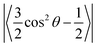Open Access Article
Open Access ArticleInvestigating the structural properties of hydrophobic solvent-rich lipid bilayers
Valeria
Zoni
,
Pablo
Campomanes
and
Stefano
Vanni
 *
*
Department of Biology, University of Fribourg, Chemin du Musée 10, 1700 Fribourg, Switzerland. E-mail: stefano.vanni@unifr.ch
First published on 28th April 2021
Abstract
In vitro reconstitutions of lipid membranes have proven to be an indispensable tool to rationalize their molecular complexity and to understand their role in countless cellular processes. However, amongst the various techniques used to reconstitute lipid bilayers in vitro, several approaches are not solvent-free, but rather contain residual hydrophobic solvents in between the two bilayer leaflets, generally as a consequence of the procedure used to generate the bilayer. To what extent the presence of these hydrophobic solvents modifies bilayer properties with respect to native, solvent-free, conditions remains an open question that has important implications for the appropriate interpretation of numerous experimental observations. Here, we thorouhgly characterize hydrophobic solvent-rich lipid bilayers using atomistic molecular dynamics simulations. Our data indicate that while the presence of hydrophobic solvents at high concentrations, such as hexadecane, has a significant effect on membrane thickness, their effects on surface properties, membrane order and lateral stress are quite moderate. Our results corroborate the validity of in vitro approaches as model systems for the investigations of biological membranes but raise a few cautionary aspects that must be considered when investigating specific membrane properties.
Introduction
Lipid membranes play an active role in the spatial and temporal modulation of biochemical processes in the cell, in large part thanks to their huge diversity in terms of lipid composition.1–3 Because of this complexity, cellular membranes can significantly vary in both their equilibrium properties as well as in their response to external stimuli such as bending, tension or interaction with proteins.1–3To understand the basic structural and material properties of lipid membranes, in vitro approaches have shown to be an invaluable tool in the last few decades.4,5 These approaches have been largely enabled by the relative ease in reconstituting lipid bilayers in aqueous medium, as a consequence of their intrinsic self-assembly properties as natural surfactants.
Amongst the various approaches used to reconstitute lipid bilayers in vitro, several of them are not solvent-free, but rather contain residual hydrophobic solvents in between the two bilayer leaflets, generally as a consequence of the procedure used to generate the bilayer.6–11 Amongst those, Black Lipid Membranes (BLMs),9 Droplet Interface Bilayers (DIBs),10 and Large-Area Model Biomembranes (LAMBs)11 are some of the most widely used.
While these techniques have advantages in terms of overall bilayer stability, for example decreasing water permeation and allowing to stabilize very large bilayers for very long times,11 the presence of residual hydrophobic solvents in the middle of the bilayer could potentially alter membrane properties and generate deviations from native-like conditions. One distinct advantage of solvent-rich lipid bilayers, however, is the relative straightforwardness in measuring their membrane capacitance,8,12,13 and hence their electrical properties.
Recently, we implemented a protocol to directly determine membrane capacitance from in silico investigations of lipid bilayers by means of molecular dynamics (MD) simulations.14 Using this approach, and based on calibration curves, we could infer the correct amount of two hydrophobic solvents that are widely used in the formation of lipid bilayers, squalene and hexadecane, by comparing the predicted data with those from experiments. We found that squalene is generally present in reconstituted bilayers at concentrations between 5 and 10 mol%, while hexadecane is much more abundant, with concentrations around 35 mol%.14
These results naturally raise doubts about whether these reconstitutions could be considered as good model systems for biological-like lipid bilayers, that are devoid of hydrophobic oils in physiological conditions,15,16 and to what extent the presence of these solvents affects bilayer properties that have been shown to be important to rationalize biological processes such as protein-membrane interactions.
To investigate this issue, here we use atomistic MD simulations, the method of choice to investigate structural and dynamical properties of lipid assemblies with atomistic resolution.17–21 With this methodology, we thoroughly analyze bilayer properties in solvent-rich lipid bilayers, and we compare it to solvent-free bilayers.
We found that while the presence of high amounts of oils, such as hexadecane, has a significant effect on membrane thickness, its effect on surface properties, membrane order and lateral stress is quite moderate. Our results corroborate the validity of such in vitro approaches as model systems for the investigations of biological membranes but raise a few cautionary aspects that must be considered when investigating specific membrane properties.
Materials and methods
Molecular dynamics simulations
Atomistic MD simulations of 100 mol% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), DOPC/Squalene (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 mol%) and DOPC/Hexadecane (68
10 mol%) and DOPC/Hexadecane (68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 32 mol%) were carried out using the software GROMACS22 as previously described in.14 In short, a DOPC bilayer containing 128 lipids was generated using the CHARMM-GUI membrane builder.23 Addition of hexadecane or squalene in the middle of the bilayer was achieved by displacing the two DOPC monolayers along the z-axis and by subsequent random insertion of the oil molecules, exclusively in a position parallel to the membrane, between the two monolayers. The systems were equilibrated for 375 ps while slowly removing constraints using the Membrane Builder six-step process.24 For all systems, the CHARMM36m force field25 was used to describe lipids while the TIP3P26 model was employed for water. Long-range electrostatic interactions were taken into account by means of the particle mesh Ewald (PME)27 algorithm with a Fourier grid space of 0.12 nm and a real space cutoff of 1.2 nm. The van der Waals interactions were truncated using the same cutoff value, and a standard smoothing function for the tail region (1.0–1.2 nm) was employed. The bonds involving hydrogen atoms were constrained using the LINCS28 and SETTLE29 algorithms, thus allowing to use of an integration time step of 2 fs. The temperature (298.15 K) and pressure (1 atm) were controlled by coupling the systems to a Nosé–Hoover chain thermostat30 with a time constant of 1 ps−1 and a Parrinello–Rahman barostat31 with a coupling constant of 5 ps−1, respectively.
32 mol%) were carried out using the software GROMACS22 as previously described in.14 In short, a DOPC bilayer containing 128 lipids was generated using the CHARMM-GUI membrane builder.23 Addition of hexadecane or squalene in the middle of the bilayer was achieved by displacing the two DOPC monolayers along the z-axis and by subsequent random insertion of the oil molecules, exclusively in a position parallel to the membrane, between the two monolayers. The systems were equilibrated for 375 ps while slowly removing constraints using the Membrane Builder six-step process.24 For all systems, the CHARMM36m force field25 was used to describe lipids while the TIP3P26 model was employed for water. Long-range electrostatic interactions were taken into account by means of the particle mesh Ewald (PME)27 algorithm with a Fourier grid space of 0.12 nm and a real space cutoff of 1.2 nm. The van der Waals interactions were truncated using the same cutoff value, and a standard smoothing function for the tail region (1.0–1.2 nm) was employed. The bonds involving hydrogen atoms were constrained using the LINCS28 and SETTLE29 algorithms, thus allowing to use of an integration time step of 2 fs. The temperature (298.15 K) and pressure (1 atm) were controlled by coupling the systems to a Nosé–Hoover chain thermostat30 with a time constant of 1 ps−1 and a Parrinello–Rahman barostat31 with a coupling constant of 5 ps−1, respectively.
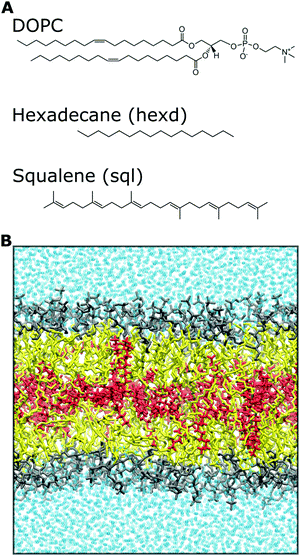
All systems (containing 128 DOPC lipids and about 77000 water molecules, plus 15 squalene or 60 hexadecane molecules) were run for at least 450 ns to collect statistics. Analyses were carried out excluding the first 50 ns, unless otherwise explicitly stated, and data points for analyses were collected every 10 ps. A representative snapshot of the molecular systems used in this study, together with the chemical structure of the molecules investigated, is shown in Fig. 1. All images were rendered using VMD32 and all plots were created using python scripts.
Bilayer analyses
| π(z) = PL(z) − PN |
For the calculation of LPPs, the program GROMACS-LS, with a grid of 0.1 nm along the z direction and a cut-off of 2.2 nm for electrostatics, was used33 (https://www.mdstress.org). For pure DOPC, the LPP was calculated over the last 200 ns of production, while, for the systems containing oils, configurations extracted from the last 300 ns of production were used for LPP calculations. The raw data have been processed through a Gaussian filter with standard deviation 1.0 using the utility “tensortools” of the GROMACS_LS program.
where θ is the average angle between the C–H vector and the bilayer normal. The calculation was performed using a tcl script (https://user.eng.umd.edu/~jbklauda/wiki/doku.php?id=scd).
Results and discussion
Hydrophobic solvents increase bilayer thickness and deplete phospholipids from the centre of the bilayer
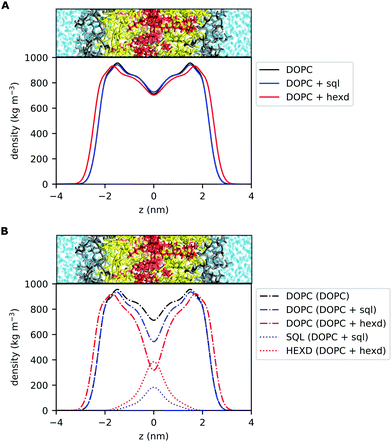
The most direct effect of the presence of residual solvents in the bilayer is an effective increase in its thickness.14 This is expected, as the membrane capacitance, that is inversely proportional to the membrane thickness,37 can be significantly lower than that of solvent free lipid bilayers when the membranes are prepared with hydrophobic oils.38 In detail, the distance between the phosphate groups of the two monolayers increases by approximately 5% (from 3.89 to 4.07 nm) in the presence of squalene and by 13% (from 3.89 to 4.38 nm) in the presence of hexadecane.14 To put this difference into some biological context, in the case of hexadecane this corresponds to roughly one entire α-helical turn (3.6 residues, 0.54 nm), a very common secondary structure conformation of transmembrane (TM) proteins. Interestingly, this difference is within the range of the observed difference between Golgi and Plasma Membrane proteins for vertebrates39 or in the range of what is sufficient to alter the localization of synthetic TM peptides in the yeast ER.40To further investigate the localization of hydrophobic solvents in the bilayer, we first compared the lateral density profiles of all non-water molecules in the bilayers. This corresponds to only DOPC molecules in the pure PL system and of DOPC + oil in the PLs + squalene and PLs + hexadecane systems (Fig. 2A). Interestingly, in the presence of squalene, the overall bilayer density profile (Fig. 2A) is quite similar to that of 100% DOPC. On the other hand, the presence of hexadecane significantly affects the density profile at almost all depths, with the sole exception of the middle of the bilayer (Fig. 2A).
Next, we computed the lateral density profiles of DOPC lipids and of the two oils (squalene, hexadecane) separately (Fig. 2B). This analysis indicates that, as expected, the two oils occupy predominantly the middle of the bilayer, i.e. the space in between the PL acyl chains (Fig. 2B). Interestingly, both squalene and hexadecane display broad tails that extend up to almost 2 nm from the centre of the bilayer (Fig. 2B).
To further characterize the molecular behaviour of oil molecules in the bilayer, we next computed their lateral diffusion and their molecular orientation. Interestingly, the presence of both squalene and hexadecane doesn’t alter the 2D diffusion of PLs in the bilayer, with DOPC diffusion coefficient being of 5.9 ± 1.6 μm2 s−1 in pure DOPC bilayers and of 6.2 ± 1.8 μm2 s−1 and 5.9 ± 4.7 μm2 s−1 in the presence of squalene and hexadecane, respectively. On the other hand, oil molecules diffuse much faster than PLs, with squalene and hexadecane having diffusion coefficients of 17.6 ± 12.1 μm2 s−1 and 114.1 ± 24.3 μm2 s−1, respectively. Of note, these results are consistent with the recent experimental observation that oil molecules diffuse faster than PLs in reconstituted solvent-rich lipid bilayers.41 On the other hand, our results seem in contrast with those recently obtained by Roke and coworkers in reconstituted lipid bilayers, who reported diffusion constants of squalene and hexadecane comparable to those of PLs.42 However, their measurements were made on large oil droplets that can be imaged with white-light microscopy, rather than on individual molecules as in our case. As the diffusion coefficient is generally inversely proportional to molecular size (e.g. in Stokes–Einstein equation), our observation that individual oil molecules diffuse much faster than individual PLs is hence in agreement with the experimental measurement of Roke and coworkers.42
As we observed that hexadecane molecules do not occupy exclusively the space between monolayers, but can also extend between the lipid acyl chains (Fig. 2B), we next characterized the molecular orientation of oil molecules, to quantify to what extent they interdigitate with DOPC acyl chains as previously suggested.42 To do so, we computed the orientation of the oil molecules with respect to the membrane normal (Fig. 3). Of note, while for both hexadecane and squalene the majority of the oil molecules lies between the two bilayer leaflets in a conformation that is perpendicular to the membrane normal (maximum at around 90 degrees in Fig. 3), hexadecane has a larger tendency to orient itself parallel to membrane normal (angles between 0 and 30 degrees) and to interdigitate with the acyl chains of the PLs.
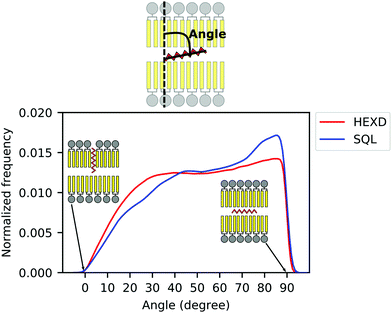 | ||
| Fig. 3 Molecular orientation of in-bilayer hydrophobic solvents. Angle between oil molecules and bilayer normal for squalene (blue) and hexadecane (red). | ||
Taken together, these data further suggest that, while squalene molecules are likely to only partially alter the property of the PL acyl chains close to the end of their tail, where they are located, hexadecane molecules are likely to impact bilayer properties closer to its interface with water, as observed from the significant difference in the overall density distribution (Fig. 2B) and molecular orientation (Fig. 3).
Hydrophobic solvents alter internal bilayer stress
To better characterize the effect of hydrophobic solvents on bilayer properties, we next computed the lateral pressure profiles (LPPs) for the three systems. In short, the LPP describes the distribution of lateral stresses across a bilayer that arise from the interactions between the lipid molecules, and it provides a direct connection between molecular (e.g. lipid–lipid interactions) and large-scale (e.g. surface tension, bending rigidity) properties of PL bilayers.As expected from the lateral density profiles in Fig. 2, the largest effect of the presence of oils in the LPPs was found in the middle of the bilayer, with the presence of hexadecane and squalene leading to a mild repulsion (positive values of the LPP) between molecules. Interestingly, this repulsion is larger for squalene (Fig. 4), despite its lower density there in comparison with hexadecane (Fig. 2B).
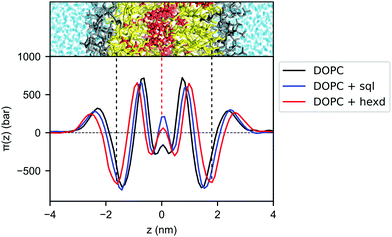 | ||
| Fig. 4 Lateral pressure profiles for DOPC bilayers with and without hydrophobic solvents. Pure DOPC (black), DOPC/sql (blue) and DOPC/hexd (red). | ||
Since the integral of the LPP corresponds to the membrane tension, this difference in pressure in the middle of the bilayer needs to be compensated elsewhere, in order to keep the bilayers tensionless, as it is the case in our simulations. In the case of squalene, this compensation takes place in the internal part of the bilayer, most notably by a decrease of the positive peaks at ±0.7 nm from the center of the bilayer, while the part of the bilayer closer to the surface (i.e. the negative peak at the level of the glycerol atom and the positive peak at the level of the polar heads) remains mostly unchanged. On the other hand, the effect of hexadecane on lateral stresses extends further away from the center of the bilayer, with an appreciable difference in the attraction between the lipids in the glycerol region (negative peaks around ±1.7 nm), consistent with the observation from the lateral density profiles (Fig. 2) that the effects of hexadecane extend closer to the bilayer surface.
Hexadecane molecules increase membrane order close to the middle of the bilayer
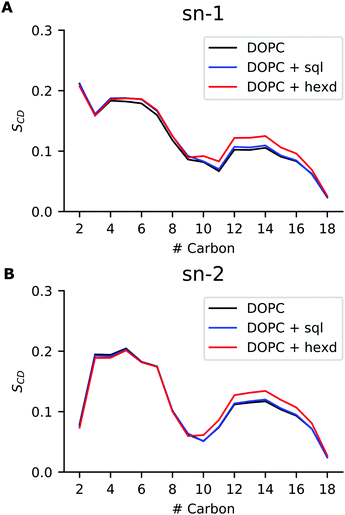
We next investigated conformational properties of lipid bilayers in the presence of hydrophobic solvents. As can be appreciated in Fig. 2B, the presence of the oils displaces the PL acyl chains, that become depleted in the middle of the bilayer. This has potential implications for structural and dynamical properties of the acyl chains. To quantify those, we next computed the acyl chain order parameters for the sn-1 and sn-2 chains of the DOPC molecules in the presence or absence of the residual solvents (Fig. 5). Order parameters provide information regarding both the overall order of the membrane and specific details of the conformations that the atoms within the lipid tails adopt. In detail, they describe the orientation of the C–H bond vector with respect to the bilayer normal (typically the z axis in a membrane simulation) averaged over all the lipids and the sampling time.As can be appreciated in Fig. 5, the presence of squalene has very minor effects on the order parameters of both the sn-1 and sn-2 chains. This is not the case for hexadecane, as its presence non-negligibly increases the order of both sn-1 and sn-2 chains starting from position 9, i.e. the site of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C double bond. This observation agrees with our previous analysis showing that hexadecane interdigitates with the acyl chains of the PLs (Fig. 3). Interestingly, however, despite the global differences in lateral density (Fig. 2A) and pressure (Fig. 4) up to the membrane surface in the presence of hexadecane, the order of the chains from position 9 up to the membrane surface doesn’t appear to be affected.
C double bond. This observation agrees with our previous analysis showing that hexadecane interdigitates with the acyl chains of the PLs (Fig. 3). Interestingly, however, despite the global differences in lateral density (Fig. 2A) and pressure (Fig. 4) up to the membrane surface in the presence of hexadecane, the order of the chains from position 9 up to the membrane surface doesn’t appear to be affected.
Hydrophobic solvents increase solvent-accessible hydrophobic surface area
Finally, we focused on the quantitative investigation of two membrane surface properties, area per lipid and lipid-packing defects, as both lateral density and pressure profiles seem to indicate that the perturbations promoted by the presence of hydrophobic solvents are mostly located in the bilayer interior.To do so, we first computed the area per lipid in the three simulations. Remarkably, despite the presence of 10 mol% squalene and 32 mol% hexadecane, the area per lipid in the presence of hydrophobic solvents increased only from 0.67 (±0.01) nm2 for pure DOPC to 0.69 (±0.01) nm2 for squalene-rich and to 0.71(±0.01) nm2 for hexadecane-rich bilayers.
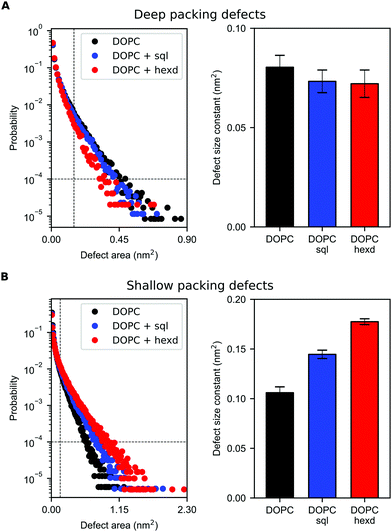
We next computed membrane-packing defects, i.e. the amount of hydrophobic surface area that is solvent-accessible, based on a cartesian algorithm developed in the software Pack-Mem.34,35 Of note, this property has been found to quantitatively correlate with membrane targeting by peripheral proteins36,43–45 and we thus use it as a proxy to evaluate whether the presence of hydrophobic solvents in reconstituted bilayers would alter the propensity of peripheral proteins to bind to them.
Remarkably, we found that the presence of squalene and hexadecane has only a very minor effect on deep (extending at least 0.1 nm below the average height of the glycerol groups of the PL) packing defects (Fig. 6A). Considering that solvent molecules display a non-negligible density up to 2 nm from the middle of the bilayer (Fig. 2B), the slight decrease in deep packing defects observed in Fig. 6A is likely due to interdigitation between the oils and the PL acyl chains,36 that prevents the formation of deep packing defects extending below the level of the glycerol atoms.
On the other hand, the presence of hydrophobic solvents increases shallow (i.e. surface-exposed, Fig. 6B) packing defects. This effect is more pronounced for hexadecane than squalene, likely as a consequence of the higher interdigitation between hexadecane molecules and PL chains, as hexadecane extends closer to the bilayer surface (Fig. 2B and 3).
Taken together, these data suggest that solvent-rich PL bilayers are a relatively good model system to investigate the binding of peripheral proteins to lipid membranes. This is particularly true for proteins able to sense deep packing defects, such as those that insert bulk hydrophobic residues.45 For proteins able to sense shallow defects, such as those inserting small hydrophobic residues,46 our simulations suggest that the presence of oil might increase their binding affinity, in particularly when non-physiological oils, such as hexadecane, are present at high concentrations.
Conclusions
In vitro reconstitutions of lipid bilayers are a powerful technique to investigate the complex role that biological membranes play in the cell. However, these reconstitutions might deviate from biological conditions, and a thorough understanding of their properties is required to infer causality from these investigations. In particular, several in vitro approaches rely on the use of hydrophobic solvents in the process of bilayer formation, and residual solvent molecules are known to remain in the bilayer.Here, we took advantage of a recent protocol to estimate hydrophobic solvent concentration in reconstituted lipid bilayers14 to investigate the structural properties of solvent-rich bilayers and compare them with solvent-free ones.
In addition to the well-known differences in membrane thickness, we found that, as expected, the largest effects on bilayer properties are found in the bilayer interior, where most of the hydrophobic solvent molecules reside. Interestingly, however, lipid order and surface properties are only marginally affected by the presence of oils, further corroborating the implicit decade-long assumption that these reconstitutions are good model systems for the investigation of protein-membrane interactions.
Interestingly, our observation that hexadecane orients itself parallel to the lipids’ acyl chains more often than squalene (Fig. 3) agrees well with decade-old considerations on the thermodynamic activity of hydrophobic solvents in reconstituted lipid bilayers.12,47 In particular, it appears that hydrophobic solvents that match well with the lipids’ acyl chain structure, such as hexadecane, can reside in bilayers at relatively high concentrations. On the other hand, hydrophobic molecules that experience a higher entropic penalty in their interactions with the lipid acyl chains, such as squalene, tend to be excluded from the bilayer.
Recently, Thiam and coworkers identified that oil molecules also have non-trivial effects on bilayer mechanics.48 For example, they found that the presence of oil molecules in the bilayer increases the critical area strain at which vesicle lysis occur, and that specific oils might alter bilayer bending rigidity.48 While our atomistic simulations are not well-adapted to the investigation of mechanical properties of solvent-rich lipid bilayers, we foresee that future coarse-grain MD simulations will help investigate the molecular origin of such mechanical effects.
For what pertains surface properties, however, we observed that hexadecane, one of the most common hydrophobic solvents in lipid bilayer reconstitutions, promotes non-negligible differences with respect to solvent-free bilayers. Most notably, we found that the presence of hexadecane increases the amount of surface-exposed hydrophobic patches, potentially leading to enhanced adsorption of peripheral proteins to the bilayer surface.
Taken together, our results highlight the power of MD simulations towards the establishment of structure–function relationships in membrane studies and posit for further integration of in silico and in vitro approaches towards the interpretation of complex processes happening at biological membranes.
Author contributions
VZ: investigation, formal analysis, writing – original draft; PC: investigation, formal analysis, validation, SV: conceptualization, supervision, writing – original draft.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Swiss National Science Foundation (grant PP00P3_163966) and by grants from the Swiss National Supercomputing Centre (CSCS) under project ID s842, s920 and s980. We acknowledge PRACE for awarding us access to Piz Daint, ETH Zurich/CSCS, Switzerland. SV and PC acknowledge support from the Novartis Foundation for medical-biological Research (17C139). SV acknowledges support from the NCCR for Bioinspired materials (51NF40-182881).Notes and references
- J. Bigay and B. Antonny, Dev. Cell, 2012, 23, 886–895 CrossRef CAS PubMed.
- G. Van Meer, D. R. Voelker and G. W. Feigenson, Nat. Cell Biol., 2008, 9, 112–124 CrossRef CAS PubMed.
- T. Harayama and H. Riezman, Nat. Rev. Mol. Cell Biol., 2018, 19, 281–296 CrossRef CAS PubMed.
- C. G. Siontorou, G.-P. Nikoleli, D. P. Nikolelis and S. K. Karapetis, Membranes, 2017, 7, 38 CrossRef PubMed.
- H. Jia and P. Schwille, Curr. Opin. Biotechnol, 2019, 60, 179–187 CrossRef CAS PubMed.
- S. H. White, Biophys. J., 1975, 15, 95–117 CrossRef CAS PubMed.
- G. J. Taylor, G. A. Venkatesan, C. P. Collier and S. A. Sarles, Soft Matter, 2015, 11, 7592–7605 RSC.
- L. C. M. Gross, A. J. Heron, S. C. Baca and M. I. Wallace, Langmuir, 2011, 27, 14335–14342 CrossRef CAS PubMed.
- D. Papahadjopoulos, Perspect. Biol. Med., 1975, 18, 575–577 CrossRef.
- H. Bayley, B. Cronin, A. Heron, M. A. Holden, W. L. Hwang, R. Syeda, J. Thompson and M. Wallace, Mol. BioSyst., 2008, 4, 1191–1208 RSC.
- P. J. Beltramo, R. Van Hooghten and J. Vermant, Soft Matter, 2016, 12, 4324–4331 RSC.
- S. H. White, Ann. N. Y. Acad. Sci., 1977, 303, 243–265 CAS.
- P. J. Beltramo, L. Scheidegger and J. Vermant, Langmuir, 2018, 34, 5880–5888 CrossRef CAS PubMed.
- V. R. Ardham, V. Zoni, S. Adamowicz, P. Campomanes and S. Vanni, J. Phys. Chem. B, 2020, 124, 8278–8286 CrossRef CAS PubMed.
- O. Adeyo, P. J. Horn, S. Lee, D. D. Binns, A. Chandrahas, K. D. Chapman and J. M. Goodman, J. Cell Biol., 2011, 192, 1043–1055 CrossRef CAS PubMed.
- V. Zoni, R. Khaddaj, P. Campomanes, A. R. Thiam, R. Schneiter and S. Vanni, eLife, 2021, 10, e62886 CrossRef PubMed.
- S. J. Marrink and D. P. Tieleman, Chem. Soc. Rev., 2013, 42, 6801–6822 RSC.
- S. J. Marrink, V. Corradi, P. C. T. Souza, H. I. Ingólfsson, D. P. Tieleman and M. S. P. Sansom, Chem. Rev., 2019, 119, 6184–6226 CrossRef CAS PubMed.
- G. Enkavi, M. Javanainen, K. Waldemar, T. Róg and I. Vattulainen, Chem. Rev., 2019, 119(9), 5607–5774 CrossRef CAS PubMed.
- T. A. Soares, S. Vanni, G. Milano and M. Cascella, J. Phys. Chem. Lett., 2017, 8, 3586–3594 CrossRef CAS PubMed.
- S. Vanni, L. Riccardi, G. Palermo and M. De Vivo, Acc. Chem. Res., 2019, 52, 3087–3096 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- S. Jo, T. Kim, V. G. Iyer and W. Im, J. Comput. Chem., 2008, 29, 1859–1865 CrossRef CAS PubMed.
- S. Jo, T. Kim and W. Im, PLoS One, 2007, 2, e880 CrossRef PubMed.
- J. Huang, S. Rauscher, G. Nawrocki, T. Ran, M. Feig, B. L. De Groot, H. Grubmüller and A. D. MacKerell, Nat. Methods, 2016, 14, 71–73 CrossRef PubMed.
- A. D. MacKerell, D. Bashford, M. Bellott, R. L. Dunbrack, J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, W. E. Reiher, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiórkiewicz-Kuczera, D. Yin and M. Karplus, J. Phys. Chem. B, 1998, 102, 3586–3616 CrossRef CAS PubMed.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- S. Miyamoto and P. A. Kollman, J. Comput. Chem., 1992, 13, 952–962 CrossRef CAS.
- D. J. Evans and B. L. Holian, J. Chem. Phys., 1985, 83, 4069–4074 CrossRef CAS.
- M. Parrinello and A. Rahman, J. Appl. Phys., 1981, 52, 7182–7190 CrossRef CAS.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- J. M. Vanegas, A. Torres-Sánchez and M. Arroyo, J. Chem. Theory Comput., 2014, 10, 691–702 CrossRef CAS PubMed.
- L. Vamparys, R. Gautier, S. Vanni, W. F. D. Bennett, D. P. Tieleman, B. Antonny, C. Etchebest and P. F. J. Fuchs, Biophys. J., 2013, 104, 585–593 CrossRef CAS PubMed.
- R. Gautier, A. Bacle, M. L. Tiberti, P. F. Fuchs, S. Vanni and B. Antonny, Biophys. J., 2018, 115, 436–444 CrossRef CAS PubMed.
- A. Bacle, R. Gautier, C. L. Jackson, P. F. J. Fuchs and S. Vanni, Biophys. J., 2017, 1417–1430 CrossRef CAS PubMed.
- C. T. Everitt and D. A. Haydon, J. Theor. Biol., 1968, 18, 371–379 CrossRef CAS PubMed.
- M. Garten, L. D. Mosgaard, T. Bornschlögl, S. Dieudonné, P. Bassereau and G. E. S. Toombes, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 328–333 CrossRef CAS PubMed.
- H. J. Sharpe, T. J. Stevens and S. Munro, Cell, 2010, 142, 158–169 CrossRef CAS PubMed.
- R. Prasad, A. Sliwa-Gonzalez and Y. Barral, Sci. Adv., 2020, 6, eaba5130 CrossRef PubMed.
- A. Santinho, V. T. Salo, A. Chorlay, S. Li, X. Zhou, M. Omrane, E. Ikonen and A. R. Thiam, Curr. Biol., 2020, 30(13), 2481–2494.e6 CrossRef CAS PubMed.
- O. B. Tarun, M. Y. Eremchev and S. Roke, Langmuir, 2018, 34, 11305–11310 CrossRef CAS PubMed.
- S. Vanni, H. Hirose, H. Barelli, B. Antonny and R. Gautier, Nat. Commun., 2014, 5, 4916 CrossRef CAS PubMed.
- C. Prévost, M. E. Sharp, N. Kory, Q. Lin, G. A. Voth, R. V. Farese and T. C. Walther, Dev. Cell, 2018, 44, 73–86.e4 CrossRef PubMed.
- S. Vanni, L. Vamparys, R. Gautier, G. Drin, C. Etchebest, P. F. J. Fuchs and B. Antonny, Biophys. J., 2013, 104, 575–584 CrossRef CAS PubMed.
- M. Pinot, S. Vanni, S. Pagnotta, S. Lacas-Gervais, L.-A. Payet, T. Ferreira, R. Gautier, B. Goud, B. Antonny and H. Barelli, Science, 2014, 345, 693–697 CrossRef CAS PubMed.
- S. H. White, Ion Channel Reconstitution, 1986, 3–35 Search PubMed.
- A. Santinho, A. Chorlay, L. Foret and A. R. Thiam, Biophys. J., 2021, 120, 607–617 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |