection of IGF2R by a quantitative mass spectrometric approach
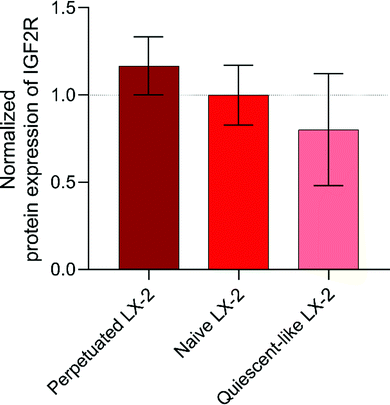
In order to provide experimental evidence of the IGF2R expression levels in LX-2 cells at different states of activation and to explain the results observed with peptide P431 proposed by Chen et al.,20 the expression of the receptor was analysed by a quantitative mass spectrometric approach. To the best of our knowledge, no information regarding the exact protein expression levels in HSCs and their comparison between the different activation-based phenotypes is available in the literature. In Fig. 9, the relative receptor expression of different phenotypes normalized to the expression of naive LX-2 cells is displayed. As indicated by the RNA-expression analysis performed by Beljaars et al., an activation-specific expression pattern of the IGF2R-peptide could be observed compared to untreated LX-2 cells.45,46 Predictably, compared to naive LX-2 cells, the perpetuated phenotype is characterized by an elevated receptor expression of approximately 16.8% (Fig. 9).The quiescent-like phenotype is defined by a decrease of approximately 19.9%. Thus, an activation-specific targeting of our peptides would demonstrate specificity not only for HSCs with respect to other cell types but also for their state of activation, and therefore determine areas with higher disease progression (activated HSCs). In LX-2 cells, IGF2R is not exclusively expressed on the cell surface, but compared to our negative controls, HepG2 and HEK293, a larger proportion of the protein should be found.45,46 Therefore, we investigated the IGF2R-expression levels of our negative controls HepG2 and HEK293. These cells were chosen based on their relatively low IGF2R RNA-expression levels derived from the Human Protein Atlas.47 However, in an apparent contradiction to the assumption, the relative protein expression of IGF2R in HepG2-cells was 7.5-fold elevated compared to naïve LX-2 cells. A possible explanation can be found in the specific cell line and in the main localization of the receptor expression. Hepatocytes are characterized by a generally increased metabolization rate positively affecting the protein translation and their release in the endoplasmic reticulum.48 Furthermore, HepG2-cells are used as a model for polarized hepatocytes but represent a well-differentiated hepatocellular carcinoma.49 It has been described that protein and gene expression is increased in an uncontrolled and unique manner with respect to cultured primary human hepatocytes.50 Therefore, it can be hypothesized that the majority of IGF2R is not expressed on the surface of the cell but in the Golgi apparatus and vesicles located in the cellular lumen.51 Thus, only a small proportion of IGF2R is located on the surface of HepG2 cells. The same explanation can be applied for HEK293 showing a 10% increase in the relative IGF2R-peptide expression with respect to naive LX-2 cells.52
Conclusions
In this study, we used an efficient in silico approach for the development of targeting ligands selective for IGF2R. Structural information of the binding site of the receptor was used for the screening of specific peptide ligands. Molecular dynamics simulations, followed by computational alanine scanning of the IGF2R/IGF2 complex, led to the identification of a putative peptide sequence (Tc) containing the most relevant amino acids for the receptor–ligand interaction (IGF2 E12-C21). The Residue Scan tool, implemented in MOE software, was then used to optimize the binding affinity by replacing amino acids at position 5 or 10 of the putative binding sequence in order to implement mutations that increase affinity without affecting proteolytic stability. Selective binding of the peptides to IGF2R-expressing cells was successfully verified for all sequences (KD ≤ 13.2 μM). With respect to Tc, the majority of mutations led to an increased receptor affinity (T1, T3, T4, T5), except for T2 where the affinity remained unchanged. In apparent contrast to the previously published P431, all sequences have shown a 2- to 4-fold increase in binding for LX-2 cells compared to the negative controls, HepG2 or HEK293 cells. They also showed superior binding compared to their associated scrambled sequences. All unlabelled sequences were tested in vitro for their proteolytic resistance by HPLC-DAD and were characterized by a stability of ≥60% intact after 24 h incubation in 50% FBS (except T3, with 58%). In view of their prospective diagnostic application, a comparison of binding affinity was performed in perpetuated and quiescent-like LX-2 cells. Increased binding was detected in the perpetuated cell state, indicating selectivity for the diagnostically relevant phenotype. In summary, the increased binding affinity of the identified peptides towards perpetuated LX-2 cells compared to other LX-2 states or other cell types, and the satisfactory proteolytic stability show that the in silico designed sequences offer a new potential strategy for the targeting of hepatic fibrosis.Conflicts of interest
The authors declare that they have no conflict of interest to report.Acknowledgements
P. L. acknowledges Lipoid GmbH for the endowment to the University of Jena. P. L. and F. W. thank the staff of the Proteomics and Mass Spectrometry Core Facility (PMSCF), Department for BioMedical Research (DBMR), University of Bern, Switzerland and of the Microscopy Imaging Centre of the University of Bern, Switzerland for technical support and scientific advice.References
- T. A. Wynn and T. R. Ramalingam, Nat. Med., 2012, 18, 1028–1040 CrossRef CAS.
- Y. J. Yoon, S. L. Friedman and Y. A. Lee, Semin. Liver Dis., 2016, 1, 87–98 CrossRef.
- L. Caballería, G. Pera, I. Arteaga, L. Rodríguez, A. Alumà and P. Ginès, Clin. Gastroenterol. Hepatol., 2018, 16, 1138–1145 CrossRef.
- L. Petitclerc, G. Sebastiani, G. Gilbert, G. Cloutier and A. Tang, J. Magn. Reson. Imaging, 2017, 45, 1276–1295 CrossRef.
- S. Petta, V. W. S. Wong, C. Cammà, J. B. Hiriart and V. de Ledinghen, Aliment. Pharmacol. Ther., 2017, 46, 617–627 CrossRef CAS.
- G. J. Ooi, S. Mgaieth, G. D. Eslick, P. R. Burton, W. W. Kemp, S. K. Roberts and W. A. Brown, Obes. Rev., 2018, 19, 281–294 CrossRef CAS.
- M. Soresi, L. Giannitrapani, M. Cervello, A. Licata and G. Montalto, World J. Gastroenterol., 2014, 20, 18131–18150 CrossRef.
- A. A. M. Shaheen, A. F. Wan and R. P. Myers, Am. J. Gastroenterol., 2007, 102, 2589–2600 CrossRef.
- M. Staderini, A. Megia-Fernandez, K. Dhaliwal and M. Bradley, Bioorg. Med. Chem., 2018, 26, 2816–2826 CrossRef CAS.
- V. P. Torchilin, Nat. Rev. Drug Discovery, 2014, 13, 813–827 CrossRef CAS.
- H. N. Abdelhamid, H.-F. Wu, D. Peer, J. M. Karp and S. Hong, Trends Biotechnol., 2013, 2, 548–556 Search PubMed.
- S. Lee, J. Xie and X. Chen, Biochemistry, 2010, 49, 1364–1376 CrossRef CAS.
- T. Higashi, S. L. Friedman and Y. Hoshida, Adv. Drug Delivery Rev., 2017, 121, 27–42 CrossRef CAS.
- P. J. Giraudi, V. J. B. Becerra, V. Marin, N. C. Chavez-Tapia and N. Rosso, Exp. Mol. Pathol., 2015, 98, 85–92 CrossRef CAS.
- S. A. Mousavi, M. S. Fønhus, G. M. Kindberg, H. Tolleshaug and T. Berg, Cell Biol. Int., 2013, 37, 703–712 CrossRef CAS.
- M. Gary-Bobo, P. Nirdé, A. Jeanjean, A. Morère and M. Garcia, Curr. Med. Chem., 2007, 14, 2945–2953 CrossRef CAS.
- J. Brown, C. Delaine, O. J. Zaccheo, C. Siebold, R. J. Gilbert and E. Y. Jones, EMBO J., 2008, 27, 265–276 CrossRef CAS.
- A. Hille-Rehfeld, Biosci. Rep., 1995, 1241, 177–194 Search PubMed.
- S. P. Surendran, R. G. Thomas, M. J. Moon and Y. Y. Jeong, Int. J. Nanomed., 2017, 12, 6997–7006 CrossRef CAS.
- Z. Chen, W. Jin, H. Liu, Z. Zhao and K. Cheng, Mol. Pharm., 2015, 12, 2180–2188 CrossRef CAS.
- J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser and C. Simmerling, J. Chem. Theory Comput., 2015, 11, 3696–3713 CrossRef CAS.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- I. S. Joung and T. E. Cheatham, J. Phys. Chem. B, 2008, 112, 9020–9041 CrossRef CAS.
- S. Joung and T. E. Cheatham, J. Phys. Chem. B, 2009, 113, 13279–13290 CrossRef.
- J.-P. Ryckaert, G. Ciccotti and H. J. C. Berendsen, J. Comput. Phys., 1977, 23, 327–341 CrossRef CAS.
- B. R. Miller, T. D. McGee, J. M. Swails, N. Homeyer, H. Gohlke and A. E. Roitberg, J. Chem. Theory Comput., 2012, 8, 3314–3321 CrossRef CAS.
- W. C. Still, A. Tempczyk, R. C. Hawley and T. Hendrickson, J. Am. Chem. Soc., 1990, 112, 6127–6129 CrossRef CAS.
- A. Contini, N. Ferri, R. Bucci, M. G. Lupo, E. Erba, M. L. Gelmi and S. Pellegrino, Pept. Sci., 2018, 110, e23089 CrossRef.
- Chemical Computing Group ULC, 2018.
- G. Valentino, C. Zivko, F. Weber, L. Brülisauer and P. Luciani, Pharmaceutics, 2019, 11, 676 CrossRef CAS.
- S. Pietkiewicz, J. H. Schmidt and I. N. Lavrik, J. Immunol. Methods, 2015, 423, 99–103 CrossRef CAS.
- P. H. Lin, R. Selinfreund, E. Wakshull and W. Wharton, Biochemistry, 1987, 26, 731–736 CrossRef CAS.
- B. P. Gray and K. C. Brown, Chem. Rev., 2014, 114, 1020–1081 CrossRef CAS.
- M. Roveri, A. Pfohl, P. Jaaks, N. Alijaj, J.-C. Leroux, P. Luciani and M. Bernasconi, Nanomedicine, 2017, 12, 1135–1151 CrossRef CAS.
- F. Wang, L. Chen, R. Zhang, Z. Chen and L. Zhu, J. Controlled Release, 2014, 196, 222–233 CrossRef CAS.
- S. Song, D. Liu, J. Peng, Y. Sun, Z. Li, J.-R. Gu and Y. Xu, Int. J. Pharm., 2008, 363, 155–161 CrossRef CAS.
- Z. Jiang, J. Guan, J. Qian and C. Zhan, Biomater. Sci., 2019, 7, 461–471 RSC.
- S. Kanki, D. E. Jaalouk, S. Lee, A. Y. C. Yu, J. Gannon and R. T. Lee, J. Mol. Cell. Cardiol., 2011, 50, 841–848 CrossRef CAS.
- C. N. Pace, G. Horn, E. J. Hebert, J. Bechert, K. Shaw, L. Urbanikova, J. M. Scholtz and J. Sevcik, J. Mol. Biol., 2001, 312, 393–404 CrossRef CAS.
- A. Walrant, A. Vogel, I. Correia, O. Lequin, B. E. S. Olausson, B. Desbat, S. Sagan and I. D. Alves, Biochim. Biophys. Acta, Biomembr., 2012, 1818, 1755–1763 CrossRef CAS.
- C. Trautwein, S. L. Friedman, D. Schuppan and M. Pinzani, J. Hepatol., 2015, 62, S15–S24 CrossRef CAS.
- T. Tsuchida and S. L. Friedman, Nat. Rev. Gastroenterol. Hepatol., 2017, 14, 397–411 CrossRef CAS.
- P. J. De Bleser, C. D. Scott, T. Niki, G. Xu, E. Wisse and A. Geerts, Hepatology, 1996, 23, 1530–1537 CrossRef CAS.
- M. E. Hemric, F. W. M. Lu, R. Shrager, J. Carey and J. M. Chalovich, J. Biol. Chem., 1993, 268, 15305–15311 CrossRef CAS.
- L. Beljaars, Hepatology, 1999, 29, 1486–1493 CrossRef CAS.
- L. Beljaars, P. Olinga, G. Molema, P. De Bleser, A. Geerts, G. M. M. Groothuis, D. K. F. Meijer and K. Poelstra, Liver, 2001, 21, 320–328 CrossRef CAS.
- M. Uhlen, P. Oksvold, L. Fagerberg, E. Lundberg, K. Jonasson, M. Forsberg, M. Zwahlen, C. Kampf, K. Wester, S. Hober, H. Wernerus, L. Björling and F. Ponten, Nat. Biotechnol., 2010, 28, 1248–1250 CrossRef CAS.
- J. Meyer and T. E. Schefter, Radiation Therapy for Liver Tumors, 2017.
- S. C. D. van IJzendoorn, M. M. P. Zegers, J. W. Kok and D. Hoekstra, J. Cell Biol., 1997, 137, 347–357 CrossRef CAS.
- A. J. Harris, S. L. Dial and D. A. Casciano, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2004, 549, 79–99 CrossRef CAS.
- T. Prabakaran, R. Nielsen, S. C. Satchell, P. W. Mathieson, U. Feldt-Rasmussen, S. S. Sørensen and E. I. Christensen, PLoS One, 2012, 7, e39975 CrossRef CAS.
- H. M. El-Shewy, M. H. Lee, L. M. Obeid, A. A. Jaffa and L. M. Luttrell, J. Biol. Chem., 2007, 282, 26150–26157 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0tb02372h |
| This journal is © The Royal Society of Chemistry 2021 |