Stimuli-responsive engineered living materials
Abstract
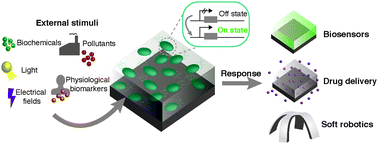
Stimuli-responsive materials are able to undergo controllable changes in materials properties in response to external cues. Increasing efforts have been directed towards building materials that mimic the responsive nature of biological systems. Nevertheless, limitations remain surrounding the way these synthetic materials interact and respond to their environment. In particular, it is difficult to synthesize synthetic materials that respond with specificity to poorly differentiated (bio)chemical and weak physical stimuli. The emerging area of engineered living materials (ELMs) includes composites that combine living cells and synthetic materials. ELMs have yielded promising advances in the creation of stimuli-responsive materials that respond with diverse outputs in response to a broad array of biochemical and physical stimuli. This review describes advances made in the genetic engineering of the living component and the processing-property relationships of stimuli-responsive ELMs. Finally, the implementation of stimuli-responsive ELMs as environmental sensors, biomedical sensors, drug delivery vehicles, and soft robots is discussed.
- This article is part of the themed collections: Soft Matter Most Popular 2021 and 2021 Soft Matter Emerging Investigators


 Please wait while we load your content...
Please wait while we load your content...