Open Access Article
Open Access ArticleCascaded pattern formation in hydrogel medium using the polymerisation approach†
Keita
Abe
 a,
Satoshi
Murata
a and
Ibuki
Kawamata
a,
Satoshi
Murata
a and
Ibuki
Kawamata
 *ab
*ab
aDepartment of Robotics, Graduate School of Engineering, Tohoku University, Japan. E-mail: keita.abe.t7@dc.tohoku.ac.jp; satoshi.murata.a4@tohoku.ac.jp; ibuki.kawamata@tohoku.ac.jp
bNatural Science Division, Faculty of Core Research, Ochanomizu University, Japan
First published on 4th June 2021
Abstract
Reaction-diffusion systems are one of the models of the formation process with various patterns found in nature. Inspired by natural pattern formation, several methods for designing artificial chemical reaction-diffusion systems have been proposed. DNA is a suitable building block to build such artificial systems owing to its programmability. Previously, we reported a line pattern formed due to the reaction and diffusion of synthetic DNA; however, the width of the line was too wide to be used for further applications such as parallel and multi-stage pattern formations. Here, we propose a novel method to programme a reaction-diffusion system in a hydrogel medium to realise a sharp line capable of forming superimposed and cascaded patterns. The mechanism of this system utilises a two-segment polymerisation of DNA caused by hybridisation. To superimpose the system, we designed orthogonal DNA sequences that formed two lines in different locations on the hydrogel. Additionally, we designed a reaction to release DNA and form a cascade pattern, in which the third line appears between the two lines. To explain the mechanism of our system, we modelled the system as partial differential equations, whose simulation results agreed well with the experimental data. Our method to fabricate cascaded patterns may inspire combinations of DNA-based technologies and expand the applications of artificial reaction-diffusion systems.
Introduction
Various types of spontaneous patterns, such as stripes, spots, and wave patterns, are widely observed in nature. For example, animal fur and fish skin patterns are formed due to inter- and intra-cellular communication via reaction and diffusion encoded in the gene expression system. Such pattern formation processes can be well represented by a mathematical model expressed as partial differential equations.1–3 Inspired by those natural patterns, artificial chemical systems built on the reaction-diffusion model can produce certain types of ordered patterns.4,5To expand various pattern formation, it is necessary to programme the reaction and diffusion by tuning various parameters of the chemical reaction systems, such as rate constants and diffusion constants. Synthetic DNA is a useful material for this purpose because of its programmability since interactions among DNA strands can be designed as hybridisation between complementary sequences of nucleotides.6 The strength of the interactions among DNA sequences can be thermodynamically predicted using efficient algorithms,7–11 and DNA sequences with desired thermodynamic properties can be computationally designed based on the prediction.12 Such designed DNA sequences can be chemically synthesised by an automated DNA synthesiser, making DNA a practical building block for artificial reaction systems.13,14 Various logic circuits proposed in DNA computing demonstrate complex information processing abilities implemented as chemical reactions.15–20 However, these systems are usually implemented in a well-mixed test tube, and thus, they are limited in utilising spatial inhomogeneity.
Recently, some research on DNA-based artificial reaction-diffusion systems has been reported where spatio-temporal pattern formations have been investigated theoretically and experimentally.21–32 As a theory, the potential of the DNA-based system was shown in computer simulation research, although it was not realistic to implement as it is.22–24 In practice, various experimental techniques to realise DNA reaction systems, such as strand displacement, photoresponsive moieties, and various DNA processing enzymes are utilized.25–29 For instance, propagating waves by amplification reactions, spiral patterns by oscillating reactions,29 and French flag patterns by activation and inhibition30 have been demonstrated. Because these reaction-diffusion systems are built-in solutions where the diffusion of molecules is considerably fast, the formed patterns are strongly subject to dissipation. To maintain the formed pattern over time, it is necessary to employ out-of-equilibrium reaction systems that are equally fast as diffusion.
In contrast, various synthetic DNA-based reaction-diffusion systems have been built in a hydrogel material. In hydrogels, the diffusion can be slowed down to the range in which we can utilise programmable reactions implemented with DNA. The properties of the DNA reaction can be adjusted by the toehold lengths and base sequences, which is useful to realise desired reaction dynamics for pattern formation. Example patterns formed in the hydrogel medium by DNA include fixed lines, metaboric lines, rings, and detected edges.25–28,41 Moreover, because the polymer network of the hydrogel medium can completely suppress the diffusion of larger molecules, the formed pattern can be maintained over time after the reaction reaches equilibrium.25 Another advantage of the hydrogel is that it can be moulded into any shape, and specific DNA species can be immobilised at designated positions in the gel.25–27,32
DNA reaction in a hydrogel medium also enables us to programme not only the reaction but also diffusion. We have proposed methods to modulate the diffusion of DNA by using reversible interactions between diffusible DNA and anchored DNA to the gel polymer.32,33 Combinations between the above methods and enzymatic reactions or DNA logic gates have been realized to construct various long-lived or immobilized patterns.25,31
The current research target has been shifted to realize hierarchical pattern formation, that is, the generated patterns in the previous reaction initiate the proceeding reactions for further pattern formation. Although such cascaded pattern formation has been theoretically proposed,22–24 experimental implementation remains quite challenging owing to the smaller reaction rate and lower yield,34,35 which results in a blurred pattern, making it difficult to distinguish several patterns in the near distance.
Here, we propose a novel method to design a DNA reaction–diffusion system for a cascaded pattern formation. Utilising a two-segment polymerisation of DNA, in which two types of single-stranded DNA alternately hybridise with each other to assemble a long double-stranded DNA, the molecule can be trapped in a narrow area in the hydrogel. This method enabled the formation of distinguishable sharper patterns compared with conventional methods. Using orthogonal DNA sequences, we created two independent patterns superimposed on the same hydrogel, which were then used to generate the next pattern using the same two-segment polymerisation strategy. The strategy was successfully demonstrated in a one-dimensional experiment, the results of which were well-modelled using partial differential equations and explained by numerical simulations.
Our method of cascaded pattern formation can be combined with other techniques for controlling chemical and physical properties of a material through DNA interactions (i.e. beads-beads interactions,53 crosslink density,48 or production of enzymes54). If we can further materialise the formed pattern (i.e. beads aggregations and gel–sol transition), it may facilitate the realisation of an artificial system that spontaneously builds its structure, similar to biological development.
Material and methods
DNA solutions
Unmodified DNA with OPC (Oligonucleotide Purification Cartridge) purification was purchased from Eurofins Genomics Japan. Fluorescent-labelled DNA with HPLC purification was purchased from Sangon Biotech Co., Ltd China, except for the FAM-labelled DNA for fluorescence recovery after photobleaching (FRAP) experiment. The exceptional DNA with OPC purification quality was also purchased from Eurofins Genomics. These DNA sequences are detailed in Fig. S1 and Table S1 (ESI†). All the DNAs were diluted in Milli-Q water to a concentration of 100 μM and stored at −30 °C.The reaction buffer contained 50 mM HEPES, 20 mM MgCl2, and 100 mM NaCl (pH 7.13). To prepare the hydrogel medium, a pre-gel solution was prepared by mixing the reaction buffer, 40% acrylamide, and 2% N,N′-methylenebis (acrylamide) to make a 10% polyacrylamide gel (C% = 5) containing the buffer. For gelation, immediately after mixing 0.1% APS and 0.1% TEMED, we applied the solution into a mould (Fig. S2, ESI†) while preventing bubbles, setting a comb, and incubating it at room temperature (25 °C) for 1 h. All reagents were purchased from FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan, except 40% acrylamide, 2% N,N′-methylenebis (acrylamide) (Bio-Rad Laboratories, Inc.), and NaCl (Sigma Life Science). The final concentration and the volume of DNA applied in the pocket of the hydrogel were 10 μM and 4 μL, respectively.
Imaging with fluorescent microscopy
We visualised the reaction field using a Nikon TE2000 U with four types of filter units for four channels of fluorescent observation (FAM, Cy5, AMCA, and Cy3). Imaging of one reaction field was performed at intervals of 10 min over 24 h. In practice, by facilitating an automated stage (Shiguma Koki, Japan), five reaction fields were imaged in parallel.Image composition
Pseudo-colour images were obtained from the grey scale images of the reaction field. Bisector pattern formation was visualised from the FAM and Cy5 images, assigning them to green and magenta channels, respectively. Stripe pattern formation was visualised by merging the FAM (green) and AMCA (blue) images (Fig. S3, ESI†). In the cascaded pattern formation, Cy5 was used for both R1 and R3 DNAs, whereas Cy3 was used for both R2 and L3 DNAs. Thus, the distribution of pair 3 was visualised from Cy5 and Cy3 images using the “Multiply” function of ImageJ.36 The resultant image was assigned to a red channel composed of a visualised image with other channels of FAM (green) and AMCA (blue) (Fig. S4, ESI†).FRAP experiment
We performed the FRAP experiment to measure the diffusion coefficients of DNA in a 10% polyacrylamide gel. The gel prepared in a tube with a volume of 10 μL was placed on a silicon chamber, covered with liquid paraffin to prevent evaporation, and photo-stimulated using a TE-3000 confocal unit attached to Olympus IX83 (Fig. S5, S6 and Table S2, ESI†).Reaction-diffusion simulation
For the reaction-diffusion simulation, we employed the executable file “rdy.exe” packaged in a reaction-diffusion simulator Ready.37 The simulations were performed in an Elite Desk800 G4 SFF (Hewlett-Packard) equipped with NVIDIA FGeForce GTX 1650 for bisector pattern formation, or DAIV-DGX760H2-M2S5 (Mouse Computer) equipped with NVIDIA GeForce RTX 2080 for stripe and cascaded pattern formations. From Visual Toolkit Image Data (vti) formatted results of simulations, numerical information such as concentration distribution was obtained using the visual toolkit (vtk) library38 of Python.Result and discussion
DNA polymerisation for bisector pattern formation
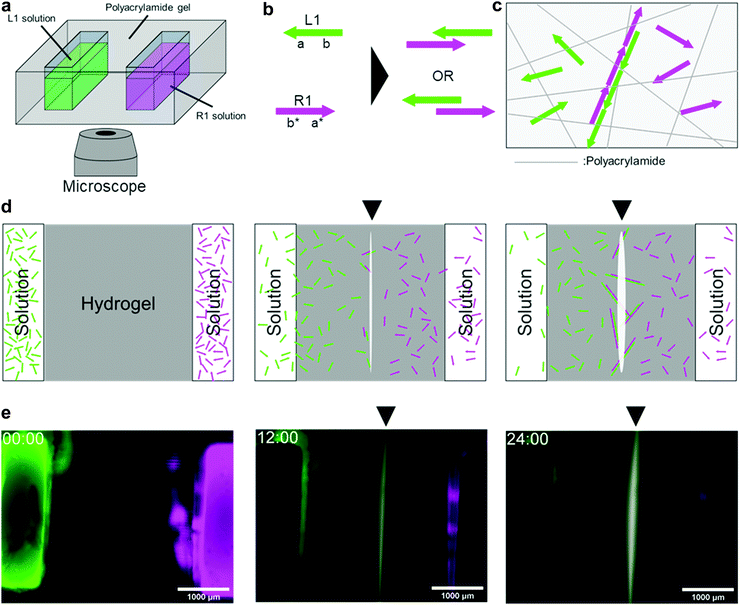
We programmed a reaction–diffusion system wherein two types of DNAs diffuse and hybridise with each other somewhere around the centre (Fig. 1). In the programme, we employed a polymerisation approach, which is a method to immobilise DNA in a hydrogel medium as a result of polymer formation by hybridisation. We implemented the programme using polyacrylamide gel and fluorescent-labelled DNA. The moulded gel with two pockets 3 mm apart, was placed on a microscope stage (Fig. 1a). We applied a 4 μL solution containing DNA into the pockets and used them as initial sources.A 46 nt of single-stranded DNA named L1 was placed in the left pocket, whereas another 46 nt single-stranded DNA named R1 was placed in the right pocket. For observation, L1 and R1 were modified with FAM (green) and Cy5 (magenta), respectively. They diffuse in the hydrogel medium (Fig. S8, ESI†), and when they encounter, they hybridise through 23 nt domains complementary to each other (Fig. 1b). This hybridisation repeats alternately as long as both DNA strands are present, forming a double-stranded DNA polymer (Fig. 1c). The polymer becomes longer as the hybridisation continues, and its diffusion coefficients become smaller following the Stokes–Einstein equation.39 Consequently, the DNA polymer was immobilised on a polyacrylamide hydrogel (Fig. 1c and d).
In 24 h of fluorescent observation, a white line appears on the bisector between the source pockets(Fig. 1e). As white is the composition of green (FAM) and magenta (Cy5), the white area represents the colocalisation of L1 (FAM-labelled) and R1 (Cy5-labelled). The white line appeared at the midpoint between the pockets because L1 and R1 had the same number of bases and diffused at approximately the same speed. This result suggests that the programme for bisector pattern formation works successfully.
Superimposed pattern formation using adjuster DNA and orthogonal DNA pairs.
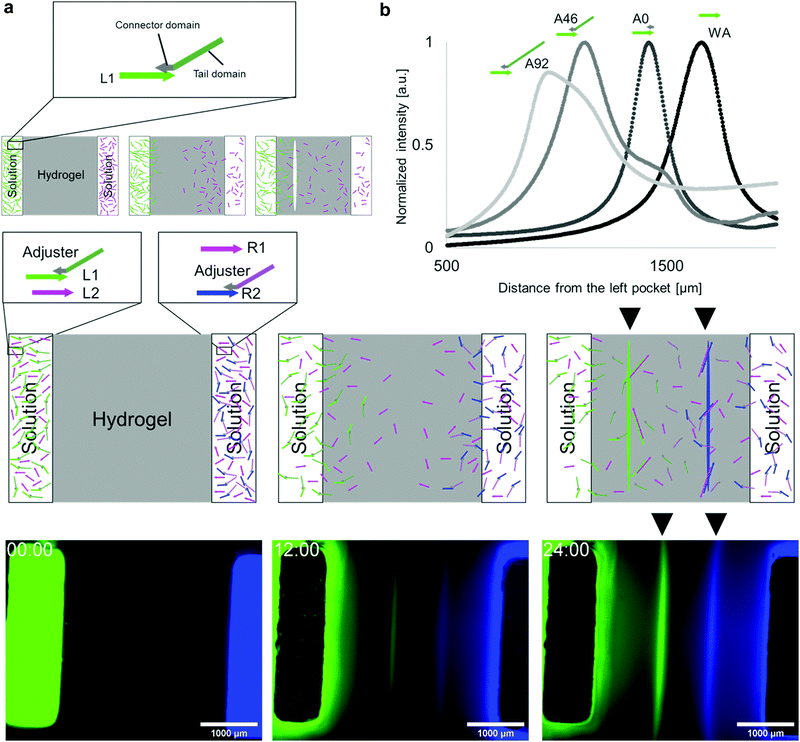
It is possible to shift the position of the white line by changing the diffusion speeds of L1 and R1. For example, when L1 diffuses slower than R1, polymerisation occurs closer to the L1 source. Therefore, a line appears on the left side of the bisector.To tune the DNA diffusion speed, we introduced adjuster DNAs. The adjuster hybridises with L1 DNA via a connector domain and slows down the diffusion because it increases the net molecular size. The adjuster also has a tail domain for tuning the molecular size (Fig. 2a). As the connector domain (15 nt) is shorter than the base pairs created by the hybridisation between L1 and R1 (23 bp), the adjuster can be released by a strand displacement reaction15,35 when the polymerisation process takes place.
We prepared three types of adjusters with 0, 46, and 92 nt tail lengths (A0, A46, and A92, respectively) and measured the line positions (Fig. 2b). The line positions from the left source, without the adjuster, A0, A46, and A92 were (1.65 ± 0.03) × 103 μm, (1.27 ± 0.07) × 103 μm, (1.05 ± 0.05) × 103 μm, and (0.98 ± 0.02) × 103 μm, respectively. Longer tails cause the line position to shift to the left as expected, which indicates the effectiveness of the diffusion tuning.
To demonstrate the multiple pattern formation superimposed in the same field, an additional DNA pair (pair 2 (L2 and R2)), which is orthogonal40 to pair 1 (L1 and R1), was designed (Fig. 2c). L1 and L2 were placed in the left source, whereas R1 and R2 were in the right source. Using the adjuster, the diffusion of L1 and R2 was set to be slower than that of R1 and L2.
L2 and R2 were modified with Cy3 and AMCA, respectively, and the distributions of the two DNA pairs were visualised by four-channel fluorescent observation. When the fluorescence images of FAM and AMCA were assigned to the green and blue channels in Fig. 2d, two lines appeared within 24 h. The results clearly show that the patterns formed by pairs 1 and 2 are formed in the left and right positions, respectively, as expected, indicating that orthogonal pairs can react independently.
Cascaded pattern formation
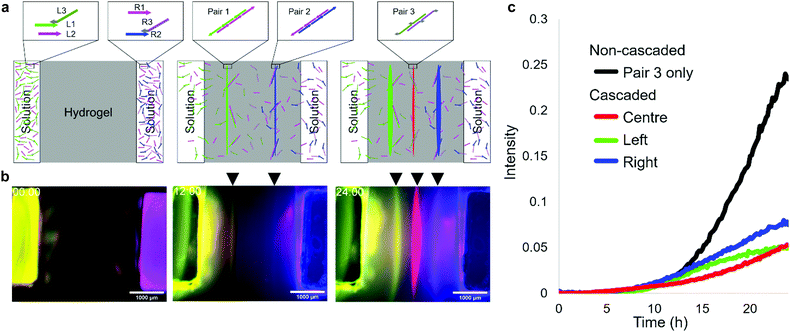
The DNA reaction–diffusion system can be cascaded in such a way that a formed pattern is used as a source of DNA in subsequent pattern formation. We designed a system that uses the previous two lines for third-line pattern formation. In the system, L1 and R2 are hybridised with adjusters L3 and R3, respectively, through 15 nt connector domains. The 46 nt tail domains of L3 and R3 have new base sequences for the third two-segment polymerization, which are orthogonal to the sequence of pairs 1 and 2 DNAs. During the polymerization of left (pair 1) and right (pair 2) lines, L3 and R3 are released and then form the centre line (pair 3) between the two (Fig. 3a).In the fluorescent observation, L1, R1, L2, R2, L3, and R3 were modified with FAM, Cy5, Cy3, AMCA, Cy3, and Cy5, respectively, and the results were composed by assigning FAM, AMCA, and (merged Cy3 and Cy5) to green, blue, and red, respectively. After 12 h, yellow (green + red) and magenta (blue + red) lines appeared, after which a red line appeared between them (Fig. 3b). The intensity of left and right lines reached 0.05 at 22.5 h and 18.5 h, respectively, then the centre line appeared at 23.5 h (Fig. 3c). In the case of a non-cascaded process using the same pair 3 as in the centre line (Fig. S7, ESI†), the intensity is reached the same level at 15 h. This experimental result indicates successful pattern formation using a cascaded reaction-diffusion system.
Simulation of non-cascaded, superimposed, and cascaded pattern formation
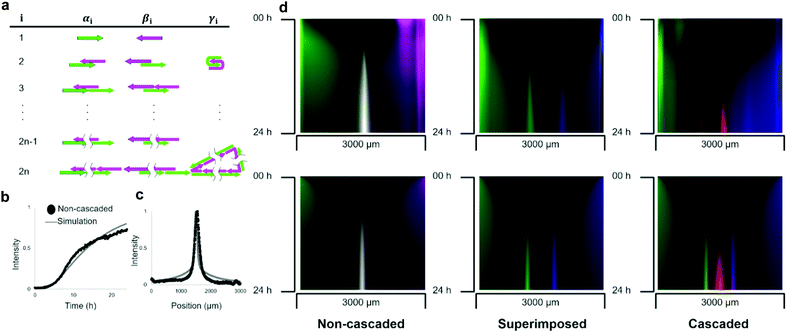
These three types of pattern formations were compared with the results of the reaction–diffusion simulation. In the simulation, the polymerisation process was modelled as a set of chemical reactions in which molecular species are intermediate polymerisation products. The products were classified into three groups according to their reactivity characterised by a left-most sticky end (Fig. 4a): the group α has 5′ end of L1, the group β has 3′ end of R1, and group γ is a circular polymer.The number of DNA strands in a polymer determines the size of the molecule. As the polymerisation process continues unlimitedly, we set N as the maximum number of strands to limit the structure enumeration. Based on the comparison with the electrophoresis results (Fig. S1, ESI†), N = 16 was used in the simulation. In the model, hybridisation of complementary single-stranded domains among intermediate structures was considered as a reaction (S5). The diffusion was modelled as a concentration-based isotropic spreading of molecules, where the diffusion coefficient of the intermediate structure was inversely related to the number of strands, which agrees with the Stokes–Einstein theory.39 The partial differential equations of the model were solved numerically.
Parameters such as rate constants and diffusion coefficients were determined by fitting the simulation curves to the experimental data (Fig. S7, ESI†). We assumed that the experimentally observed fluorescence intensity is proportional to the sum of the concentrations of intermediate structures multiplied by the number of fluorescent-labelled DNA. To fit the simulation, we compared the time development of the non-cascaded pattern between the experiment and simulation (Fig. 4b). For simplicity, a comparison was made only for the position that gives the concentration peak at 24 h. The obtained hybridisation constant was kh = 3.5 × 10−4 M−1 s−1, the diffusion coefficient of single-stranded DNA was D1 = 20 μm2 s−1, and the rate constant of intramolecular hybridisation was kc = 3.0 × 10−3 s−1. We think the value of diffusion coefficient is reasonable because it has the same order of magnitude with the values obtained by FRAP analysis (Table S2, ESI†). For example, the relative error between the experiment and simulation results that gives 0.5 fluorescent intensities was less than 9%.
To compare the pattern formed after 24 h in the experiment and simulation, the geometric means of the fluorescent intensities of L1 and R1 were plotted (Fig. 4c). As expected, the simulation results agree with the experimentally formed pattern. Quantitatively, the positions of the peaks were 1.55 and 1.50 mm in the experiment and simulation, respectively, giving a relative error of less than 4%. We further quantified the relative error of the half-width of the curve, which was 31% (1.8 × 102 and 1.4 × 102 μm in the experiment and simulation, respectively).
To simulate the superimposed and cascaded pattern formations, we created similar models. In the case of the superimposed pattern, almost the same polymerisation process takes place, except for the reaction of displacing adjuster strands. The rate constant of the hybridisation reaction in the model, which changed to this displacement reaction, is substituted with ks from kh. The kymographs of the fluorescence distribution between the pockets show that the simulation successfully agreed with the experiments (Fig. 4d).
Conclusions
We have proposed a simple but novel method that utilises the DNA polymerisation approach to form sharp line patterns in a hydrogel medium. The programmability of the position of the formed lines was demonstrated by implementing reaction–diffusion systems employing adjuster DNA strands.Our polymerisation approach, which requires three DNA strands, is concise compared with the conventional methods that form similar line patterns based on DNA reaction–diffusion systems with seven26 and eleven strands.25 The half-widths of the lines formed by our method were significantly sharper than those of the other methods. The value was 181 μm (6% of the distance between two sources) compared to 842 μm, which is 55% of the source distance in the conventional method.25
These improvements made it possible to design orthogonal DNA sequences to form multiple patterns in parallel and in sequence. As demonstrations, we succeeded in realising superimposed and cascaded pattern formations in which two lines appeared simultaneously, and an additional line appeared between them.
To validate the polymerisation approach results in pattern formation, we modelled and simulated a DNA reaction–diffusion system based on partial differential equations. The agreement between the simulation and experimental results suggests that the pattern formation behaviour is ascribed to the designed reaction and diffusion of DNA.
As the polymerisation approach is driven by DNA hybridisation reactions, this method is compatible with other DNA-based systems such as enzymatic reactions42,43 and DNA hydrogel.41,44–48 For example, if sufficient DNA can be supplied, gel–sol transition of DNA material may be possible.55 As potential applications of this cascaded pattern formation, we can anticipate building artificial systems that autonomously construct their own structures, mimicking the pattern formation process seen in the development of living organisms.49 In the morphogenesis of flies, stripe patterns are formed step by step from a simple concentration gradient, and body segments are materialised based on this pattern.50 Inspired by these processes, we assume that the cascaded pattern formation can be combined with other technologies that control the concentration of other functional molecules, such as enzymes, and change the physical properties of the hydrogel medium.51,52
Author contributions
K. A., S. M., and I. K. conceived and developed the project. K. A. performed experiments and computer simulations. K. A., S. M., and I. K. analyzed and interpret the data and wrote the manuscript.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by JSPS KAKENHI Grant Numbers JP18K18144, JP19KK0261, JP20H05971, JP20H00618, JP20K20979, JP20H05969, JP19J20990.References
- A. M. Turing, Philos. Trans. R. Soc. London, 1952, B237, 37–72 CAS.
- S. Kondo and R. Asai, Nature, 1995, 376(6543), 765 CrossRef CAS PubMed.
- S. Kondo and T. Miura, Science, 2010, 329(5999), 1616–1620 CrossRef CAS PubMed.
- A. Adamatzky and B. L. Costello, Phys. Lett. A, 2003, 309.5-6, 397–406 CrossRef.
- A. Sirimungkala, H. D. Försterling, V. Dlask and R. J. Field, J. Phys. Chem. A, 1999, 103(8), 1038–1043 CrossRef CAS.
- J. D. Watson and F. H. Crick, Nature, 1953, 171.4356, 737–738 CrossRef PubMed.
- J. Santalucia, H. T. Allawi and P. A. Seneviratne, Biochemistry, 1996, 35.11, 3555–3562 CrossRef PubMed.
- J. Santalucia, Proc. Natl. Acad. Sci. U. S. A., 1998, 95.4, 1460–1465 CrossRef PubMed.
- N. Sugimoto, S. Nakano, M. Yoneyama and K. Honda, Nucleic Acids Res., 1996, 24(22), 4501–4505 CrossRef CAS PubMed.
- D. Y. Zhang and E. Winfree, J. Am. Chem. Soc., 2009, 131(47), 17303–17314 CrossRef CAS PubMed.
- M. Zuker, Nucleic Acids Res., 2003, 31.13, 3406–3415 CrossRef PubMed.
- J. N. Zadeh, C. D. Steenberg, J. S. Bois, B. R. Wolfe, M. B. Pierce, A. R. Khan and N. A. Pierce, J. Comput. Chem., 2011, 32(1), 170–173 CrossRef CAS PubMed.
- X. Song and J. Reif, ACS Nano, 2019, 13(6), 6256–6268 CrossRef CAS PubMed.
- C. Zhang, L. Ge, Y. Zhuang, Z. Shen, Z. Zhong, Z. Zhang and X. You, Sci. China Inf. Sci., 2019, 62(6), 61301 CrossRef.
- D. Soloveichik, G. Seelig and E. Winfree, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(12), 5393 CrossRef CAS PubMed.
- A. Okamoto, K. Tanaka and I. Saito, J. Am. Chem. Soc., 2004, 126(30), 9458–9463 CrossRef CAS PubMed.
- X. Chen, J. Am. Chem. Soc., 2011, 134(1), 263–271 CrossRef PubMed.
- Q. Lulu and E. Winfree, Science, 2011, 332.6034, 1196–1201 Search PubMed.
- N. Shimada, K. Saito, T. Miyata, H. Sato, S. Kobayashi and A. Maruyama, Adv. Funct. Mater., 2018, 28(17), 1707406 CrossRef.
- K. M. Cherry and L. Qian, Nature, 2018, 559(7714), 370–376 CrossRef CAS PubMed.
- K. Abe and S. Murata, New Gener Comput., 2020, 38, 379–393 CrossRef.
- D. Scalise and R. Schulman, Technology, 2014, 2(01), 55–66 CrossRef.
- I. Kawamata, S. Yoshizawa, F. Takabatake, K. Sugawara and S. Murata, International Conference on Unconventional Computation and Natural Computation. Springer, Cham, 2016. pp. 168–181 Search PubMed.
- I. Kawamata, T. Hosoya, F. Takabatake, K. Sugawara, S. M. Nomura, T. Isokawa, F. Peper, M. Hagiya and S. Murata, 2016 Fourth International Symposium on Computing and Networking (CANDAR). IEEE, 2016. pp. 215–221 Search PubMed.
- K. Abe, I. Kawamata, S. M. Nomura and S. Murata, Mol. Syst. Des. Eng., 2019, 4.3, 639–643 RSC.
- J. Zenk, D. Scalise, K. Wang, P. Dorsey, J. Fern, A. Cruz and R. Schulman, RSC Adv., 2017, 7(29), 18032–18040 RSC.
- S. Chen and G. Seelig, Soft Matter, 2020, 16(14), 3555–3563 RSC.
- S. M. Chirieleison, P. B. Allen, Z. B. Simpson, A. D. Ellington and X. Chen, Nat. Chem., 2013, 5(12), 1000 CrossRef CAS PubMed.
- A. Padirac, T. Fujii, A. Estévez-Torres and Y. Rondelez, J. Am. Chem. Soc., 2013, 135(39), 14586–14592 CrossRef CAS PubMed.
- A. S. Zadorin, Y. Rondelez, G. Gines, V. Dilhas, G. Urtel, A. Zambrano and A. Estévez-Torres, Nat. Chem., 2017, 9(10), 990 CrossRef CAS PubMed.
- G. Urtel, A. Estevez-Torres and J. C. Galas, Soft Matter, 2019, 15(45), 9343–9351 RSC.
- T. Hosoya, I. Kawamata, S. M. Nomura and S. Murata, New Gener. Comput., 2019, 37(1), 97–111 CrossRef.
- T. Rodjanapanyakul, F. Takabatake, K. Abe, I. Kawamata, S. M. Nomura and S. Murata, Phys. Rev. E, 2018, 97(5), 052617 CrossRef PubMed.
- B. Yurke, A. J. Turberfield, A. P. Mills, F. C. Simmel and J. L. Neumann, Nature, 2000, 406(6796), 605–608 CrossRef CAS PubMed.
- N. Srinivas, J. Parkin, G. Seelig, E. Winfree and D. Soloveichik, Science, 2017, 358(6369), eaal2052 CrossRef PubMed.
- C. T. Rueden, J. Schindelin, M. C. Hiner, B. E. DeZonia, A. E. Walter, E. T. Arena and K. W. Eliceiri, BMC Bioinf., 2017, 18(1), 1–26 CrossRef PubMed.
- T. Hutton, R. Munafo, A. Trevorrow, T. Rokicki and D. Wills, Ready, a cross-platform implementation of various reaction-diffusion systems, https://github.com/GollyGang/ready.
- https://vtk.org/ .
- A. Einstein, [AdP 17, 549 (1905)], Ann. Phys., 2005, 14.S1 1, 182–193 CrossRef.
- T. Kitajima, M. Takinoue, K. I. Shohda and A. Suyama, International Workshop on DNA-Based Computers. Springer, Berlin, Heidelberg, 2007. pp. 119–129 Search PubMed.
- P. J. Dorsey, D. Scalise and R. Schulman, Angew. Chem., 2021, 133.1, 342–348 CrossRef.
- K. Montagne, R. Plasson, Y. Sakai, T. Fujii and Y. Rondelez, Mol. Syst. Biol., 2011, 7.1, 466 CrossRef PubMed.
- T. Fujii and Y. Rondelez, ACS Nano, 2013, 7.1, 27–34 CrossRef PubMed.
- F. Li, D. Lyu, S. Liu and W. Guo, Adv. Mater., 2020, 32(3), 1806538 CrossRef CAS.
- D. Kandatsu, K. Cervantes-Salguero, I. Kawamata, S. Hamada, S. M. Nomura, K. Fujimoto and S. Murata, ChemBioChem, 2016, 17(12), 1118–1121 CrossRef CAS PubMed.
- D. C. Lin, B. Yurke and N. A. Langrana, J. Biomech. Eng., 2004, 126.1, 104–110 CrossRef PubMed.
- Z. Zhu, C. Wu, H. Liu, Y. Zou, X. Zhang, H. Kang and W. Tan, Angew. Chem., Int. Ed., 2010, 49(6), 1052–1056 CrossRef CAS PubMed.
- A. Cangialosi, C. Yoon, J. Liu, Q. Huang, J. Guo, T. D. Nguyen and R. Schulman, Science, 2017, 357(6356), 1126–1130 CrossRef CAS PubMed.
- L. Wolpert, T. Cheryll and A. M. Arias, Principles of Development, Oxford University Press, USA, 2015 Search PubMed.
- D. St Johnston and C. Nüsslein-Volhard, Cell, 1992, 68(2), 201–219 CrossRef CAS PubMed.
- M. Lovrak, W. E. Hendriksen, C. Maity, S. Mytnyk, V. van Steijn, R. Eelkema and J. H. van Esch, Nat. Commun., 2017, 8, 15317 CrossRef CAS PubMed.
- M. Lovrak, W. E. Hendriksen, M. T. Kreutzer, V. van Steijn, R. Eelkema and J. H. van Esch, Soft Matter, 2019, 20, 20 Search PubMed.
- W. Zhao, W. Chiuman, M. A. Brook and Y. Li, ChemBioChem, 2007, 8(7), 727–731 CrossRef CAS PubMed.
- Y. J. Chen, B. Groves, R. A. Muscat and G. Seelig, Nat. Nanotechnol., 2015, 10(9), 748–760 CrossRef CAS PubMed.
- G. Zanchetta, Curr. Opin. Colloid Interface Sci., 2019, 40, 1–13 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sm00296a |
| This journal is © The Royal Society of Chemistry 2021 |