Open Access Article
Open Access ArticleA naphthalimide-based peptide conjugate for concurrent imaging and apoptosis induction in cancer cells by utilizing endogenous hydrogen sulfide†
Narendra
Singh‡
a,
Swati
Sharma‡
a,
Ramesh
Singh‡
 b,
Swati
Rajput‡
c,
Naibedya
Chattopadhyay
b,
Swati
Rajput‡
c,
Naibedya
Chattopadhyay
 c,
Deepshikha
Tewari
c,
Deepshikha
Tewari
 *d,
Khashti Ballabh
Joshi
*d,
Khashti Ballabh
Joshi
 *b and
Sandeep
Verma
*b and
Sandeep
Verma
 *a
*a
aDepartment of Chemistry and Centre for Nanosciences, Indian Institute of Technology Kanpur, U.P. 208016, India. E-mail: sverma@iitk.ac.in
bDepartment of Chemistry, School of Chemical Science and Technology, Dr. Harisingh Gour Vishwavidyalaya (A Central University), Sagar, M.P. 470003, India. E-mail: kbjoshi77@gmail.com; kbjoshi@dhsgsu.ac.in
cCSIR-Central Drug Research Institute, Sector 10, Jankipuram Extension, Sitapur Road, Lucknow 226031, Uttar Pradesh, India
dAdvance Imaging Center, Centre for Nanosciences, Indian Institute of Technology Kanpur, U.P. 208016, India. E-mail: dtewari@iitk.ac.in
First published on 17th November 2021
Abstract
The excessive production of endogenous hydrogen sulfide (H2S) in cancer cells leads to enhanced tumor growth and metastasis. On the other hand, decreased endogenous H2S suppresses tumor growth. The reported approaches for inhibiting tumor growth are selective silencing of the tumor-promoting genes and pharmacological inhibition of these proteins. To enhance the antitumor efficacy of frontline chemotherapeutic agents, herein, we synthesized a highly sensitive endogenous H2S responsive fluorescent probe, i.e., a hydrogen sulfide-sensing naphthalimide-based peptide conjugate (HSNPc), which showed selective inhibition of proliferation of cancer cells due to apoptosis induction. Furthermore, HSNPc suppressed the glycolytic reserve, a critical energy source for the proliferation of cancer cells. HSNPc also decreased the Young's modulus of HeLa cells compared to the control cells, which demonstrated a direct relation between cell apoptosis and cell stiffness. Taken together, we demonstrated the dual function of detection and killing of cancer cells by HSNPc that can be likened to a theranostic role.
Introduction
Hydrogen sulfide (H2S) is an important signaling molecule that regulates proliferation, apoptosis and angiogenesis. These cellular events are critical regulators of growth and metastasis of tumors.1–5 Malignant cells overexpress cystathionine β-synthase (CBS), cystathionine γ-lyase (CSE) and 3-mercaptopyruvate sulfur transferase (3-MST) to produce increased H2S that in turn promotes growth, survival and metastasis of these cells.6,7 Several studies reported that overexpression of H2S producing enzymes leads to an enhanced endogenous H2S concentration which then acts in an autocrine mode to promote proliferation, angiogenesis and bioenergetic reprogramming of the cancer cells.4,8An emerging strategy for treating cancer is targeting their “unique” property of producing endogenous H2S that allows differentiation between healthy and malignant tissue.9 Consequently, there is an active search for theranostic agents to exploit the H2S sensing ability and in this context, several detection methods10–15 have been developed in recent years to sense endogenous H2S.16–20 Some fluorescent probes based on naphthalimide,21,22 rhodamine,23 fluorescein,24,25 and cyanine,26,27 show good sensitivity and selectivity among others but there are very few reported schemes that can sense endogenous H2S and impede cancer cell proliferation at the same time.28,29
Despite significant advancement in our understanding of cancer biology and therapeutics, diagnosis and treatment are two separate arms of the clinical management of cancer. Discovery and development of theranostic agents could integrate these two arms and potentially revolutionize cancer management by early detection and clearance of cancer cells without impacting normal cells. Also, cell penetration is the biggest hurdle for drug efficacy and biodistribution. To combat these problems, we have rationally designed a naphthalimide conjugated diphenylalanine-based short peptide prone to cellular uptake30 and membrane permeability.31 Peptide-based agents are suitable for theranostic purposes given their biocompatibility in human and mammalian systems. Such peptides with cell-penetrating properties have been evaluated to transport diverse cargoes into cells, tissues, and organs.32
The peptide–naphthalimide fluorescent pendant was further equipped with a H2S sensing probe to make this peptide activatable at the orthogonal site to impart high sensitivity to endogenous H2S (Scheme 1) thereby enabling it to selectively image and simultaneously kill the cancer cells. 1,8-Naphthalimide derivatives such as amonafide and33 mitonafide are emerging theranostic agents with potential of cell growth inhibition as well as cellular imaging. These molecules intercalate with DNA and inhibit topoisomerase and cause cell apoptosis.33–35 Capping of the hydroxy group of naphthalimide is the biggest advantage over non-capped naphthalimide-based anticancer molecules, as the H2S-sensing group makes it proactive and highly selective. Therefore, HSNPc is a novel and efficient platform possessing the selective ability for cell apoptosis and cancer cell imaging.
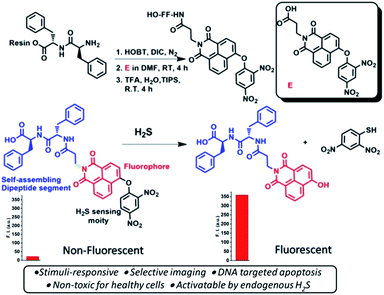 | ||
| Scheme 1 Schematics of the synthesis of the hydrogen sulfide responsive fluorescent peptide conjugate. Bottom: represents the non-fluorescent peptide conjugate turning fluorescent upon H2S treatment. | ||
Results and discussion
A H2S-sensing naphthalimide-based peptide conjugate (HSNPc) was synthesized by both, solution phase36–38 and solid-phase peptide synthesis using standard protocols.39 4-Bromo naphthalic anhydride was used as a precursor molecule. Conjugation of β-alanine to this anhydride followed by substitution of bromine with the dinitrobenzene probe at the fourth position of 4-bromo naphthalic anhydride were carried out in the solution phase. A separate segment of the pre-synthesized diphenylalanine peptide was added to beta-alanine. The molecule was then purified by preparative HPLC and characterized by 1H NMR, 13C NMR and HRMS (ESI).Our data revealed that the HSNPc is highly sensitive towards H2S to the extent that it could be seen by the naked eye in daylight. To further assess the H2S sensitivity of HSNPc, we treated its 1 mM solution with 10 mM solution of sodium sulfide (Na2S) and observed a color transformation from colorless to yellow color. The color transformation was also observed under short UV light and long UV light. Under short UV light, it was greenish-yellow and under long UV light, it was fluorescent yellow (ESI Fig. S5†). To confirm and characterize these observations, a time-dependent HPLC analysis was performed by treating 1 mM HSNPc with 10 mM Na2S solution40 and the resultant spectra were recorded over time. The retention time of HSNPc was observed to be ∼17.17 min as a control. Notably, two new peaks emerged for Na2S-treated HSNPc solution. The first peak was at 12.38 min and another was at 10.85 min, while the parent peak at 17.17 min decreased in intensity with time. The peak at 12.38 min belongs to the peptide linked 1,8 naphthalimide moiety and that at 10.85 min belongs to the 2,4-dinitrobenzenethiol group. We also observed that with 10 equivalents of Na2S, almost 99% HSNPc was degraded into two fragments (Scheme 1) within 45 min (ESI Fig. S6†).
To validate the above observations, UV-Vis titration experiments were performed. A bathochromic shift was observed when HSNPc solution was titrated with Na2S solution. The absorption maximum (λmax) at 350 nm was shifted to 440 nm due to the ipso attack of the nucleophile, HS− ions, on the dinitrobenzene ring followed by cleavage of the 2,4-dinitrobenzene group from HSNPc (Fig. 1A, ESI Fig. S7 and S8†). The absorbance of 2,4-dinitrobenzenethiol was found centered at 418 nm.37 Therefore, the absorbance of the free naphthalimide conjugated peptide segment and 2,4-dinitrobenzenethiol likely overlapped to produce a broad-range peak at 440 nm (ESI Fig. S9†). The time-course study revealed that the 350 nm peak diminished with the concurrent increase in the 440 nm peak when Na2S was added to the HSNPc solution (Fig. 1A). Fluorescence spectroscopy data also corresponded well with the observed UV-Vis results (Fig. 1B).
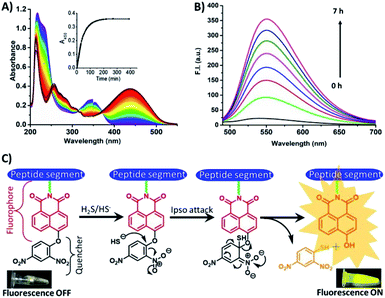 | ||
| Fig. 1 Time-dependent UV-Vis and fluorescence spectra of HSNPc, showing sensitivity in the presence of H2S. (A) UV-Vis, (B) fluorescence and (C) fluorescence ON and OFF mechanisms. | ||
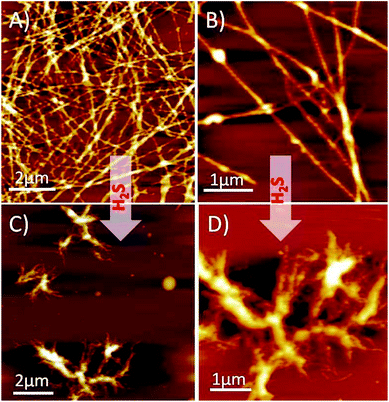
Next, we performed solution-phase studies to determine the morphology of HSNPc. A 200 μM freshly prepared solution of HSNPc in ethanol systems self-assembled into a long and thin fibrous assembly (Fig. 2A). However, at a higher magnification, these fibers appeared long and twisted with fibrous assembly (Fig. 2B) which is likely due to the non-covalent interactions where aromatic π–π interactions played an instrumental role. However, in the presence of H2S, the morphology of HSNPc was altered to fibrous “bush-like” assembly (Fig. 2C and D), thus suggesting the effect of H2S on the molecular transformation of the peptide structure.
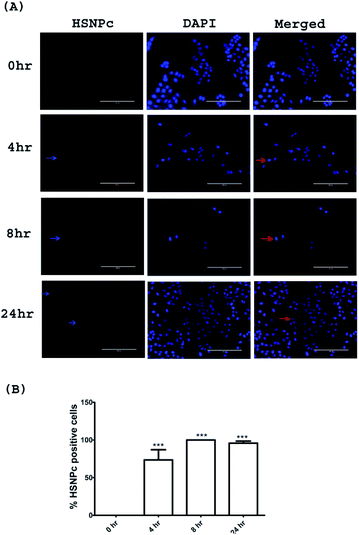
Since, unlike normal (non-malignant) cells, cancer cells are known to have activated H2S production machinery, we studied the effect of HSNPc on HeLa cells, a line derived from cervical cancer. As shown in Fig. 3, the peptide was detected in the nuclei from 4 h to 24 h. Human mesenchymal stem cells (hMSCs) were used as the representative non-malignant/non-transformed cells. Unlike in HeLa cells, a fluorescence signal was not detectable in hMSCs when treated with HSNPc (Fig. 4, topmost panels). Next, to assess whether endogenous H2S promotes nuclear localization of HSNPc, we treated hMSCs with Na2S (a HS− donor) in the presence of the peptide. In the presence of Na2S (80 μM and higher), a fluorescence signal was detectable in the nuclei albeit extra-nuclear fluorescence was also detectable. From these data, it is reasonable to conclude that HSNPc is a H2S-dependent fluorophore that could be developed as an imaging agent for cancer cells.
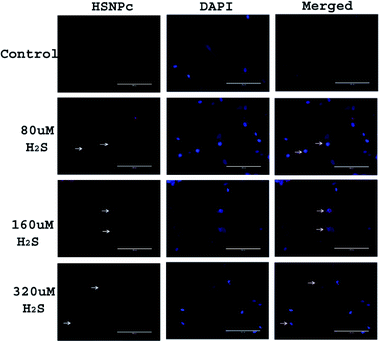 | ||
| Fig. 4 Representative images of hMSCs with indicated treatments. Images were taken at 20× (bar length 200 μm). Arrow show the HSNPc positive cells. | ||
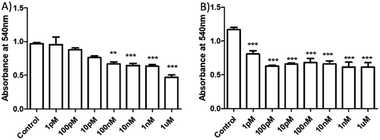
Having demonstrated the sensitivity of HSNPc to H2S, we next assessed the effect of this peptide on HeLa cells and hMSCs. HeLa cells and hMSCs were treated with HSNPc (10−12 M to 10−6 M) for 48 h. HSNPc was found to have significant cytotoxicity at almost every concentration in HeLa cells (IC50: 25 nM) but not in hMSCs (Fig. 5 and S13, ESI†).
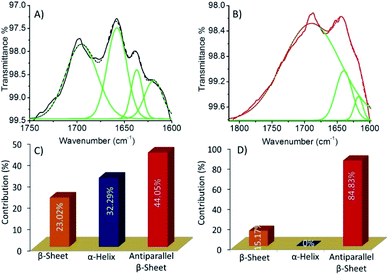
The reason for cytotoxicity of HSNPc in HeLa cells is the presence of the 1,8-naphthalimide moiety, a well-known DNA targeting anticancer agent which binds DNA by a combination of interactions such as hydrogen bonding, electrostatic and intercalation.41HSNPc appeared to have been activated in the presence of endogenous H2S in HeLa cells and produced the peptide-linked 1,8-naphthalimide moiety (Scheme 1, fluorescent probe) but failed in hMSCs due to the lack of H2S production machinery. Therefore, the cellular toxicity of the peptide-linked 1,8-naphthalimide moiety (control) in HeLa cells and human mesenchymal stem cells was also studied by the MTT assay which revealed that the control peptide has no cellular toxicity in either cells (Fig. S14†). The observed toxicity to cancer cells by HSNPc compared to the control peptide could be attributed to factors including unique peptide nano-assemblies (Fig. 2) which are known to target cancer cells,42 its stimulus (endogenous H2S)-responsive cancer cell-specific biodistribution, and less physical deactivation to name a few. The reported literature revealed that the presence of α-helix or β-sheet type secondary structures can play an important role during cellular uptake in which the helicity of peptides is more important for their cell-penetrating abilities.43,44 Therefore, we studied the secondary structure of both the molecules by using the FT-IR study (Fig. 6). The quantitative analysis of the FT-IR spectra45 revealed that HSNPc fibrous structures contributed 44.05% to anti-parallel β-sheets, 23.02% to β-sheets and 32.29% to α-helix (Fig. 6A, C and Table S1†) whereas the morphologies of the control peptide showed 84.83% for anti-parallel β-sheets and 15.17% for β-sheets (Fig. 6B, D and Table S2†). Taken together, it is concluded that the unique peptide nano-assemblies of HSNPc facilitated its higher cellular uptake and therefore perhaps shows greater cellular toxicity in cancer cells.
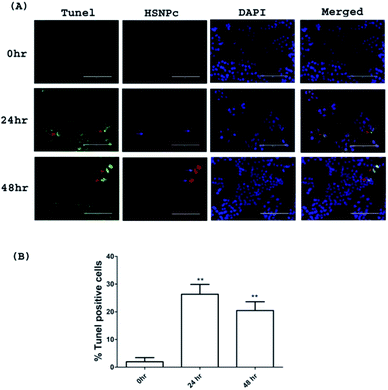
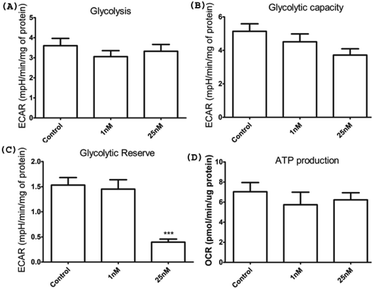
Given the loss of viability of HeLa cells upon HSNPc treatment, we surmised the induction of apoptosis as the cause of this effect. Fig. 7 shows that HSNPc induced apoptosis of HeLa cells at both 24 and 48 h when assessed by quantitative TUNEL staining. We next studied the mechanism of apoptosis of HeLa cells by using HSNPc. Cancer cells are known to be dependent on aerobic glycolysis for deriving energy to proliferate. We, therefore, studied the energy metabolism of HeLa cells in response to HSNPc. Prior to this experiment, we investigated the possible mitochondrial localization of the peptide using a mitotracker which showed its distribution in the mitochondria (ESI Fig. S12†). However, our study revealed no change in ATP production by HSNPc (Fig. 8D). Among the glycolysis parameters (Fig. 8A–C), only the glycolytic reserve was significantly decreased by the peptide (Fig. 8C, ESI Fig. S12†). These data suggested that consistency with the energy metabolism is characteristic of cancer cells HeLa cells are critically dependent on glycolysis and depletion of glycolytic reserve likely deprived the cells from carrying out energy-demanding events such as proliferation. No change in ATP production is indicative of the lack of mitochondrial involvement in energy production by HeLa cells.
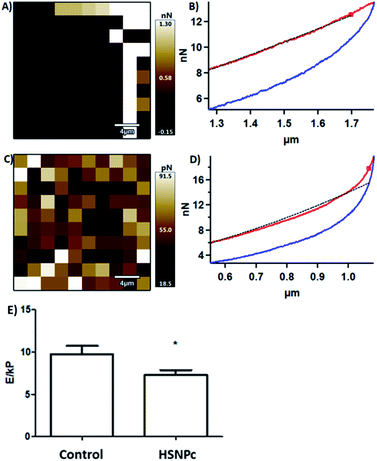
Toseland et al. reported that cisplatin-induced DNA damage is associated with cell stiffness.46 The damage in the DNA caused by a DNA nick by HSNPc and detected by TUNEL staining (Fig. 7) could affect the stiffness of HeLa cells. Cell biomechanics importantly regulates the function of the cells,47 and stiffness is reported to alter the cell migration and invasion, which is particularly relevant for cancer metastasis.48,49 These reports prompted us to measure HeLa cell biomechanics in response to HSNPc using atomic force microscopy (AFM). HeLa cell lines were adhered to an 18 mm glass slide and were treated with 25 mM HSNPc for 24 h. To determine the Young's modulus, AFM measurements were recorded after exposing a group of 50 cells to 10 nN force. Hertz model fitted force–distance curves were used to calculate the Young's modulus (‘E’). Fig. 9 shows the force–distance curve of the control and HSNPc-treated HeLa cells. ‘E’ of HSNPc treated HeLa and control cells was respectively 7.26 ± 0.6 kP and 9.76 ± 0.95 kP thus demonstrating a significant decrease in the cell stiffness after 24 h treatment (*p < 0.05). AFM-determined cell stiffness showed a decrease in the Young's modulus which could inhibit cell motility and metastasis. Kim et al. studied the effect of paclitaxel on HeLa cell stiffness and concluded that reduced cell stiffness is related to apoptosis.48 Moreover, drug-resistant breast and ovarian cancer cells are stiffer than deformable cells.50–53HSNPc by decreasing the stiffness could therefore reduce the risk of metastasis and chemotherapeutic resistance of cancer cells.
Conclusions
In summary, we have developed a naphthalimide-based novel bifunctional fluorescent probe for cancer cell imaging and concomitant induction of apoptosis. Our designed probe is successfully activated by endogenously produced H2S of cancer cells and could be applied for specific imaging of cancer cells. Apart from cell imaging, the probe induced apoptosis of cancer cells. Such unique properties of the probe make it an ideal candidate for precise cancer diagnosis and treatment. We anticipate that the H2S sensing peptide-conjugated fluorescent probe such as the one described here holds potential for becoming a theranostic agent for effective cancer management.Experimental
Reagents and kits
Cell culture supplements were from Sigma-Aldrich, India. TUNEL assay kit was from Invitrogen (#C10617), mitotracker from Invitrogen (#M7514), and Seahorse XF Cell Mito Stress Test Kit and Seahorse XF Glycolysis Stress Test Kit from Agilent Technologies.Cell culture
HeLa cells were maintained in DMEM high glucose medium supplemented with 10% fetal bovine serum (FBS). Cells at 90% confluency were trypsinized and seeded for experiments. For microscopic experiments, the cells were seeded in a chamber slide with 40% confluency and treated with HSPNc for the mentioned time period and after 24 or 48 h, the cells were washed with phosphate buffered saline (PBS) twice, fixed with 10% formalin for 15 min, co-stained with DAPI (1 μg μL−1) for 15–20 min and mounted with mounting medium. Fluorescence microscopy was performed at 20×. For mitochondrial localization, the cells were incubated with 200 nM mitotracker green in 1% DMEM high glucose and incubated for 30 min followed by fluorescence microscopy. hMSCs were maintained in DMEM F12 medium supplemented with 10% FBS. Cells were trypsinized and seeded for localization of HSNPc after endogenous H2S treatment. Cells were treated with 25 nM HSNPc for 24 h and before termination H2S was added at different concentrations (20 μM to 320 μM).TUNEL assay
HeLa cells were plated on a chamber slide and treated with 25 nM HSNPc for 24 and 48 h. Click-iT™ Plus TUNEL Assay kit (C10617, Invitrogen) was used for in situ detection of apoptotic cells according to the manufacturer's protocols.54Assessment of bioenergetics parameters in HeLa cells
HeLa cells were plated on 24-well Seahorse V7-PS Flux plates (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well). After 24 h, the cells were treated with the vehicle or HSNPc (final concentration 25 nM) for 48 h. The detailed protocol was described before.55
000 cells per well). After 24 h, the cells were treated with the vehicle or HSNPc (final concentration 25 nM) for 48 h. The detailed protocol was described before.55
Data availability
The information about detailed experiments and related data can be found in ESI file.†Author contributions
NS, SV and KBJ conceptualized the study. Synthesis and characterization were done by NS and supervised by KBJ. NS, SS, RS, and SR performed the experiments with cell cultures and performed the image analysis under the guidance of DT and NC. NS and SS performed the AFM force mapping and spectroscopy in the supervision of KBJ and DT. RS performed the AFM imaging and spectroscopy and discussed the overall results. KBJ, SV, DT and NC supervised all the experimental parts and discussed the overall results with all the co-authors. The initial draft of the manuscript was written by NS and SS that was further commented and refined by KBJ, DT and SV. All authors contributed to the data analysis and its compilation and agreed with the final form of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
SV would like to thank the J. C. Bose Fellowship (SERB, India) and DST Nanomission for financial support. This work was also partially supported by the IIT Kanpur Stem Cell Research Facility. NS, SS, RS and SR thank CSIR-UGC, MHRD, UGC and DBT for pre-doctoral senior and junior research fellowships, respectively. DT acknowledges DST, India for the research grant. Prof. S. Ganesh, Department of Biological Sciences and Engineering – Indian Institute of Technology Kanpur, is acknowledged for gifting HeLa cells and some reagent support. We acknowledge the Sophisticated Instrument Centre (SIC)-Dr. Harisingh Gour Vishwavidyalaya (A Central University) Sagar, India for AFM facility.Notes and references
- C. Szabo, Nat. Rev. Drug Discovery, 2016, 15, 185–203 CrossRef CAS PubMed.
- C. Szabo, Cells, 2021, 10, 220 CrossRef CAS PubMed.
- S. Sen, B. Kawahara, D. Gupta, R. Tsai, M. Khachatryan, S. Roy-Chowdhuri, S. Bose, A. Yoon, K. Faull, R. Farias-Eisner and G. Chaudhuri, Free Radical Biol. Med., 2015, 86, 228–238 CrossRef CAS PubMed.
- C. Szabo, C. Coletta, C. Chao, K. Modis, B. Szczesny, A. Papapetropoulos and M. R. Hellmich, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12474–12479 CrossRef CAS PubMed.
- M. Li, Y. Liu, Y. Deng, L. Pan, H. Fu, X. Han, Y. Li, H. Shi and T. Wang, Oncol. Rep., 2021, 45, 68 CrossRef CAS PubMed.
- N. L. Kanagy, C. Szabo and A. Papapetropoulos, Am. J. Physiol., 2017, 312, C537–C549 CrossRef PubMed.
- C. Köhn, G. Dubrovska, Y. Huang and M. Gollasch, Int. J. Biomed. Sci., 2012, 8, 81–86 Search PubMed.
- M. R. Hellmich and C. Szabo, Hydrogen Sulfide and Cancer, in Handbook of Experimental Pharmacology, 2015, pp. 233–241 Search PubMed.
- X. Wang, L. An, Q. Tian and K. Cui, RSC Adv., 2019, 9, 33578–33588 RSC.
- A. R. Lippert, E. J. New and C. J. Chang, J. Am. Chem. Soc., 2011, 133, 10078–10080 CrossRef CAS PubMed.
- R. Peltier, G. Chen, H. Lei, M. Zhang, L. Gao, S. S. Lee, Z. Wang and H. Sun, Chem. Commun., 2015, 51, 17273–17276 RSC.
- T. Jia, C. Fu, C. Huang, H. Yang and N. Jia, ACS Appl. Mater. Interfaces, 2015, 7, 10013–10021 CrossRef CAS PubMed.
- C.-G. Dai, X.-L. Liu, X.-J. Du, Y. Zhang and Q.-H. Song, ACS Sens., 2016, 1, 888–895 CrossRef CAS.
- G.-J. Mao, T.-T. Wei, X.-X. Wang, S. Huan, D.-Q. Lu, J. Zhang, X.-B. Zhang, W. Tan, G.-L. Shen and R.-Q. Yu, Anal. Chem., 2013, 85, 7875–7881 CrossRef CAS PubMed.
- S. Gong, E. Zhou, J. Hong and G. Feng, Anal. Chem., 2019, 91, 13136–13142 CrossRef CAS PubMed.
- R. Wang, X. Gu, Q. Li, J. Gao, B. Shi, G. Xu, T. Zhu, H. Tian and C. Zhao, J. Am. Chem. Soc., 2020, 142, 15084–15090 CrossRef CAS PubMed.
- Y. Qian, J. Karpus, O. Kabil, S.-Y. Zhang, H.-L. Zhu, R. Banerjee, J. Zhao and C. He, Nat. Commun., 2011, 2, 495 CrossRef PubMed.
- V. S. Lin, A. R. Lippert and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7131–7135 CrossRef CAS PubMed.
- L. Zhang, H. Zhu, M. Li and X. Gu, Chem. Commun., 2015, 51, 13135–13137 RSC.
- L. Zhang, X. E. Zheng, F. Zou, Y. Shang, W. Meng, E. Lai, Z. Xu, Y. Liu and J. Zhao, Sci. Rep., 2016, 6, 18868 CrossRef CAS PubMed.
- S.-A. Choi, C. S. Park, O. S. Kwon, H.-K. Giong, J.-S. Lee, T. H. Ha and C.-S. Lee, Sci. Rep., 2016, 6, 26203 CrossRef CAS PubMed.
- J. Liu, X. Liu, S. Lu, L. Zhang, L. Feng, S. Zhong, N. Zhang, T. Bing and D. Shangguan, Analyst, 2020, 145, 6549–6555 RSC.
- M. D. Hammers, M. J. Taormina, M. M. Cerda, L. A. Montoya, D. T. Seidenkranz, R. Parthasarathy and M. D. Pluth, J. Am. Chem. Soc., 2015, 137, 10216–10223 CrossRef CAS PubMed.
- J. Lv, F. Wang, J. Qiang, X. Ren, Y. Chen, Z. Zhang, Y. Wang, W. Zhang and X. Chen, Biosens. Bioelectron., 2017, 87, 96–100 CrossRef CAS PubMed.
- X. Jin, S. Wu, M. She, Y. Jia, L. Hao, B. Yin, L. Wang, M. Obst, Y. Shen, Y. Zhang and J. Li, Anal. Chem., 2016, 88, 11253–11260 CrossRef CAS PubMed.
- H. Wang, D. Yang, R. Tan, Z. J. Zhou, R. Xu, J. F. Zhang and Y. Zhou, Sens. Actuators, B, 2017, 247, 883–888 CrossRef CAS.
- Z. Chen, X. Mu, Z. Han, S. Yang, C. Zhang, Z. Guo, Y. Bai and W. He, J. Am. Chem. Soc., 2019, 141, 17973–17977 CrossRef CAS PubMed.
- K. N. Bobba, G. Saranya, P. T. Sujai, M. M. Joseph, N. Velusamy, A. Podder, K. K. Maiti and S. Bhuniya, ACS Appl. Bio Mater., 2019, 2, 1322–1330 CrossRef CAS.
- C. Ge, X. Di, S. Han, M. Wang, X. Qian, Z. Su, H.-K. Liu and Y. Qian, Chem. Commun., 2021, 57, 1931–1934 RSC.
- E. J. Sayers, K. Cleal, N. G. Eissa, P. Watson and A. T. Jones, J. Controlled Release, 2014, 195, 55–62 CrossRef CAS PubMed.
- R. Perkins and V. Vaida, J. Am. Chem. Soc., 2017, 139, 14388–14391 CrossRef CAS PubMed.
- R. M. Johnson, S. D. Harrison and D. Maclean, Methods Mol. Biol., 2011, 683, 535–551 CrossRef CAS PubMed.
- Y. Wang, J. Zhang, M. Li, M. Li, S. Xie and C. Wang, Chem. Biol. Drug Des., 2017, 89, 670–680 CrossRef CAS PubMed.
- G.-B. Liang, J.-H. Wei, H. Jiang, R.-Z. Huang, J.-T. Qin, H.-L. Wang, H.-S. Wang and Y. Zhang, Eur. J. Med. Chem., 2021, 210, 112951 CrossRef CAS PubMed.
- Z. Chen, X. Liang, H. Zhang, H. Xie, J. Liu, Y. Xu, W. Zhu, Y. Wang, X. Wang, S. Tan, D. Kuang and X. Qian, J. Med. Chem., 2010, 53, 2589–2600 CrossRef CAS PubMed.
- N. Singh, R. Singh, S. Sharma, K. Kesharwani, K. B. Joshi and S. Verma, New J. Chem., 2021, 45, 153–161 RSC.
- N. Singh, R. Singh, M. Shukla, G. Kaul, S. Chopra, K. B. Joshi and S. Verma, ACS Infect. Dis., 2020, 6, 2441–2450 CrossRef CAS PubMed.
- S. Sharma, C. Kulkarni, M. M. Kulkarni, R. Ali, K. Porwal, N. Chattopadhyay, D. Tewari and S. Verma, Chem. Commun., 2020, 56, 3043–3046 RSC.
- R. Ali, H. A. Pal, R. Hameed, A. Nazir and S. Verma, Chem. Commun., 2019, 55, 10142–10145 RSC.
- M. D. Hartle and M. D. Pluth, Chem. Soc. Rev., 2016, 45, 6108–6117 RSC.
- S. Banerjee, E. B. Veale, C. M. Phelan, S. A. Murphy, G. M. Tocci, L. J. Gillespie, D. O. Frimannsson, J. M. Kelly and T. Gunnlaugsson, Chem. Soc. Rev., 2013, 42, 1601 RSC.
- D. M. Raymond and B. L. Nilsson, Chem. Soc. Rev., 2018, 47, 3659–3720 RSC.
- M. Oba, Y. Nagano, T. Kato and M. Tanaka, Sci. Rep., 2019, 9, 1349 CrossRef PubMed.
- E. Eiríksdóttir, K. Konate, Ü. Langel, G. Divita and S. Deshayes, Biochim. Biophys. Acta, Biomembr., 2010, 1798, 1119–1128 CrossRef PubMed.
- R. Singh, N. K. Mishra, N. Singh, P. Rawal, P. Gupta and K. B. Joshi, New J. Chem., 2020, 44, 9255–9263 RSC.
- Á. dos Santos, A. W. Cook, R. E. Gough, M. Schilling, N. A. Olszok, I. Brown, L. Wang, J. Aaron, M. L. Martin-Fernandez, F. Rehfeldt and C. P. Toseland, Nucleic Acids Res., 2021, 49, 340–353 CrossRef PubMed.
- H. Huang, C. Dai, H. Shen, M. Gu, Y. Wang, J. Liu, L. Chen and L. Sun, Int. J. Mol. Sci., 2020, 21, 6248 CrossRef CAS PubMed.
- K. S. Kim, C. H. Cho, E. K. Park, M.-H. Jung, K.-S. Yoon and H.-K. Park, PLoS One, 2012, 7, e30066 CrossRef CAS PubMed.
- G. Runel, N. Lopez-Ramirez, J. Chlasta and I. Masse, Cells, 2021, 10, 887 CrossRef CAS PubMed.
- S. Park, J. Cancer Prev., 2018, 23, 87–92 CrossRef PubMed.
- M. Lekka, Bionanoscience, 2016, 6, 65–80 CrossRef PubMed.
- S. Sharma, C. Santiskulvong, L. A. Bentolila, J. Rao, O. Dorigo and J. K. Gimzewski, Nanomedicine, 2012, 8, 757–766 CrossRef CAS PubMed.
- Y. H. Seo, Y. Jo, Y. J. Oh and S. Park, Biol. Pharm. Bull., 2015, 38, 389–395 CrossRef CAS PubMed.
- H. Yin, H. Zhang, Y. Kong, C. Wang, Y. Guo, Y. Gao, L. Yuan, X. Yang and J. Chen, Apelin protects auditory cells from cisplatin-induced toxicity in vitro by inhibiting ROS and apoptosis, Elsevier Ireland Ltd, 2020, vol. 728 Search PubMed.
- K. Porwal, S. Pal, K. Dev, S. P. China, Y. Kumar, C. Singh, T. Barbhuyan, N. Sinha, S. Sanyal, A. K. Trivedi, R. Maurya and N. Chattopadhyay, J. Nutr. Biochem., 2017, 44, 22–34 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc04030h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |