Disentangling collective motion and local rearrangements in 2D and 3D cell assemblies†
Abstract
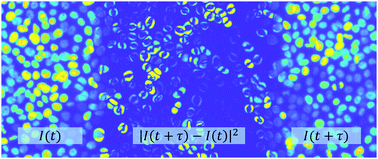
The accurate quantification of cellular motility and of the structural changes occurring in multicellular aggregates is critical in describing and understanding key biological processes, such as wound repair, embryogenesis and cancer invasion. Current methods based on cell tracking or velocimetry either suffer from limited spatial resolution or are challenging and time-consuming, especially for three-dimensional (3D) cell assemblies. Here we propose a conceptually simple, robust and tracking-free approach for the quantification of the dynamical activity of cells via a two-step procedure. We first characterise the global features of the collective cell migration by registering the temporal stack of the acquired images. As a second step, a map of the local cell motility is obtained by performing a mean squared amplitude analysis of the intensity fluctuations occurring when two registered image frames acquired at different times are subtracted. We successfully apply our approach to cell monolayers undergoing a jamming transition, as well as to monolayers and 3D aggregates that exhibit a cooperative unjamming-via-flocking transition. Our approach is capable of disentangling very efficiently and of assessing accurately the global and local contributions to cell motility.
- This article is part of the themed collections: Soft Matter Editor-in-Chief's Choice and 2021 Soft Matter Emerging Investigators


 Please wait while we load your content...
Please wait while we load your content...