Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Cationic helicenes as selective G4 DNA binders and optical probes for cellular imaging†
Peter A.
Summers‡
 a,
Ajesh P.
Thomas‡
a,
Ajesh P.
Thomas‡
 a,
Timothy
Kench
a,
Jean-Baptiste
Vannier
bc,
Marina K.
Kuimova
a,
Timothy
Kench
a,
Jean-Baptiste
Vannier
bc,
Marina K.
Kuimova
 *a and
Ramon
Vilar
*a and
Ramon
Vilar
 *a
*a
aDepartment of Chemistry, Molecular Sciences Research Hub, Imperial College London, 82 Wood Lane, White City Campus, W12 0BZ, UK. E-mail: m.kuimova@imperial.ac.uk; r.vilar@imperial.ac.uk; Tel: +44 (0)20 7594 1967 Tel: +44 (0)20 7594 8558
bTelomere Replication and Stability Group, Medical Research Council – London Institute of Medical Sciences, London, W12 0NN, UK
cInstitute of Clinical Sciences, Faculty of Medicine, Imperial College London, London, W12 0NN, UK
First published on 15th October 2021
Abstract
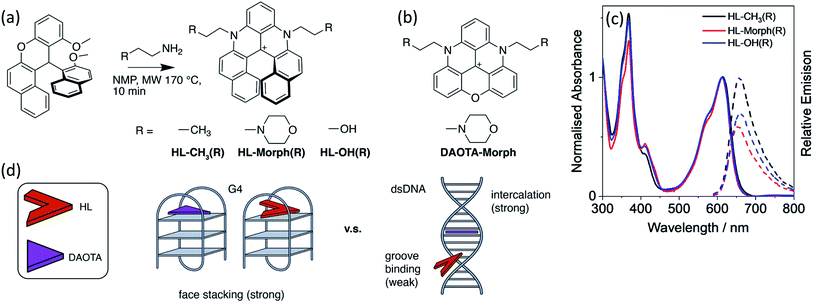
The important role that G-quadruplex DNA (G4 DNA) structures play in regulating biological processes is becoming widely recognised. These structures have also been proposed to be attractive drug targets. Therefore, there has been significant interest in developing small molecules that can selectively bind to G4 DNA over other topologies. In this paper we investigate the interaction between DNA and helical compounds (helicenes) based on a central carbocation trisubstituted with aromatic rings. We show that the non-planar structure of these helicenes results in a significantly reduced affinity for dsDNA when compared to their planar analogues, whilst maintaining a high affinity for G4 DNA. Additionally, the right- and left-handed enantiomers of one of these helicenes recognise the chiral DNA environments of G4 and dsDNA differently. We show that upon DNA binding the helicenes display a fluorescence switch-on effect, which we have successfully used for cellular imaging in live and fixed U2OS cells, staining mitochondria and the nucleus, respectively.
Introduction
It is increasingly recognised that non-canonical DNA structures (i.e. non-duplex DNA) have important biological functions in living organisms.1 One such structure is the guanine-quadruplex (G4), a tetra-stranded helical assembly that forms in guanine-rich sequences of DNA. Bioinformatic studies as well as growing experimental evidence indicate that G4s are involved in a number of biological processes including telomere maintenance, replication and regulation of gene expression.2,3 Because of their proposed biological roles, G4s have been intensively studied as potential targets for the development of drugs, particularly for cancer.4While G4s are thermodynamically stable and form readily in vitro, their presence in a cellular environment is proposed to be transient. This is due to the double stranded structure being the predominant topology in coiled DNA as well as the presence of dedicated helicases to unfold G4 structures.5 However, the formation and stability of G4s can be significantly enhanced in the presence of small molecules that preferentially bind to these tetra-stranded structures over other DNA topologies.6–9 Thus, G4 DNA binders can shift the duplex–quadruplex equilibrium and prevent helicases from resolving G4s.5 It has also been shown that small molecules can modify the interaction between G4s and proteins, particularly in the telomeres.10 This, together with the proposed biological roles of G4s, has prompted the development of a large number of small molecules designed to bind selectively to G4s over other topologies.6,8,9,11 Some of these molecules have been shown to trigger a number of biological responses in cells that are consistent with G4 stabilisation, while others have been successfully used as optical probes to visualise and detect G4 structures in vitro and, in a few cases, in live cells.12–16
Most G4 DNA binders are based on planar, polyaromatic molecules featuring positively charged substituents. As discussed extensively elsewhere, the planar core binds to the guanine tetrads via π–π interactions, while the substituents provide means to increase solubility and DNA affinity (e.g. with protonated amines), as well as selectivity for G4s over duplex DNA (dsDNA).6,8,9,11 While this strategy has yielded some very strong G4 DNA binders, alternative non-planar structural motifs have been explored with the aim of improving selectivity for a specific G4 topology, not only over duplex DNA, but also over other G4 topologies.8 One such strategy has been to exploit the explicit chiral environment formed in both duplex and G4 DNA, designing enantiomerically specific DNA binding molecules with stereoselectivity originating from octahedral metal-centres or from steric hindrance. For example, Thomas and co-workers studied the DNA binding properties of the di-ruthenium complex [{Ru(bipy)2}2(tpphz)]4+ where each of the two metal centres is in a chiral octahedral environment. They showed that the ΛΛ isomer has ca. 40 times higher affinity for human telomeric G4 DNA than the ΔΔ isomer.17 Another class of chiral G4 DNA binders are metallohelicenes, where two octahedral metal centres are bridged by three chelating ligands. For example, Qu and co-workers reported that one of the two enantiomers of a di-nickel metallohelice recognised HTelo G4 DNA with high affinity and selectivity over duplex DNA.18–20 Binding of related di-iron metallohelices to right and left-handed HTelo G4 DNA was recently investigated. The Δ, and Λ-enantiomers showed selectivity for right, and left-handed G4 DNA, respectively.21 Organic helical molecules such as foldamers have also been shown to bind G4s with high affinity and selectivity.22 It was proposed that the binding mode of these helical structures is different to that displayed by planar compounds – and likely to involve interactions with the backbone of G4 DNA. The M enantiomer of a cyclic helicene showed selectivity for B-DNA over the P isomer [Kd(P)/Kd(M) = 2.0], whereas the reverse trend was observed when binding Z-DNA [Kd(P)/Kd(M) = 0.3].23 Following on from this, a series of related chiral helicenes (with varying dihedral angles) showed enantioselective recognition of the M isomer to neighbouring G4s in the telomeric region.24
We have previously reported that a triangulenium, DAOTA-Morph (also known as DAOTA-M2 – see Fig. 1), has good affinity for DNA and switches on its fluorescence upon binding.12,15 Its binding affinity towards different DNA topologies is ca. 2-fold higher for G4 than for duplex DNA (Kd values ca. 1.7 μM and 1.0 μM for duplex and G4 DNA structures respectively).12,25 Interestingly, the fluorescence lifetime of this probe is highly dependent on the DNA topology it binds to, which has allowed us to use DAOTA-Morph to probe the formation of G4 structures via fluorescence lifetime imaging microscopy (FLIM) in live cells.12,15
With the aim of improving further the selectivity of this type of probe for specific G4 structures over other DNA topologies, herein we present studies with the helical analogues of trianguleniums, namely cationic helicenes. This type of compound, previously reported by Lacour and co-workers,26 can often be resolved into their stereoisomers and they are emissive over a wide range of wavelengths, depending on bridging atoms and substituents. These helical trianguleniums derivatives have been used as dyes for cellular imaging and shown to accumulate in mitochondria of live cells, and in one report, they were shown to bind to duplex DNA.27 Herein we report the synthesis of the new racemic (R) helicences HL-OH(R) and HL-Morph(R) (the latter being a direct analogue of the planar DAOTA-Morph compound which has been successfully used as a lifetime-based optical probe for G4s12,15) and their DNA binding properties, which are compared with those of the previously reported helicene HL-CH3(R).26,28–32 Further, the racemic mixture of HL-OH(R) was partially resolved into the corresponding M and P stereoisomers. We show that these compounds have high selectivity for G4 DNA over duplex DNA; this is particularly the case for the HL-OH(M) stereoisomer. We also show that in live cells HL-Morph(R) is cell permeable and localises in mitochondria, whereas in fixed cells nuclear staining of DNA is possible, as confirmed using FLIM.
Results and discussion
Design and synthesis of helicenes as DNA binders
As stated above, the planar aromatic molecule DAOTA-Morph [Fig. 1(b)] binds to dsDNA and G4 DNA with similar affinities.12 We aimed to reduce the probe's affinity for dsDNA, whilst maintaining strong G4 binding, thus resulting in improved selectivity for G4 over dsDNA. To achieve this, we designed and synthesised a cationic helicene molecule HL-Morph(R) [Fig. 1(a)] in which steric hindrance induced by extension of the aromatic system, causes distortion away from a planar structure. This nonplanar helical topology should in principle be prevented from intercalating into adjacent bases in duplex DNA, favouring a weak groove binding arrangement [Fig. 1(d)]. On the other hand, the helical molecule still has the appropriate structural features to bind to G4 DNA via end-stacking. We also synthesised HL-CH3(R) as a control, and HL-OH(R) which we were able to partially separate into the enantiomers HL-OH(P) and HL-OH(M) as confirmed by CD spectroscopy and chiral HPLC (see below).These helicenes were synthesised using a multi-step synthetic strategy based on that previously reported by Lacour and co-workers for similar helical molecules (Fig. 1 and S1†).26 The key intermediate 11-methoxy-12-(2-methoxynaphthalen-1-yl)-12H-benzo[a]xanthen-12-ylium tetrafluoroborate [1 in Fig. S1†] was prepared in bulk.26 This was heated with the corresponding amine in NMP to form the cationic racemic mixtures of the corresponding diaza-helicenes, namely HL-CH3(R), HL-Morph(R) and HL-OH(R). The crude compounds were purified by flash chromatography and characterised by 1H NMR and 13C NMR spectroscopy, and ESI-MS (Fig. S2–S7†).
HL-CH3(R), HL-Morph(R) and HL-OH(R) binding affinities to ctDNA and c-Myc
We first set out to investigate the binding affinities of HL-CH3(R), HL-Morph(R), and HL-OH(R) to dsDNA (ctDNA) and G4 DNA (c-Myc). As indicated above, DAOTA-Morph, the planar aromatic analogue of HL-Morph(R), binds to ctDNA and G4 DNA with similar binding affinities.12 By distorting the aromatic structure of DAOTA-Morph to make HL-Morph(R), we aimed to maintain strong π–π stacking to the G4 quartet, whilst reducing affinity for dsDNA. These helicenes are fluorescent and their emission intensity in aqueous buffered media is switched-on upon DNA binding. Therefore, we were able to study the DNA binding affinity of HL-CH3(R), HL-Morph(R) and HL-OH(R) through titrations with ctDNA and c-Myc G4 DNA (see Fig. 2 and S8, S9†). The UV/visible spectra undergo a red-shift in absorption maximum during addition of DNA [Fig. 2(a)], as the fluorescence intensity increases [Fig. 2(b)].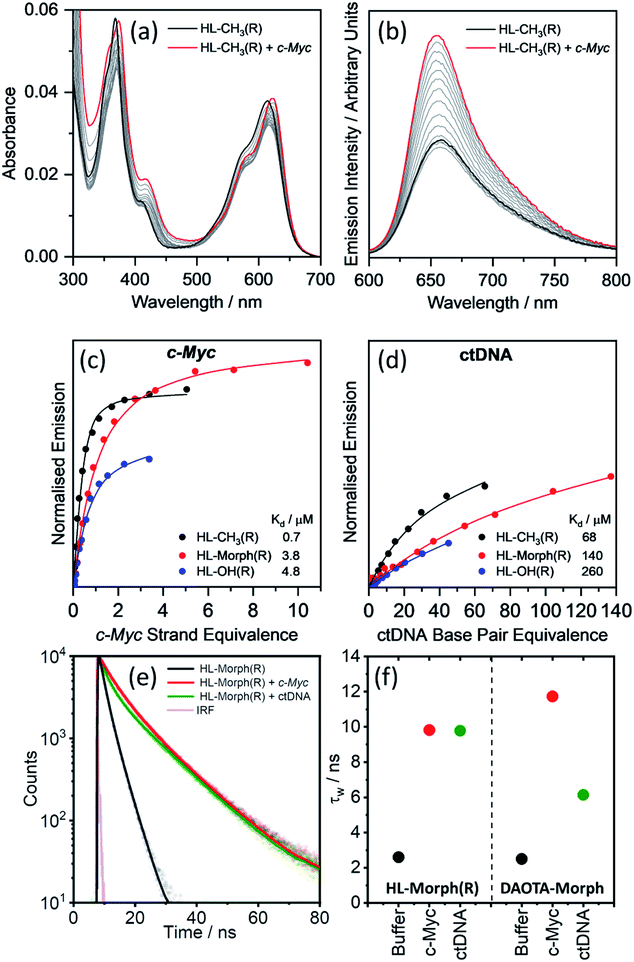 | ||
| Fig. 2 Binding affinities of HL-CH3(R), HL-Morph(R), and HL-OH(R) to c-Myc and ctDNA. (a) Absorption and (b) emission spectra of HL-CH3(R) (4 μM) during addition of c-Myc G4 DNA (0–5 strand equivalence). Difference in integrated emission (λex = 580 nm, λem = 600–700 nm) during titration with (c) c-Myc and (d) ctDNA. Integrated intensities are normalised against the absorption at the excitation wavelength. The solid line is the best fit of a simple binding model (see Methods section and Fig. S10† for binding models used) to solve for KH and kΔGH. For all titration data recorded for HL-CH3(R), HL-Morph(R), and HL-OH(R) see Fig. S8 and S9(e) and (f)† fluorescence lifetime analysis of HL-Morph(R) in buffered aqueous solution and bound to different oligonucleotide topologies. (e) Time resolved fluorescence decays of HL-Morph(R) (2.5 μM, black trace) and following the subsequent additions of G4 (c-Myc, 10 strand equivalence, red dots), and dsDNA (ctDNA, 140 base pair equivalence, green dots). Solid lines are bi-exponential fits of the decay traces. (f) Variation in average lifetime (τw) of the bi-exponential fits in (e). Results are presented alongside the corresponding τw values recorded for DAOTA-Morph, adapted from ref. 12. All experiments performed in 10 mM lithium cacodylate buffer (pH 7.3) with 100 mM KCl. | ||
Fitting the fluorescence titrations' data [see Methods section and Fig. S10† for binding models used], the dissociation constants for c-Myc [Kd = 0.7–4.8 μM, Fig. 2(c)] are comparable to those displayed by DAOTA-Morph (Kd = 0.8 μM for myc2345). However, those for ctDNA [Kd = 68–260 μM, Fig. 2(d)] show that the interaction of these helicenes with dsDNA is at least 50 times weaker than that displayed by DAOTA-Morph [Kd = 1.3 μM]. G4 binding was confirmed using a fluorescent intercalator displacement (FID) assay,33 with all three helicene complexes showing a similar ability to displace TO from c-Myc (Fig. S9†).
We next investigated the fluorescence lifetime (τw, a concentration independent parameter) of the helicenes upon binding to DNA. When free in aqueous buffer at pH 7.3, the helicenes' τw ranges between 2.6 and 5.1 ns (Fig. S12†) while, upon DNA binding, a significant increase in lifetime to between 8 and 12 ns is observed (see Fig. 2 and S12†). Although the helicenes' fluorescence lifetime cannot be used to discriminate between different DNA topologies (unlike DAOTA-Morph12,15), the significant increase of τw when bound to DNA was useful for cellular imaging studies in order to confirm that the dyes are bound to DNA in cellular organelles (see below).
Binding modes of HL Morph(R) and DAOTA-Morph to dsDNA
Given that HL-Morph(R) is ca. 100 times weaker binder towards ctDNA than DAOTA-Morph (Table 1), we investigated if this difference could be a result of a different binding mode. Structural perturbations of DNA induced by dye binding can be monitored using CD spectroscopy and used to investigate the dye binding mode. The characteristic CD spectrum of ctDNA (in its B form) shows an increase in the band at 277 nm upon DAOTA-Morph binding [Fig. S13(a) and (b)†]. Similar spectral changes have been observed following the intercalation of planar aromatic dyes into dsDNA, tentatively assigned to the unwinding of the helical DNA structure to accommodate the intercalated dye.34,35 Conversely, when the same number of HL-Morph(R) molecules per base pair are bound to ctDNA, a small decrease in band intensity at 277 nm is observed [Fig. S13(a) and (b)†]. This minor change in the CD signal is characteristic of groove binding which results in minimal disruption to the double helical structure.36,37 We next studied the accessibility of ctDNA bound DAOTA-Morph and HL-Morph(R) to fluorescence quenching by iodide. If intercalated, the proximity of base pairs above and below the dye should protect against quenching, whereas this shielding will be less for groove binding dyes which are still exposed to the solvent environment. Once bound, DAOTA-Morph is almost completely protected from quenching by iodide [KSV = 21.8 (free) and 3.1 (bound) M−1], whereas for HL-Morph(R) the quenching is in fact enhanced [KSV = 36.1 (free) and 48.7 (bound) M−1, Fig. S13(c)†]. This seemingly unusual enhancement in quenching when bound to dsDNA has been observed for other positively charged, weak groove binding dyes,36,38 which was assigned to weakened electrostatic HL-Morph(R)-dsDNA interaction through increasing ionic strength of the solvent upon the addition of KI. As a result, HL-Morph(R) is released into solution allowing more efficient quenching. Molecular docking studies provided further evidence that DAOTA-Morph intercalates into dsDNA whilst HL-Morph(R) interacts via groove binding. These studies showed that HL-Morph(R) is too wide to fit into an intercalation site (Fig. S14†), whereas DAOTA-Morph fits well. Taken together, this evidence indicates that distortion of the aromatic surface in HL-Morph(R) away from a planar structure did indeed result in a change in binding mode, which favours a weak groove binding mode over intercalation into dsDNA. This in turn accounts for the high selectivity of the helicenes for G4 DNA (over ctDNA) confirming our original hypothesis.| c-Myc | ctDNA |
c-Myc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ctDNA ctDNA |
|||
|---|---|---|---|---|---|
| K d | Switch-on | K d | Switch-on | Selectivitya | |
| a Selectivity values calculated as Kd ctDNA/Kdc-Myc. b Values for DAOTA-Morph binding to myc2345 and ctDNA from ref. 12. | |||||
| HL-CH3(R) | 0.7 | 1.8 | 68 | 1.8 | 97 |
| HL-Morph(R) | 3.8 | 2.7 | 140 | 2.6 | 37 |
| HL-OH(R) | 4.7 | 1.9 | 260 | 1.9 | 55 |
| DAOTA-Morph | 0.8 | ca. 5 | 1.3 | ca. 2.5 | 1.6 |
HL-OH(P) and HL-OH(M) binding affinities to c-Myc, BCL2, HTG4 and ctDNA
As discussed in the previous section, HL-CH3(R), HL-Morph(R), and HL-OH(R) show high selectivity for G4 over dsDNA (Table 1), so we therefore investigated if the individual enantiomers (P and M) would show different DNA binding properties. We were able to partially resolve HL-OH(R) (0% ee) into HL-OH(P) (32% ee) and HL-OH(M) (96% ee) as evidenced by chiral HPLC [Fig. S22b†]; attempts to resolve HL-Morph(R) were unsuccessful. The binding affinities of the partially-resolved sample of HL-OH(P) and HL-OH(M) towards c-Myc, BCL2, and HTG4 quadruplexes, as well as ctDNA, were determined (Fig. 3). To improve the accuracy of fitting, titrations of HL-OH(R), HL-OH(P) and HL-OH(M) were fitted simultaneously to a competitive binding model (see Methods section and Fig. S10† for details) to solve for Kd(P) and Kd(M).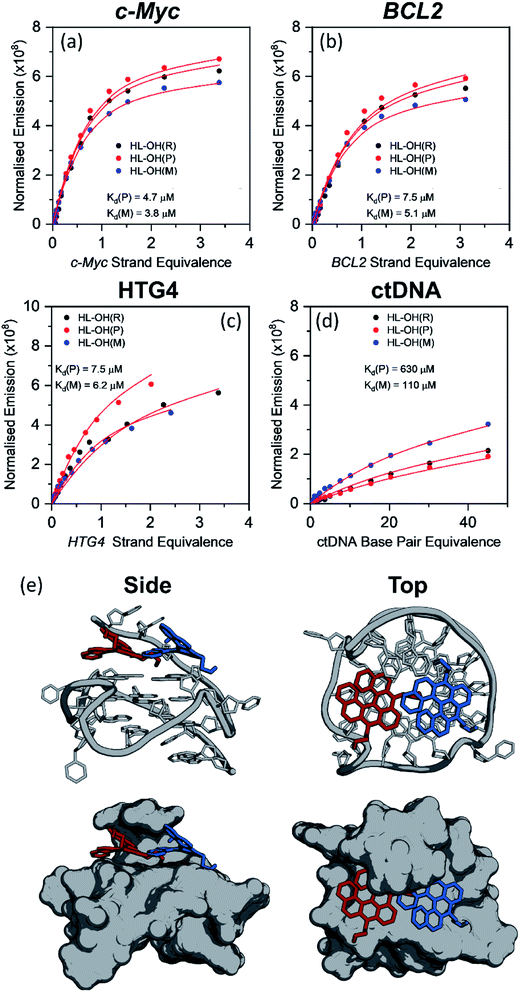 | ||
| Fig. 3 Plots showing the competitive binding of HL-OH(R) (6 μM, 0% ee), HL-OH(P) (6 μM, 32% ee), and HL-OH(M) (6 μM, 96% ee) to (a) c-Myc, (b) BCL2 (c) HTG4 and (d) ctDNA. Data points are the difference in integrated emission during titration, normalised against the absorption at the excitation wavelength. The solid red line is a simultaneous best fit of all three titrations to a competitive binding model (Fig. S9†), to solve for KP, KM, kΔGP and kΔGM. All experiments were performed in 10 mM lithium cacodylate buffer (pH 7.3) with 100 mM KCl. For all titration data see Fig. S13–S16.† (e) Lowest energy orientation following molecular docking of HL-OH(P) (red), and HL-OH(M) (blue) to c-Myc. The 5′ overhang creates a chiral pocket for the helicene to bind in. | ||
HL-OH(M) consistently binds more strongly to both G4 and dsDNA than HL-OH(P). For c-Myc [Kd = 4.7 and 3.8 μM for HL-OH(P) and HL-OH(M), respectively, Fig. 3 and S15†] and HTG4 [Kd = 7.5 and 6.2 μM for HL-OH(P) and HL-OH(M), respectively, Fig. 3 and S17†] this difference is small, presumably because the open face of these quadruplex structures presents a modestly chiral environment for binding.39 Using the published NMR-structure of c-Myc incorporating a G4-binder,39 we performed docking studies on the ligand free structure [Fig. 3(e)]. This confirmed the independent biding preferences for both HL-OH(P) and HL-OH(M) into a chiral pocket caused by the 5′ overhang of the G4. We next investigated binding of HL-OH(P) and HL-OH(M) to a mixed parallel/antiparallel quadruplex structure that forms in the promoter region of the BCL2 gene, as it contains loop regions that interact with both G-tetrad faces, potentially forming an increased chiral environment for binding.40 Indeed, the difference in binding between HL-OH(P) and HL-OH(M) [Kd = 7.5 μM and 5.1 μM, respectively, Fig. 3 and S14†] is slightly increased when interacting with BCL2, as compared to c-Myc and HTG4.
A much bigger difference in affinity between the two isomers was observed when binding to ctDNA [Fig. 3 and S18†]. HL-OH(P) [Kd = 630 μM, Fig. 3(c)] shows a reduced interaction with ctDNA compared to HL-OH(M) [Kd = 110 μM, Fig. 3(c)], with selectivity for c-Myc over ctDNA of 134 and 29, respectively. As binding to BCL2 is weaker, the selectivity over ctDNA is decreased for both HL-OH(P) and HL-OH(M) (84 and 22, respectively) compared to c-Myc.
Given the large difference in binding to ctDNA between the P and M isomers, we investigated if this difference could be observed by CD spectroscopy when HL-Morph(R) is bound to a large excess of ctDNA. Racemic HL-Morph(R) shows no bands in the CD spectrum, however, once added to a large excess of ctDNA, negative CD bands at 314, 374 and 472 nm develop [Fig. S19(a)†]. The appearance of a CD signal in a region outside of any ctDNA absorption implies the enrichment of one isomer bound to ctDNA compared to one isomer free in solution. Based on the measured spectra of free HL-OH(M), and HL-OH(M) bound to ctDNA, we calculated CD spectra that would be expected if either the M or the P isomers had the stronger association constant [Fig. S19(b)†]. The spectrum expected for the M isomer strongly bound to ctDNA, closely matches the experimental spectrum, confirming the preference of the M isomer in binding to ctDNA.
HL-Morph(R) staining in live and fixed U2OS cells
We next investigated how HL-Morph(R) stains live and fixed U2OS cells. We chose HL-Morph(R) due to low toxicity to live cells (Fig. S20†), synthetic ease and the ability to compare against our previous results with structurally related lifetime-based G4 probe, DAOTA-Morph.12 At the concentrations used in our live cell experiments (10 μM, 24 h), the cytotoxicity of HL-Morph(R) is negligible. Similarly to other previously reported helicene compounds,27aHL-Morph(R) accumulates in mitochondria of live cells, confirmed by co-localisation with mitotracker green [MTG, Fig. 4(a)]. Again, this contrasts with DAOTA-Morph, which localises predominantly in the nucleus of live U2OS cells.12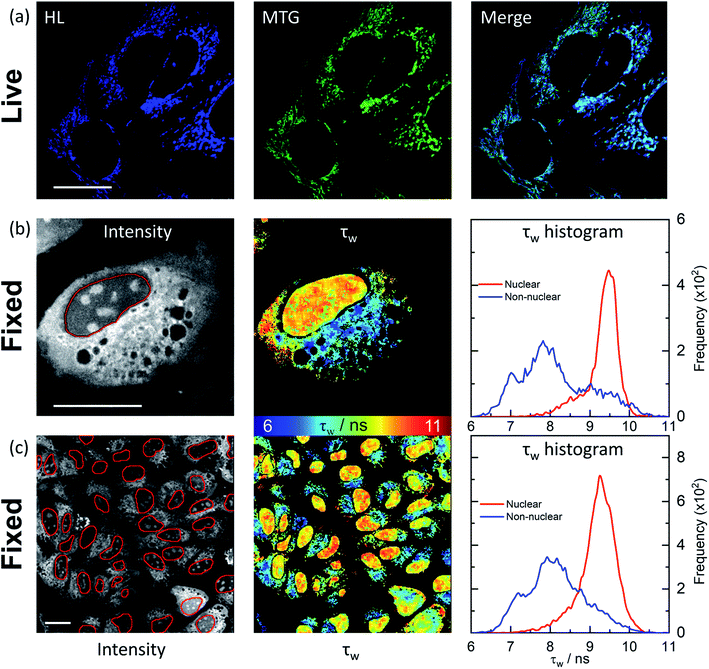 | ||
| Fig. 4 Confocal and FLIM images of HL-Morph(R) incubated with live and fixed U2OS cells. (a) Confocal images (1024 × 1024 resolution) of helicene fluorescence (left: HL, 10 μM, 24 h, λex = 633 nm, λem = 650–790 nm) and co-incubated with Mito-Tracker Green (middle: MTG, 50 nM, 45 min, λex = 488 nm, λem = 500–600 nm). Right: image merge confirms co-localisation. (b) and (c): FLIM analysis of fixed U2OS cells stained with HL-Morph(R) (20 μM, 0.5 h, λex = 640 nm, λem = 650–790 nm). Left: fluorescence intensity images recorded at (b) 512 × 512 (c) and 256 × 256 resolution, red lines represent the nuclear segmentation used for the FLIM analysis. Middle: corresponding FLIM maps. Right: histogram of fluorescence lifetime distribution. For χ2 maps see Fig. S21.† Scale bars: 20 μm. | ||
In fixed U2OS cells, HL-Morph(R) passes the nuclear membrane, revealing both nuclear and non-nuclear staining [Fig. 4(b) and (c)]. Recording FLIM maps of these fixed cells, intensity weighted average lifetimes recorded within the nucleus are represented by a histogram with a peak maximum at ca. 9.5 ns, consistent with in vitro experiments for HL-Morph(R) bound to DNA [9.8 ns regardless of topology, Fig. 2(f)]. We note that significantly higher fluorescence intensity can be observed in the nucleoli [Fig. 4(b), left panel], although the concentration and topology-independent lifetime measurement does not reveal any reason for this.
Conclusions
The vast majority of G4 DNA binders reported in the literature are based on planar polyaromatic systems. While planar molecules have high affinities to G4 DNA due to efficient π–π end-stacking, they also tend to intercalate efficiently into duplex DNA. Herein we have demonstrated that by breaking the planarity of polyaromatic systems, it is possible to generate much more selective G4 binders. More specifically, we have shown that the fluorescent helicene compounds, HL-CH3(R), HL-Morph(R), and HL-OH(R) have high selectively for G4 over duplex DNA, and further selectivity can be introduced through partial chiral resolution of HL-OH(R) into HL-OH(P) and HL-OH(M). The distorted core of the helicene compound reduces affinity for dsDNA compared to the planar analogue DAOTA-Morph, whist maintaining strong affinity for G4 DNA [selectivity = 134 for HL-OH(P)]. We also show that HL-OH(P) and HL-OH(M) bind differently to G4 and dsDNA topologies, with HL-OH(M) consistently displaying a higher affinity. We have also used the increased fluorescence intensity and lifetime of HL-Morph(R) upon DNA binding to enable cellular imaging studies. In live U2OS cells this helicene accumulates in the mitochondria, whereas in fixed cells, HL-Morph(R) passes the nuclear membrane and binds to DNA, as confirmed using fluorescence lifetime imaging microscopy, FLIM.Methods
General synthetic procedures
All chemicals were purchased from commercial sources and used as received, unless stated otherwise. 1H and 13C NMR spectra were recorded using either a 400 or 500 MHz Bruker Avance Ultrashield NMR spectrometer at 296 K. Spectra were referenced internally by using the residual solvent (1H δ = 3.34 and 13C δ = 49.86 for CD3OD-d4 relative to SiMe4). ESI-MS spectra were recorded by Dr L. Haigh (Imperial College London) on a Bruker Daltronics Esquire 3000 spectrometer. HL-CH3(R) was synthesised and characterised according to published procedures.26Synthesis of HL-Morph(R)
Compound 1 (0.1 g, 0.198 mmol) and 4-(2-aminoethyl)morpholine (0.644 g, 4.96 mmol) were mixed with anhydrous 1-methyl-2-pyrrolidinone (NMP) (1 mL) under an argon atmosphere in a microwave (MW) tube. This reaction mixture was stirred in a MW synthesizer for 10 min at 170 °C then allowed to cool down to room temperature. CH2Cl2 (5 mL) was added to the reaction mixture and was washed with a 1 M aqueous solution of HBF4 (2 × 2 mL). The organic phase was separated and dried over anhydrous Na2SO4 and evaporated under vacuum. The resulting crude material was purified using flash chromatography to yield HL-Morph(R) (0.015 g, 11%). 1H NMR (400 MHz, CD3OD): δ 8.43 (d, 3J = 12 Hz, 2H), 8.34 (t, 3J = 8 Hz, 1H), 8.21 (d, 3J = 12 Hz, 2H), 7.98 (d, 3J = 8 Hz, 2H), 7.92 (d, 3J = 8 Hz, 2H), 7.36 (t, 3J = 8 Hz, 2H), 7.10 (d, 3J = 8 Hz 2H), 6.77 (t, 3J = 8 Hz, 2H), 5.23–5.17 (m, 3J = 8 Hz, 2H), 5.16–4.98 (m, 3J = 8 Hz, 2H), 3.70–3.55 (m, 8H), 3.07 (t, 3J = 8 Hz 4H), 2.65 (t, 3J = 8 Hz, 8H). 13C NMR (100 MHz, CD3OD): δ 144.8, 143.8, 140.6, 140.5, 137.0, 131.7, 131.1, 130.5, 129.5, 129.2, 124.7, 118.4, 117.9, 109.5, 68.8, 57.0, 55.9. ESI-MS-m/z calculated for C39H39N4O2+ = 595.31 a.m.u.; found = 595.46 a.m.u.Synthesis of HL-OH(R)
Compound 1 (0.1 g, 0.198 mmol) and freshly distilled 2-amino ethanol (0.352 g, 4.96 mmol) was mixed with anhydrous 1-methyl-2-pyrrolidinone (NMP) (1 mL) under an argon atmosphere in a MW tube. This reaction mixture was stirred in a MW synthesiser for 10 min at 170 °C then allowed to cool down to room temperature. CH2Cl2 (5 mL) was added to the reaction mixture and was washed with a 1 M aqueous solution of HBF4 (2 × 2 mL). The organic phase was separated, dried over anhydrous Na2SO4, and evaporated under vacuum. The resulting crude material was purified using flash chromatography to yield the desired helicene HL-OH(R) (0.018 g, 16%). 1H NMR (500 MHz, CD3OD): δ 8.40 (d, 3J = 10 Hz, 2H), 8.28 (t, 3J = 5 Hz, 3H), 8.03 (d, 3J = 5 Hz, 2H), 7.89 (d, 3J = 10 Hz, 2H), 7.32 (t, 3J = 7.5 Hz, 2H), 7.15 (d, 3J = 5 Hz, 2H), 6.77 (t, 3J = 7.5 Hz, 2H), 5.20–5.16 (m, 2H), 5.02–4.97 (m, 2H), 4.34–4.28 (m, 4H). 13C NMR (125 MHz, CD3OD): δ 144.61, 140.13, 139.50, 135.97, 130.96, 130.58, 129.57, 128.66, 128.40, 124.17, 117.56, 117.41, 109.00, 60.32, 52.62, ESI-MS-m/z calculated for C31H25N2O2+ = 457.19 a.m.u.; found = 457.19 a.m.u.Partial enantiomeric resolution of HL-OH(R) into HL-OH(P) and HL-OH(M)
Partial enantiomeric resolution was achieved by adopting a reported protocol, using the enantiomeric phosphorous complex, [Me2NH2][Λ-BINPHAT].26HL-OH(R) (18 mg, 0.033 mmol) and [Me2NH2][Λ-BINPHAT] (33.80 mg, 0.039 mmol) were dissolved in a CH2Cl2/acetone (1/1 2 mL) mixture in a vial. The solution was stirred for 30 min at RT after which time the solvent was removed under reduced pressure. The solid obtained was dissolved in 3 mL acetone and kept overnight at 0 °C. The solid deposited at the bottom of the vial was separated from the mother liquor. The solvent of the latter was removed in a rotary evaporator to yield a second solid. CH2Cl2 was added to each solid fraction (the original precipitate and the solid obtained after evaporation) followed by HPF6 (9 mg) and 0.6 mL KPF6 (0.1 M solution in water) and stirred for 1 h. The organic layers were separated and purified using flash chromatography.To confirm the enantiomeric resolution of HL-OH(R), samples were analysed by chiral HPLC as follows: a 100 μM stock solution of the corresponding compound (i.e.HL-OH(R), HL-OH(P) or HL-OH(M)) was prepared in MeOH. 1.5 equivalents of NaBH4 were added, causing an immediate colour change from blue to colourless – due to reaction of the helicene carbocation as described previously.26 The solution was left for 6 h to allow for full NaBH4 hydrolysis. Next, the corresponding solution was diluted to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 hexane
50 hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH before injection onto a Chiralpak AD-H 250 × 4.6 mm column. An isocratic 60
MeOH before injection onto a Chiralpak AD-H 250 × 4.6 mm column. An isocratic 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 hexane
40 hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) isopropanol gradient was run and the peaks corresponding to each stereoisomer integrated to give an ee of 96% for HL-OH(M) and 32% for HL-OH(P) (see Fig. S22(b)† for chromatograms). The two enantiomers were analysed by CD spectroscopy and the spectra are shown in Fig. S22(a).†
isopropanol gradient was run and the peaks corresponding to each stereoisomer integrated to give an ee of 96% for HL-OH(M) and 32% for HL-OH(P) (see Fig. S22(b)† for chromatograms). The two enantiomers were analysed by CD spectroscopy and the spectra are shown in Fig. S22(a).†
General methods for spectroscopic and biophysical studies
Stock solutions of HL-CH3(R), HL-Morph(R), HL-OH(R), HL-OH(P) and HL-OH(M) were prepared in DMSO and the concentration determined in CH2Cl2 using the molar extinction coefficient 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 596 M−1 cm−1 published for HL-CH3(R) at 616 nm.26 The enantiomeric excess (ee) of HL-OH(P) (ee = 32%) and HL-OH(M) (ee = 96%) were calculated using chiral HPLC [Fig. S22(b)†] as described above, and confirmed by CD spectroscopy in CH2Cl2 [Fig. S22(a)†] using the published Δε values for HL-CH3(P) (107 Δε, 96% ee) and HL-CH3(M) (−87.3 Δε, 92% ee).26c-Myc (5′-TGAGGGTGGGTAGGGTGGGTAA-3′) BCL2 (5′-GGGCGCGGGAGGAATTGGGCGGG-3′) and HTG4 (5′-AGGGTTAGGGTTAGGGTTAGGG -3′) were purchased from Eurogentec. A two-nucleotide mutated form of the BCL2 gene was used, namely BCL2Mid, as it favours formation of a well-defined single structure.40 This modified structure is referend to as BCL2 throughout this manuscript. The oligonucleotides were dissolved in 10 mM lithium cacodylate buffer at pH 7.3. KCl was added to a final concentration of 100 mM, then annealed at 95 °C for 10 min. Calf thymus DNA (ctDNA, Sigma) were dissolved in the same cacodylate buffer, and KCl added to a final concentration of 100 mM. All oligonucleotide concentrations were determined in salt free buffer (before any annealing) using the molar extinction coefficients 228
596 M−1 cm−1 published for HL-CH3(R) at 616 nm.26 The enantiomeric excess (ee) of HL-OH(P) (ee = 32%) and HL-OH(M) (ee = 96%) were calculated using chiral HPLC [Fig. S22(b)†] as described above, and confirmed by CD spectroscopy in CH2Cl2 [Fig. S22(a)†] using the published Δε values for HL-CH3(P) (107 Δε, 96% ee) and HL-CH3(M) (−87.3 Δε, 92% ee).26c-Myc (5′-TGAGGGTGGGTAGGGTGGGTAA-3′) BCL2 (5′-GGGCGCGGGAGGAATTGGGCGGG-3′) and HTG4 (5′-AGGGTTAGGGTTAGGGTTAGGG -3′) were purchased from Eurogentec. A two-nucleotide mutated form of the BCL2 gene was used, namely BCL2Mid, as it favours formation of a well-defined single structure.40 This modified structure is referend to as BCL2 throughout this manuscript. The oligonucleotides were dissolved in 10 mM lithium cacodylate buffer at pH 7.3. KCl was added to a final concentration of 100 mM, then annealed at 95 °C for 10 min. Calf thymus DNA (ctDNA, Sigma) were dissolved in the same cacodylate buffer, and KCl added to a final concentration of 100 mM. All oligonucleotide concentrations were determined in salt free buffer (before any annealing) using the molar extinction coefficients 228![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 M−1 cm−1 (strand for c-Myc), 227
700 M−1 cm−1 (strand for c-Myc), 227![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 M−1 cm−1 (strand for BCL2), 228
300 M−1 cm−1 (strand for BCL2), 228![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 M−1 cm−1 (strand for HTG4) and 13
500 M−1 cm−1 (strand for HTG4) and 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 M−1 cm−1 (base pair for CT-DNA). Concentrations of G4 and dsDNA are expressed as per strand, and per base pair, respectively.
200 M−1 cm−1 (base pair for CT-DNA). Concentrations of G4 and dsDNA are expressed as per strand, and per base pair, respectively.
Fluorescence spectra were recorded using a Fluoromax-4 spectrofluorimeter (Jobin-Yvon; Horiba). Absorbance spectra were recorded using an 8453 UV-Visible Spectroscopy System (Agilent). Circular Dichroism (CD) spectra were recorded using either a J-810 (JASCO) or V100 (Chirascan) CD spectrophotometer.
Time-correlated single photon counting (TCSPC)
Time-resolved fluorescence decays were obtained using an IBH 5000F (Jobin Ybon, Horiba) time-correlated single photon counting (TCSPC) device equipped with a 635 nm NanoLED as an excitation source (pulse width <200 ps, HORIBA) with a 100 ns time window and 4096 time bins. Decays were detected at λem = 655 nm (±4 nm) after passing through a 645 nm long pass filter to remove any scattered excitation pulse. Decays were accumulated to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 counts at the peak of fluorescence decay. A neutral density filter was used for the instrument response function (IRF) measurements using a Ludox solution, detecting the emission at the excitation wavelength. Decay traces were fitted by iterative reconvolution to the equation I(t) = I0(α1e−t/τ1 + α2e−t/τ2) where α1 and α2 are variables normalised to unity. The intensity-weighted average lifetime (τw) was calculated using the equation:
000 counts at the peak of fluorescence decay. A neutral density filter was used for the instrument response function (IRF) measurements using a Ludox solution, detecting the emission at the excitation wavelength. Decay traces were fitted by iterative reconvolution to the equation I(t) = I0(α1e−t/τ1 + α2e−t/τ2) where α1 and α2 are variables normalised to unity. The intensity-weighted average lifetime (τw) was calculated using the equation: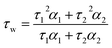 | (1) |
A prompt shift was included in the fitting to take into account differences in the emission wavelength between the IRF and measured decay. The goodness of fit was judged by consideration of the deviations from the model via a weighted residuals plot.
Oligonucleotide titrations
The helicene under study (2–6 μM) was dissolved in 10 mM lithium cacodylate buffer (pH 7.3) supplemented with 100 mM KCl and the UV/visible/fluorescence/CD spectra and/or TCSPC lifetime recorded (where applicable). Increasing amounts of the oligonucleotide under study were added, maintaining a constant concentration of the helicene compound. After each addition, the mixture was left to equilibrate for >30 s (a time determined to be sufficient for equilibrium to occur) before the corresponding photophysical measurements were recorded. Fluorescence spectra (λex = 580 nm, λex = 590–850 nm) were integrated between 600 and 800 nm and the integrated intensity normalised against the absorption at the excitation wavelength. Titrations are plotted as ΔF, the difference between the normalised emission, and the normalised emission of the free helicene (first point of the titration). For HL-OH(M) in aqueous buffer, broadening of the absorbance spectral bands can be observed, consistent with compound aggregation. In this case, ΔF was calculated as the difference between the normalised emission, and the normalised emission from the first addition of oligonucleotide (second point of the titration).Titration curves were fitted to either a simple binding (which assumes the 2 stereoisomers are equivalent)41,42 or competitive binding (which assumes independent binding for each stereoisomer)43,44 models using a modified form of the MatLab script reported previously (Fig. S10†).41,42 Based on the titration data, a binding stoichiometry of two compounds to one G-quadruplex and two compounds per five base pairs for ctDNA was used allowing for direct comparison with DAOTA-Morph.12 No co-operativity between binding sites and no oligonucleotide fluorescence response was assumed. Titrations of racemic mixtures of HL-CH3(R), HL-Moph(R) and HL-OH(R) were fitted independently to the simple binding model to solve for the association constant (KH) and the fluorescence change on binding (kΔGH). Titrations with HL-OH(R) (0% ee), HL-OH(P) (32% ee) and HL-OH(M) (96% ee) were fitted simultaneously to the competitive binding model solve for KP, KM, kΔGP and kΔGM. In the case of ctDNA, a complete titration was not possible due to a low binding affinity, which resulted in a less accurate fit. To account for this, the values of kΔGP and kΔGH were fixed from the results of competitive binding to c-Myc. Reported Kd values are the reciprocal of the association constant. The fluorescence ‘switch-on’ values for HL-CH3(R), HL-Morph(R), and HL-OH(R) were calculated as F/F0, where F is calculated from the asymptotic value of the binding fit, and F0 is the initial point on the binding curve, before the addition of oligonucleotide. N.B., this value includes correction for absorption at the excitation wavelength. The selectivities of G4 over dsDNA was calculated as ratios of Kd values (Table 1).
Thiazole orange fluorescence indicator displacement assay (TO-FID)
Experiments were carried out using a BMG CLARIOstar® Microplate reader with Greiner Bio-One half volume (100 μl well) plates using a method adapted from the literature.33 Fluorescence titrations (λex = 475 nm, λex = 520 nm) were carried out using c-Myc G4 DNA (100 mM KCl, 10 mM Tris-HCl, pH 7.3). The final concentrations in the plate were 1 μM G4 DNA, 2 μM thiazole orange (TO) and 0–20 μM helicene (0, 0.94, 1.25, 1.88, 2.50, 3.75, 5.0, 7.5, 10, 15, 20). Sample preparation was carried out by first preparing double concentration stocks of helicene (40 and 30 μM) and a DNA/TO mixture (2 μM and 4 μM respectively). Helicene concentrations were prepared using serial dilutions at 50 μL per well, which was followed by the addition of 50 μL of the DNA/TO mixture and gently shaken for 5 minutes. Percentage displacement curves were calculated from the measured fluorescence intensity (F), using: displacement![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) = 100 − [(F/F0) × 100], where F0 is TO fluorescence from the probe bound to c-Myc without added helicene. Displacements were fitted to a Hill function which was used to calculate the DC50.
= 100 − [(F/F0) × 100], where F0 is TO fluorescence from the probe bound to c-Myc without added helicene. Displacements were fitted to a Hill function which was used to calculate the DC50.
General cell culture
Human bone osteosarcoma epithelial cells (U2OS, from ATCC) were grown in high glucose Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) at 37 °C with 5% CO2 in humidified air.Cytotoxicity of HL-Morph(R) and HL-CH3(R)
Cytotoxicity of HL-Morph(R) and HL-CH3(R) in U2OS cell lines were investigated using the MTS assay. The cells were equally distributed (5 × 103 cells per well) in a 96 well plate in a DMEM medium containing 10% FBS and incubated for 12 h under standard condition. The culture media was removed, and fresh media added with compounds at required concentrations (20, 10, 5, 2.5, 1.25 and 0.625 μM) in triplicate. Wells were maintained without compound (only cells) and without cells (only culture medium) as 100% and 0% viability controls, respectively. After 24 h incubation the cells were treated with MTS/PMS solution and incubated for another 4 h before taking an absorption reading using a plate reader.Confocal imaging
1024 × 1024 resolution fluorescence images were collected using an inverted confocal laser scanning microscope (Leica SP5 II). MitoTracker Green (MTG) emission (500–600 nm) was collected following one-photon excitation from an internal microscope laser at 488 nm, and helicene emission collected at (650–790 nm) following excitation from an internal microscope laser at 633 nm.Fixed cell experiments
Cells were seeded on chambered coverglass (1.5 × 104 cells, 250 μL, 0.8 cm2) for 48 h. Cells were washed (x3) in ice cold PBS before incubation in ice cold paraformaldehyde (PFA, 4% in PBS) solution for 10 min, and a further wash (x3) with ice cold PBS. Fixed cells were further treated with HL-Morph(R) (20 μM, 0.5 h, 21 °C) in PBS before being left under PBS. Cells were left under PBS for imaging to limit the effect of refractive index of the fixation medium on the florescence lifetime.45Fluorescence lifetime imaging microscopy (FLIM)
FLIM was performed through time-correlated single-photon counting (TCSPC), using an inverted confocal laser scanning microscope (Leica SP5 II) and a SPC-830 single-photon counting card (Becker & Hickl GmbH). A pulsed diode laser (Becker & Hickl GmbH, 640 nm, 20 MHz) was used as the excitation source, with a PMC-100-1 photomultiplier tube (Hamamatsu) detector. Fluorescence emission (650–790 nm) was collected through an Airy 1 pinhole for an acquisition time sufficient to obtain signal strength suitable for decay fitting. For all live cell imaging, cells were mounted (on chambered coverglass slides) in the microscope stage, heated by a thermostat (Lauda GmbH, E200) to 37 (±0.5)°C, and kept under an atmosphere of 5% CO2 in air. A 100× (oil, NA = 1.4) objective was used to collect images at either 256 × 256 or 512 × 512 pixel resolution, as stated in the text. The IRF used for deconvolution was recorded using reflection of the excitation beam from a glass cover slide.Lifetime data were fitted using the FLIMfit software tool developed at Imperial College London (v5.1.1, Sean Warren, Imperial College London) to a bi-exponential function, and the intensity-weighted lifetime (τw) calculated using eqn (1). 5 × 5 and 9 × 9 square binning was used to increase signal strength for images recorded at 256 × 256 and 512 × 512 resolution, respectively. A scatter parameter was added to the decay fitting to account for scattered excitation light. Before fitting, a mask was applied to the images to analyse individual cell nuclei staining, or extra-nuclear staining. A threshold was applied to the average of each nucleus to require a minimum of 175 at the peak of the decay and a goodness-of-fit measured by χ2 of less than 2.
Molecular docking
Molecular docking was performed using AutodocVina.46 Ligand structures of isolated compounds were minimised in Gaussian using Density Functional Theory (DFT), the B3LYP functional, and a 6-31G(d,p) basis. Compounds were docked into the lowest energy form of c-Myc (PDB ID:5W77),39 or dsDNA (PDB ID:1Z3F),47 already stripped of their bound ligands. A grid box encompassing the entire quadruplex was used for blind docking. The lowest energy solution was used and the docked structures were visualised using PyMol v2.3.4.Data availability
The essential spectroscopic characterisation and analytical data is included in the ESI.† Additional data is available from the corresponding authors upon reasonable request.Author contributions
J. B. V., M. K. K. and R. V. conceptualised the study and acquired funding. P. A. S., A. P. T., J. B. V., M. K. K. and R. V. designed the methodology and performed the investigation. A. P. T. synthesised and characterised the compounds. P. A. S. and A. P. T. performed the formal analysis. T. K. performed the TO displacement experiments and enantiomeric resolution by HPLC. P. A. S., A. P. T., M. K. K. and R. V. co-wrote and edited the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The Engineering and Physical Sciences Research Council (EPSRC) of the UK is thanked for financial support including a studentship to T. K., and a fellowship for M. K. K. (EP/I003983/1). Imperial College London is thanked for support from the Excellence Fund for Frontier Research. Vannier lab's work is supported by the London Institute of Medical Sciences (LMS), which receives its core funding from UKRI (MRC) and by an ERC Starter Grant (637798; MetDNASecStr). We thank Dr Jochen Brandt for useful discussions.References
- D. Varshney, J. Spiegel, K. Zyner, D. Tannahill and S. Balasubramanian, Nat. Rev. Mol. Cell Biol., 2020, 21, 459–474 CrossRef CAS PubMed.
- D. Rhodes and H. J. Lipps, Nucleic Acids Res., 2015, 43, 8627–8637 CrossRef CAS PubMed.
- R. Hänsel-Hertsch, M. Di Antonio and S. Balasubramanian, Nat. Rev. Mol. Cell Biol., 2017, 18, 279–284 CrossRef PubMed.
- S. Neidle, Nat. Rev. Chem., 2017, 1, 10 CrossRef.
- O. Mendoza, A. Bourdoncle, J. B. Boule, R. M. Brosh and J. L. Mergny, Nucleic Acids Res., 2016, 44, 1989–2006 CrossRef CAS PubMed.
- D. Monchaud and M. P. Teulade-Fichou, Org. Biomol. Chem., 2008, 6, 627–636 RSC.
- S. Neidle, J. Med. Chem., 2016, 59, 5987–6011 CrossRef CAS PubMed.
- A. R. Duarte, E. Cadoni, A. S. Ressurreicao, R. Moreira and A. Paulo, ChemMedChem, 2018, 13, 869–893 CrossRef CAS PubMed.
- R. Vilar, in Metallo-Drugs: Development and Action of Anticancer Agents, ed. H. Sigel, A. Sigel, E. Freisinger and R. K. O. Sigel, De Gruyter, Berlin, 2018, ch. 12, pp. 325–351 Search PubMed.
- H. Tahara, K. Shin-ya, H. Seimiya, H. Yamada, T. Tsuruo and T. Ide, Oncogene, 2006, 25, 1955–1966 CrossRef CAS PubMed.
- D. Monchaud, in Annu. Rep. Med. Chem., ed. S. Neidle, Academic Press, Cambridge Massachusetts, 2020, ch. 5, pp. 133–160 Search PubMed.
- A. Shivalingam, M. A. Izquierdo, A. L. Marois, A. Vyšniauskas, K. Suhling, M. K. Kuimova and R. Vilar, Nat. Commun., 2015, 6, 8178 CrossRef CAS PubMed.
- M. Di Antonio, A. Ponjavic, A. Radzevicius, R. T. Ranasinghe, M. Catalano, X. Y. Zhang, J. Z. Shen, L. M. Needham, S. F. Lee, D. Klenerman and S. Balasubramanian, Nat. Chem., 2020, 12, 832–837 CrossRef CAS PubMed.
- L.-Y. Liu, W. Liu, K.-N. Wang, B.-C. Zhu, X.-Y. Xia, L.-N. Ji and Z.-W. Mao, Angew. Chem., Int. Ed., 2020, 59, 9719–9726 CrossRef CAS PubMed.
- P. A. Summers, B. W. Lewis, J. Gonzalez-Garcia, R. M. Porreca, A. H. M. Lim, P. Cadinu, N. Martin-Pintado, D. J. Mann, J. B. Edel, J. B. Vannier, M. K. Kuimova and R. Vilar, Nat. Commun., 2021, 12, 162 CrossRef CAS PubMed.
- T. Y. Tseng, W. W. Chen, I. T. Chu, C. L. Wang, C. C. Chang, M. C. Lin, P. J. Lou and T. C. Chang, Sci. Rep., 2018, 8, 16082 CrossRef PubMed.
- T. Wilson, P. J. Costa, V. Felix, M. P. Williamson and J. A. Thomas, J. Med. Chem., 2013, 56, 8674–8683 CrossRef CAS PubMed.
- H. J. Yu, X. H. Wang, M. L. Fu, J. S. Ren and X. G. Qu, Nucleic Acids Res., 2008, 36, 5695–5703 CrossRef CAS PubMed.
- B. L. Xu, C. Q. Zhao, Y. Chen, H. Tateishi-Karimata, J. S. Ren, N. Sugimoto and X. G. Qu, Chem.–Eur. J., 2014, 20, 16467–16472 CrossRef CAS PubMed.
- H. S. Qin, C. Q. Zhao, Y. H. Sun, J. S. Ren and X. G. Qu, J. Am. Chem. Soc., 2017, 139, 16201–16209 CrossRef CAS PubMed.
- C. Q. Zhao, H. L. Song, P. Scott, A. D. Zhao, H. Tateishi-Karimata, N. Sugimoto, J. S. Ren and X. G. Qu, Angew. Chem., Int. Ed., 2018, 57, 15723–15727 CrossRef CAS PubMed.
- S. Muller, K. Laxmi-Reddy, P. V. Jena, B. Baptiste, Z. Y. Dong, F. Godde, T. Ha, R. Rodriguez, S. Balasubramanian and I. Huc, ChemBioChem, 2014, 15, 2563–2570 CrossRef PubMed.
- Y. Xu, Y. X. Zhang, H. Sugiyama, T. Umano, H. Osuga and K. Tanaka, J. Am. Chem. Soc., 2004, 126, 6566–6567 CrossRef CAS PubMed.
- K. Shinohara, Y. Sannohe, S. Kaieda, K. Tanaka, H. Osuga, H. Tahara, Y. Xu, T. Kawase, T. Bando and H. Sugiyama, J. Am. Chem. Soc., 2010, 132, 3778–3782 CrossRef CAS PubMed.
- A. Shivalingam, A. Vysniauskas, T. Albrecht, A. J. P. White, M. K. Kuimova and R. Vilar, Chem.–Eur. J., 2016, 22, 4129–4139 CrossRef CAS PubMed.
- F. Torricelli, J. Bosson, C. Besnard, M. Chekini, T. Burgi and J. Lacour, Angew. Chem., Int. Ed., 2013, 52, 1796–1800 CrossRef CAS PubMed.
- (a) C. Bauer, R. Duwald, G. M. Labrador, S. Pascal, P. M. Lorente, J. Bosson, J. Lacour and J. D. Rochaix, Org. Biomol. Chem., 2018, 16, 919–923 RSC; (b) O. Kel, A. Fürstenberg, N. Mehanna, C. Nicolas, B. Laleu, M. Hammarson, B. Albinsson, J. Lacour and E. Vauthey, Chem.–Eur. J., 2013, 19, 7173–7180 CrossRef CAS PubMed.
- J. Bosson, G. M. Labrador, S. Pascal, F. A. Miannay, O. Yushchenko, H. D. Li, L. Bouffier, N. Sojic, R. C. Tovar, G. Muller, D. Jacquemin, A. D. Laurent, B. Le Guennic, E. Vauthey and J. Lacour, Chem.–Eur. J., 2016, 22, 18394–18403 CrossRef CAS PubMed.
- H. D. Li, A. Wallabregue, C. Adam, G. M. Labrador, J. Bosson, L. Bouffier, J. Lacour and N. Sojic, J. Phys. Chem. C, 2017, 121, 785–792 CrossRef CAS.
- R. Duwald, S. Pascal, J. Bosson, S. Grass, C. Besnard, T. Burgi and J. Lacour, Chem.–Eur. J., 2017, 23, 13596–13601 CrossRef CAS PubMed.
- Z. Jarolimova, J. Bosson, G. M. Labrador, J. Lacour and E. Bakker, Electroanalysis, 2018, 30, 650–657 CrossRef CAS.
- G. M. Labrador, C. Besnard, T. Burgi, A. I. Poblador-Bahamonde, J. Bosson and J. Lacour, Chem. Sci., 2019, 10, 7059–7067 RSC.
- E. Largy, F. Hamon and M. P. Teulade-Fichou, Anal. Bioanal. Chem., 2011, 400, 3419–3427 CrossRef CAS PubMed.
- D. Sarkar, P. Das, S. Basak and N. Chattopadhyay, J. Phys. Chem. B, 2008, 112, 9243–9249 CrossRef CAS PubMed.
- M. Monnot, O. Mauffret, E. Lescot and S. Fermandjian, Eur. J. Biochem., 1992, 204, 1035–1039 CrossRef CAS PubMed.
- D. Sahoo, P. Bhattacharya and S. Chakravorti, J. Phys. Chem. B, 2010, 114, 2044–2050 CrossRef CAS PubMed.
- B. Jana, S. Senapati, D. Ghosh, D. Bose and N. Chattopadhyay, J. Phys. Chem. B, 2012, 116, 639–645 CrossRef CAS PubMed.
- C. V. Kumar, R. S. Turner and E. H. Asuncion, J. Photochem. Photobiol., A, 1993, 74, 231–238 CrossRef CAS.
- D. R. Calabrese, X. Chen, E. C. Leon, S. M. Gaikwad, Z. Phyo, W. M. Hewitt, S. Alden, T. A. Hilimire, F. He, A. M. Michalowski, J. K. Simmons, L. B. Saunders, S. L. Zhang, D. Connors, K. J. Walters, B. A. Mock and J. S. Schneekloth, Nat. Commun., 2018, 9, 4229 CrossRef PubMed.
- J. X. Dai, D. Chen, R. A. Jones, L. H. Hurley and D. Z. Yang, Nucleic Acids Res., 2006, 34, 5133–5144 CrossRef CAS PubMed.
- P. Thordarson, Chem. Soc. Rev., 2011, 40, 1305–1323 RSC.
- P. Thordarson, Chem. Soc. Rev., 2011, 40, 5922–5923 RSC.
- I. Jarmoskaite, I. AlSadhan, P. P. Vaidyanathan and D. Herschlag, eLife, 2020, 9, e57264 CrossRef CAS PubMed.
- Z. X. Wang, FEBS Lett., 1995, 360, 111–114 CrossRef CAS PubMed.
- L. Joosen, M. A. Hink, T. W. J. Gadella and J. Goedhart, J. Microsc., 2014, 256, 166–176 CrossRef CAS PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS.
- A. Canals, M. Purciolas, J. Aymani and M. Coll, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2005, 61, 1009–1012 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc04567a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |