Open Access Article
Open Access ArticleCoumarin luciferins and mutant luciferases for robust multi-component bioluminescence imaging†
Zi
Yao‡
a,
Donald R.
Caldwell‡
d,
Anna C.
Love‡
a,
Bethany
Kolbaba-Kartchner
ef,
Jeremy H.
Mills
 ef,
Martin J.
Schnermann
ef,
Martin J.
Schnermann
 *d and
Jennifer A.
Prescher
*d and
Jennifer A.
Prescher
 *abc
*abc
aDepartment of Chemistry, University of California, Irvine, CA, USA. E-mail: jpresche@uci.edu
bDepartment of Molecular Biology & Biochemistry, University of California, Irvine, CA, USA
cDepartment of Pharmaceutical Sciences, University of California, Irvine, CA, USA
dChemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD, USA. E-mail: martin.schnermann@nih.gov
eSchool of Molecular Sciences, Arizona State University, Tempe, AZ, USA
fThe Biodesign Center for Molecular Design and Biomimetics, Arizona State University, Tempe, AZ, USA
First published on 2nd August 2021
Abstract
Multi-component bioluminescence imaging requires an expanded collection of luciferase–luciferin pairs that emit far-red or near-infrared light. Toward this end, we prepared a new class of luciferins based on a red-shifted coumarin scaffold. These probes (CouLuc-1s) were accessed in a two-step sequence via direct modification of commercial dyes. The bioluminescent properties of the CouLuc-1 analogs were also characterized, and complementary luciferase enzymes were identified using a two-pronged screening strategy. The optimized enzyme-substrate pairs displayed robust photon outputs and emitted a significant portion of near-infrared light. The CouLuc-1 scaffolds are also structurally distinct from existing probes, enabling rapid multi-component imaging. Collectively, this work provides novel bioluminescent tools along with a blueprint for crafting additional fluorophore-derived probes for multiplexed imaging.
Introduction
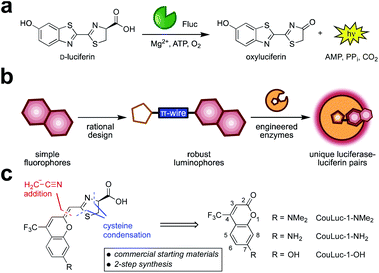
Bioluminescent enzymes (luciferases) are among the most popular reporters for imaging biological processes in vitro, in live cells, and in animal models.1,2 Luciferases generate light via the oxidation of small molecule luciferins (Fig. 1a). Since no external light source is needed, bioluminescent probes offer high signal-to-noise ratios in heterogeneous environments.3–5 The remarkable sensitivity, combined with the broad dynamic range, has made bioluminescence a go-to imaging technique for tracking cell movements, proliferation, and numerous other features in living organisms.6–8While widely used, bioluminescence has been slow to transition to imaging multiple targets simultaneously, owing to a lack of distinguishable probes.9,10 Multiplexed imaging is possible using spectrally resolved luciferase–luciferin pairs.11–14 Indeed, several luciferin analogs have been developed that emit different colors of light.15–17 The emission profiles can be further tuned with engineered luciferases18,19 or luciferase–fluorescent probe fusions.20–23 In many cases, though, the broad spectral profiles and overlapping emission maxima preclude routine multiplexing in vivo. Sensitive imaging in whole organisms further requires >650 nm light, as these wavelengths are significantly less absorbed by tissue.24 However, few bioluminescent probes emit substantial numbers of photons in the requisite far-red to near-infrared (NIR) range. The perceived color of emission in traditional bioluminescence detection also changes with the depth of the source, complicating the assignment of different colored probes.25
Multiplexed bioluminescence imaging can also be achieved via substrate resolution—using luciferases that recognize different luciferin structures (i.e., orthogonal pairs).10 Light is produced when complementary enzymes and substrates react, but minimized in all other cases.26–28 A handful of such orthogonal probes have been co-opted for dual imaging in vivo, but applications in deep tissue remain challenging.11,29–31 This is due to a shortage of luciferins with both sufficient red emission and distinct molecular architectures. Candidate luciferins must also be sufficiently bright, bioavailable, and easy to synthesize—criteria that few existing probes meet.32 Consequently, a set of three or more red-shifted probes for routine multicomponent imaging remains elusive.
We surmised that unique classes of orthogonal, NIR-emitting luciferins could be developed using fluorescent scaffolds as guides (Fig. 1b). Our approach was inspired by previous reports of luciferin analogs comprising entirely new heterocycles,33,34 including benzothiophene,35 quinoline,36 and coumarin derivatives.37 We also took cues from recent efforts to develop red-shifted luciferins by extending the pi-conjugation38–40 of the scaffold and restricting conformational flexibility (e.g., AkaLumine and infraluciferin).41–43 Luciferins produced from these seminal studies emitted far-red light, but many were poor substrates for firefly luciferase (Fluc). In some cases, light emission was recouped, although extensive enzyme engineering was required (e.g., generating Akaluc and related mutants).38,43,44
We focused on a new class of luciferins (CouLuc-1s) comprising both an elongated pi-system and a 4-trifluoromethylcoumarin unit (Fig. 1c). The coumarin fluorophore is a well-established imaging agent, and structurally distinct from heterocycles found in existing red-shifted luciferins.45,46 Additionally, coumarins have been incorporated into other motifs to achieve bright NIR emission.47–51 The small size of the coumarin core would also likely require only minimal enzyme engineering to identify complementary luciferases. Given the unique structure of the CouLuc-1s, we further anticipated that the analogs could be used for multi-component imaging with other red-emitting probes, including AkaLumine/Akaluc and furimazine/Antares.
Here we detail the synthesis and evaluation of the CouLuc-1 probes. We developed a two-step route to bridge the fluorescent coumarin heterocycle with the key thiazoline unit necessary for luciferin bioluminescence. The resulting conjugates displayed red-shifted emission. Complementary luciferases were identified via a parallel engineering approach. The resulting luciferase–luciferin pairs provided robust light outputs that were suitable for multiplexed imaging. Overall, these efforts provide a new class of easily accessible, long-wavelength bioluminescent pairs with significant promise for orthogonal imaging in vivo.
Results and discussion
Design and synthesis of coumarin-linked luciferins
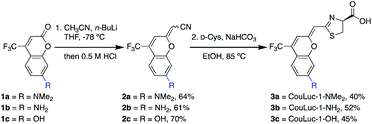
We set out to prepare CouLuc-1 analogs bearing different electron donors (–NMe2, –NH2, and OH) at C7 of the coumarin heterocycle. These modifications are commonly found in coumarin fluorophores and are known to modulate the excited state properties.45 A C4 trifluoromethyl group was also included, as this motif is known to both red-shift the emission and increase the fluorescence quantum yield of the parent coumarin.52Retrosynthetic analysis of the CouLuc-1 analogs revealed a key disconnection in the alkene linkage between the coumarin and thiazoline (Fig. 1c). We envisioned that if the olefin bridge could be installed with a nitrile handle, subsequent cysteine condensation would afford the desired luciferins in a highly concise two-step sequence. Syntheses of related pi-extended or red-shifted luciferins typically require 5–10 steps.28,41,43,53,54 While methods to directly modify the carbonyl group on coumarins are rare, recent reports suggested that thiolactones are viable intermediates for synthesizing pi-extended coumarins.47,48,55 Additionally, direct thiazoline formation via cysteine condensation has traditionally been limited to benzothiazole or otherwise activated nitriles; non-activated scaffolds require more steps.15,40
With these synthetic challenges in mind, we set out to develop olefination conditions for installing a cyanomethylene moiety onto commercial coumarin starting materials. After examining several strategies, we found that cyanomethyl anions generated in situ from acetonitrile and n-BuLi react readily with coumarins 1a–c.56 Subsequent treatment of the addition products with 0.5 M HCl afforded the desired cyanomethylene coumarins 2a–c as mixtures of E/Z isomers in good yield (Scheme 1). Exposing 2a–c (as a mixture of isomers) to D-cysteine and NaHCO3 in ethanol generated CouLuc-1 analogs 3a–c in 3–5 days. Following cycloaddition, a single isomer was formed, and the geometry was verified by 2D NOSEY (Fig. S1†). Overall, the route provided access to 3a–c in 26–32% yield in just two steps. This approach is among the shortest luciferin syntheses to date from readily available starting materials. To highlight the scalability of the route, we also developed a chromatography-free procedure to access 3a (Fig. S2 and S3†). Quick access to large quantities of luciferin is necessary for efficient identification of complementary luciferases.
In vitro characterization with native luciferase
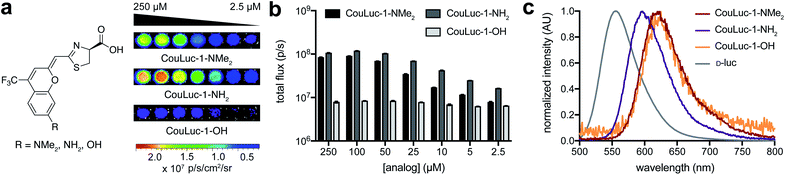
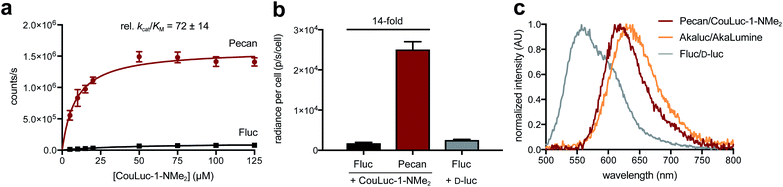
With the CouLuc-1 analogs in hand, we first examined their bioluminescent properties. All three analogs produced light when combined with Fluc and the necessary cofactors (Fig. 2a). Scaffolds with amino (CouLuc-1-NH2) or dimethylamino (CouLuc-1-NMe2) substituents exhibited stronger photon outputs than the hydroxy variant (CouLuc-1-OH, Fig. 2b). Similar boosts in brightness have been observed with AkaLumine and related luciferins.15,42 Compared to the native Fluc substrate (D-luc), though, the CouLuc-1 derivatives produced lower levels of light (∼1000-fold dimmer, Fig. S4†). The reduced brightness was partly attributed to low binding affinities between Fluc and the coumarin analogs (Fig. S5†). Similar trends have been observed with other sterically26,57 and electronically modified luciferins33,34 exhibiting comparable emission values (Fig. S6†). While low, the photon outputs achieved with Fluc and the CouLuc-1 derivatives provided a clear starting point for evolving brighter luciferases.In terms of color, CouLuc-1-NMe2 and –OH exhibited peak emission wavelengths (λmax) at 620 and 625 nm, respectively. CouLuc-1-NH2 was slightly blue-shifted, with λmax = 597 nm. Compared to D-luc, all analogs were red-shifted by 30–60 nm at 25 °C (Fig. 2c). Nearly 40% of the emitted photons from CouLuc-1-NMe2 and CouLuc-1-OH were >650 nm. For comparison, Fluc/D-luc emits only 5% of photons >650 nm in vitro (8% in cellulo58). For Akaluc/AkaLumine, a popular red-shifted luciferase–luciferin pair, the value is 44% (in cellulo at 25 °C). We further compared the CouLuc-1 bioluminescence spectra to the corresponding fluorescence spectra. In aqueous media, the fluorescence readouts exhibited similar trends, with the observed maxima red-shifted by 70–100 nm compared to D-luc (Table S1†). This strong agreement between CouLuc-1 fluorescence and bioluminescence is consistent with other pi-extended and amino luciferins.28,38,59
Engineering complementary luciferases for CouLuc-1s
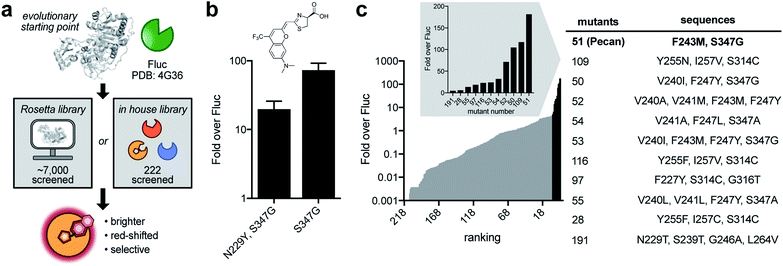
After establishing the CouLuc-1s as viable luminophores, we set out to improve photon outputs by engineering the luciferase enzyme (Fig. 3a). The ideal mutants would exhibit improved turnover, maintain red-shifted emission, and be selective for the coumarin scaffolds. We initially focused on CouLuc-1-NMe2 due to its robust activity with Fluc and desirable spectral properties. We used an established two-pronged approach to identifying mutants from library screens via (1) in silico design and (2) a semi-rational strategy.38 Both methods have been used to generate complementary enzymes for synthetic luciferins.26,38,44 In the first approach, we sculpted the luciferase active site using RosettaDesign. This strategy is useful for identifying mutations unique to an analog without significant prior knowledge or evolutionary starting point.60,61 To computationally identify mutations suited for the CouLuc-1 scaffold, we used the Rosetta-Match algorithm62,63 to dock CouLuc-1-NMe2 with existing Fluc crystal structures.64 The model oriented the coumarin heterocycle toward the pocket normally adjacent to C4′ on D-luc. In this configuration, the dimethylamino substituent was predicted to clash with the backbone of some active site residues (Fig. S7†), likely resulting in diminished turnover. We next employed RosettaDesign62,63 to resolve this clash and optimize the packing interaction between the coumarin luciferin and surrounding residues. From this analysis, a total of 41 sites were mutated to create a complementary active site for CouLuc-1-NMe2 (Fig. S8†). These residues were then ranked for targeting based on their proximity and known biochemical data.27,65–67 Twenty sites were ultimately selected for randomization via combinatorial codon mutagenesis (Fig. S8†).The resulting Rosetta-inspired library was introduced into bacteria, and the transformed colonies were sprayed with CouLuc-1-NMe2. Out of ∼7000 colonies screened on plate, ∼150 were light-emitting. These colonies were collected, and the mutants were verified in two secondary screens (Fig. S9†). Variants with >10-fold improved photon output over Fluc were carried forward. Unique sequences were then validated in a second bacterial cell assay. From this workflow, two hits were identified (Fig. 3b). Intriguingly, both variants contained a S347G mutation. We also screened the Rosetta-based library with CouLuc-1-OH. In this case, three hits were identified, with the point mutant S347G providing the highest photon outputs (Fig. S10†). While only a subset of the Rosetta library was screened, the frequency of the S347G mutation among the ‘hits’ suggested that this residue is beneficial for CouLuc-1 processing. S347G is also known to stabilize the open conformation of the luciferase active site68,69 and process luciferins with steric bulk at C4′.26,29
In parallel with the Rosetta approach, we screened CouLuc-1-NMe2 against a focused library of 222 characterized Fluc mutants.29 The luciferases comprise mutations confined to the luciferin binding pocket and exhibit unique preferences for sterically and electronically modified luciferins.26,38 Screens of this library could provide additional information on residues underlying substrate specificity for the CouLuc-1 scaffolds. All three analogs were subjected to the focused library. Eleven hits were identified for CouLuc-1-NMe2 (Fig. 3c). A similar number of mutants were found to exhibit enhanced light emission for the –NH2 and –OH analogs (Fig. S11†). The brightest mutants from these screens also comprised the S347G mutation, reinforcing the notion that residue 347 plays a pivotal role in CouLuc-1 processing. Another mutation common to multiple hits was F243M, a residue previously shown to aid in processing bulky luciferin analogs.29,57
In vitro and in cellulo characterization of lead mutant
From the screening hits, we selected the F243M/S347G mutant (dubbed Pecan) for additional characterization with CouLuc-1-NMe2. Pecan was particularly attractive for orthogonal probe development as it has been previously used in vivo.29,31 When CouLuc-1-NMe2 was incubated with purified Pecan, more intense light emission (77-fold) was observed compared to Fluc (Fig. S12†). The boost in light output was likely due to the enhanced binding affinity of CouLuc-1s with Pecan, as revealed by kinetic analyses (Fig. 4a and S13†). Importantly, robust emission in the NIR region was maintained. Approximately 30% of photons produced by CouLuc-1-NMe2 were >650 nm, and the emission spectra for the other analogs were similarly red-shifted (Fig. S14†).After examining Pecan/CouLuc-1-NMe2in vitro, we evaluated the pair in mammalian cells. Pecan and Fluc were transiently expressed in HEK293 cells. The cells were incubated with either the CouLuc-1 analogs or D-luc. Peak photon outputs for each enzyme-substrate combination were measured and normalized to a common transfection marker (GFP). As shown in Fig. 4b and S15†, Pecan-expressing cells treated with CouLuc-1-NMe2 emitted 14-fold more photons than Fluc-expressing cells. Similar improvements were observed when Pecan-expressing cells were incubated with either CouLuc-1-NH2 or CouLuc-1-OH (Fig. S16†). The robust emission from CouLuc-1 analogs was also recapitulated in Pecan-expressing DB7 cells (Fig. S17†). Pecan/CouLuc-1-NMe2 provided larger photon outputs than native Fluc/D-luc under standard cellular imaging conditions (Fig. 4b and S15†). Achieving such bright emission with a minimally modified luciferase is notable. Only two mutations to the Fluc scaffold were necessary. In our previous work to identify a complementary luciferase for a pi-extended luciferin, four rounds of engineering were required. The “winning” enzymes comprised 5–7 mutations and provided photon outputs that were only ∼2% of Fluc/D-luc emission.38 Pecan/CouLuc-1-NMe2 also compares favorably to Akaluc/AkaLumine, a popular red-shifted probe, in terms of photon output and color (Fig. 4c and S18†). Akaluc comprises 28 mutations and was identified after 21 rounds of evolution.44 Pecan/CouLuc-1-NMe2 also emits significant numbers of NIR photons (32% of photons >650 nm) in cells, on par with other state-of-the-art bioluminescent tools (Fig. S19†). Collectively, these data suggest that the Pecan/CouLuc-1-NMe2 is immediately useful for routine imaging.
Multicellular imaging with pecan and CouLuc-1-NMe2
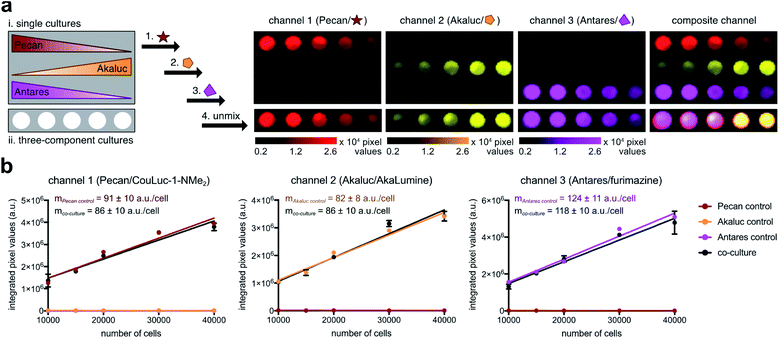
The optical properties of the CouLuc-1 analogs coupled with their unique structures made them promising candidates for multiplexed imaging. As noted earlier, resolving luciferases by substrate requires unique luciferin architectures. The more structurally diverse the luciferins, the better they can be distinguished by engineered enzymes.29 Substrate unmixing and image processing algorithms can rapidly detect unique enzyme-substrate pairings within complex mixtures.31,70 Multiple classes of orthogonal probes are available for multicomponent imaging via these methods, but only a few exhibit the necessary optical properties (NIR bioluminescence) for sensitive imaging.Given the unique structures and robust emission of Pecan/CouLuc-1-NMe2, we reasoned that this pair would be useful for multiplexing with other red-shifted probes. We were drawn to Akaluc/AkaLumine44 and Antares/furimazine71 as these pairs have been recently used to monitor tumor-immune interactions in vivo.30 Pecan and Akaluc derive from the insect luciferase family and are thus inherently orthogonal to Antares, which uses a different mechanism for light emission. The CouLuc-1 scaffold is also structurally distinct from AkaLumine, suggesting that Pecan and Akaluc could be readily differentiated based on their substrate preference. Indeed, minimal crosstalk was observed when Pecan was treated with AkaLumine or when the CouLuc-1 luciferin was added to Akaluc (Fig. S20†). The high level of orthogonality highlights the unique chemical space occupied by the coumarin analogs.
To test the multiplexed strategy, we plated Pecan-, Akaluc-, and Antares-expressing DB7 cells in varying ratios. Cell cultures containing a single reporter were also plated as controls (Fig. 5a). Each substrate was added sequentially, and an image was acquired after each administration. The resulting images were analyzed using a linear unmixing algorithm,31,72 and the relative abundance of each cell type was false colored in a composite image (Fig. 5a). Each luciferase-expressing cell was readily discerned, and the unmixed signals correlated with the number of cells present (Fig. 5b). The entire triple component imaging study was completed within 30 min, a notable improvement over traditional methods that require substrate clearance (multiple days). Altogether, these results suggest that Pecan/CouLuc-1-NMe2 can be readily integrated with other engineered probes for rapid, multicellular imaging. Pecan, Akaluc, and Antares also constitute the first readily distinguishable, bioluminescent triplet probe set with NIR emission.
Conclusions
Despite the growing number of luciferin analogs, only a few are both chemically distinct and red-shifted. We have developed and characterized a new panel of structurally unique and near infrared-emitting luciferins based on a modified coumarin scaffold. The substrates were synthesized in just two steps without chromatographic purification. The CouLuc-1 analogs were found to emit light with Fluc. While the emission levels were weak compared to existing bioluminescent systems, mutant luciferases were identified that afforded enhanced outputs. The brightest luciferase-CouLuc-1 pair exhibited luminescent signals outperforming native bioluminescent probes. Such robust emission suggests that the CouLuc-1 luciferins can be immediately adopted for biological imaging.The unique structural and optical features of the CouLuc-1 analogs are well suited for multiplexed imaging. We demonstrated that the probes and their complementary enzymes could be used for rapid imaging of multiple targets. We also identified an easily distinguishable triplet set of NIR bioluminescent tools. Combinations of such red-emitting probes are necessary for applications in tissue and other scattering environments. More broadly, our approach to accessing novel luminophores from simple fluorophores could spur the development of an expanded set of bioluminescent tools. Future work will also investigate whether more fluorophore–luminophore hybrids can be accessed using the olefination strategy.
Author contributions
Z. Y., D. R. C., M. J. S. and J. A. P. conceived the project idea. Z. Y., D. R. C., A. C. L., B. K.-K. performed the experiments. D. R. C synthesized and characterized the luciferin analogs. B. K.-K. and J. H. M. performed Rosetta analyses. Z. Y. and A. C. L. performed the luciferase engineering and characterization experiments, along with the imaging studies. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
Z. Y., D. R. C., A. C. L., M. J. S., and J. A. P. are on a provisional patent application, filed through the NCI and UCI based on the results described here.Acknowledgements
This work was supported by the U.S. National Institutes of Health (R01 GM107630 to J. A. P.) and the Intramural Research Program of the National Institutes of Health (NIH), NCI-CCR. Z. Y. was supported by the National Science Foundation via the BEST IGERT (DGE-1144901) program and a Graduate Research Fellowship (DGE-1321846). A. L. was supported by the National Science Foundation via the BEST IGERT (DGE-1144901). We thank Dr. Joseph Barchi, NCI Center for Cancer Research (NCI-CCR) for NMR assistance and Dr. James Kelley, NCI-CCR, for mass spectrometry analysis. Some experiments were performed at the Laser Spectroscopy labs (LSL) at UCI. We thank Caroline Brennan for assisting with image processing, Anastasia Ionkina for cell line generation, along with other members of the Prescher lab for helpful discussions. We also thank members of the Weiss and Martin laboratories (UCI) for providing equipment and reagents.References
- M. A. Paley and J. A. Prescher, MedChemComm, 2014, 5, 255–267 RSC.
- L. Mezzanotte, M. van 't Root, H. Karatas, E. A. Goun and C. W. G. M. Löwik, Trends Biotechnol., 2017, 35, 640–652 CrossRef CAS PubMed.
- J. A. Prescher and C. H. Contag, Curr. Opin. Chem. Biol., 2010, 14, 80–89 CrossRef CAS PubMed.
- N. Thorne, J. Inglese and D. S. Auld, Chem. Biol., 2010, 17, 646–657 CrossRef CAS PubMed.
- X. Ji, S. T. Adams Jr and S. C. Miller, in Methods Enzymol., ed. D. M. Chenoweth, Academic Press, 2020, vol. 640, pp. 165–183 Search PubMed.
- H.-W. Yeh and H.-W. Ai, Annu. Rev. Anal. Chem., 2019, 12, 129–150 CrossRef CAS.
- A. C. Love and J. A. Prescher, Cell Chem. Biol., 2020, 27, 904–920 CrossRef CAS PubMed.
- Z. Yao, B. S. Zhang and J. A. Prescher, Curr. Opin. Chem. Biol., 2018, 45, 148–156 CrossRef CAS PubMed.
- C. M. Rathbun and J. A. Prescher, Biochemistry, 2017, 56, 5178–5184 CrossRef CAS PubMed.
- S. J. Williams and J. A. Prescher, Acc. Chem. Res., 2019, 52, 3039–3050 CrossRef CAS PubMed.
- G. Zambito, M. P. Hall, M. G. Wood, N. Gaspar, Y. Ridwan, F. F. Stellari, C. Shi, T. A. Kirkland, L. P. Encell, C. Löwik and L. Mezzanotte, iScience, 2021, 24, 101986 CrossRef CAS PubMed.
- M. Aswendt, S. Vogel, C. Schäfer, A. Jathoul, M. Pule and M. Hoehn, Neurophotonics, 2019, 6, 025006 CAS.
- L. Mezzanotte, I. Que, E. Kaijzel, B. Branchini, A. Roda and C. Löwik, PLoS One, 2011, 6, e19277 CrossRef CAS PubMed.
- C. L. Stowe, T. A. Burley, H. Allan, M. Vinci, G. Kramer-Marek, D. M. Ciobota, G. N. Parkinson, T. L. Southworth, G. Agliardi, A. Hotblack, M. F. Lythgoe, B. R. Branchini, T. L. Kalber, J. C. Anderson and M. A. Pule, eLife, 2019, 8, e45801 CrossRef PubMed.
- S. Iwano, R. Obata, C. Miura, M. Kiyama, K. Hama, M. Nakamura, Y. Amano, S. Kojima, T. Hirano, S. Maki and H. Niwa, Tetrahedron, 2013, 69, 3847–3856 CrossRef CAS.
- H.-w. Yeh, Y. Xiong, T. Wu, M. Chen, A. Ji, X. Li and H.-W. Ai, ACS Chem. Biol., 2019, 14, 959–965 CrossRef CAS PubMed.
- D. K. Sharma, S. T. Adams, K. L. Liebmann, A. Choi and S. C. Miller, Org. Lett., 2019, 21, 1641–1644 CrossRef CAS PubMed.
- B. R. Branchini, D. M. Ablamsky, J. M. Rosenman, L. Uzasci, T. L. Southworth and M. Zimmer, Biochemistry, 2007, 46, 13847–13855 CrossRef CAS PubMed.
- B. R. Branchini, D. M. Ablamsky, M. H. Murtiashaw, L. Uzasci, H. Fraga and T. L. Southworth, Anal. Biochem., 2007, 361, 253–262 CrossRef CAS PubMed.
- K. Suzuki, T. Kimura, H. Shinoda, G. Bai, M. J. Daniels, Y. Arai, M. Nakano and T. Nagai, Nat. Commun., 2016, 7, 13718 CrossRef CAS PubMed.
- A. Takai, M. Nakano, K. Saito, R. Haruno, T. M. Watanabe, T. Ohyanagi, T. Jin, Y. Okada and T. Nagai, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 4352–4356 CrossRef CAS PubMed.
- F. X. Schaub, M. S. Reza, C. A. Flaveny, W. Li, A. M. Musicant, S. Hoxha, M. Guo, J. L. Cleveland and A. L. Amelio, Cancer Res., 2015, 75, 5023–5033 CrossRef CAS PubMed.
- J. Hiblot, Q. Yu, M. D. B. Sabbadini, L. Reymond, L. Xue, A. Schena, O. Sallin, N. Hill, R. Griss and K. Johnsson, Angew. Chem., Int. Ed., 2017, 56, 14556–14560 CrossRef CAS PubMed.
- B. W. Rice and C. H. Contag, Nat. Biotechnol., 2009, 27, 624–625 CrossRef CAS PubMed.
- R. Weissleder and V. Ntziachristos, Nat. Med., 2003, 9, 123–128 CrossRef CAS PubMed.
- K. A. Jones, W. B. Porterfield, C. M. Rathbun, D. C. McCutcheon, M. A. Paley and J. A. Prescher, J. Am. Chem. Soc., 2017, 139, 2351–2358 CrossRef CAS PubMed.
- S. T. Adams, D. M. Mofford, G. S. K. K. Reddy and S. C. Miller, Angew. Chem., Int. Ed., 2016, 55, 4943–4946 CrossRef CAS PubMed.
- D. M. Mofford, G. R. Reddy and S. C. Miller, J. Am. Chem. Soc., 2014, 136, 13277–13282 CrossRef CAS PubMed.
- C. M. Rathbun, W. B. Porterfield, K. A. Jones, M. J. Sagoe, M. R. Reyes, C. T. Hua and J. A. Prescher, ACS Cent. Sci., 2017, 3, 1254–1261 CrossRef CAS PubMed.
- Y. Su, J. R. Walker, Y. Park, T. P. Smith, L. X. Liu, M. P. Hall, L. Labanieh, R. Hurst, D. C. Wang, L. P. Encell, N. Kim, F. Zhang, M. A. Kay, K. M. Casey, R. G. Majzner, J. R. Cochran, C. L. Mackall, T. A. Kirkland and M. Z. Lin, Nat. Methods, 2020, 17, 852–860 CrossRef CAS PubMed.
- C. M. Rathbun, A. A. Ionkina, Z. Yao, K. A. Jones, W. B. Porterfield and J. A. Prescher, ACS Chem. Biol., 2021, 16, 682–690 CrossRef CAS PubMed.
- G. Zambito, N. Gaspar, Y. Ridwan, M. P. Hall, C. Shi, T. A. Kirkland, L. P. Encell, C. Löwik and L. Mezzanotte, Mol. Imaging Biol., 2020, 22, 1523–1531 CrossRef CAS PubMed.
- D. C. McCutcheon, M. A. Paley, R. C. Steinhardt and J. A. Prescher, J. Am. Chem. Soc., 2012, 134, 7604–7607 CrossRef CAS PubMed.
- B. S. Zhang, K. A. Jones, D. C. McCutcheon and J. A. Prescher, ChemBioChem, 2018, 19, 470–477 CrossRef CAS PubMed.
- C. C. Woodroofe, P. L. Meisenheimer, D. H. Klaubert, Y. Kovic, J. C. Rosenberg, C. E. Behney, T. L. Southworth and B. R. Branchini, Biochemistry, 2012, 51, 9807–9813 CrossRef CAS PubMed.
- B. R. Branchini, in Methods Enzymol., Academic Press, 2000, vol. 305, pp. 188–195 Search PubMed.
- H. Takakura, K. Sasakura, T. Ueno, Y. Urano, T. Terai, K. Hanaoka, T. Tsuboi and T. Nagano, Chem.–Asian J., 2010, 5, 2053–2061 CrossRef CAS PubMed.
- Z. Yao, B. S. Zhang, R. C. Steinhardt, J. H. Mills and J. A. Prescher, J. Am. Chem. Soc., 2020, 142, 14080–14089 CrossRef CAS PubMed.
- N. Kitada, T. Saitoh, Y. Ikeda, S. Iwano, R. Obata, H. Niwa, T. Hirano, A. Miyawaki, K. Suzuki, S. Nishiyama and S. A. Maki, Tetrahedron Lett., 2018, 59, 1087–1090 CrossRef CAS.
- A. P. Jathoul, H. Grounds, J. C. Anderson and M. A. Pule, Angew. Chem., Int. Ed., 2014, 53, 13059–13063 CrossRef CAS PubMed.
- Y. Ikeda, T. Nomoto, Y. Hiruta, N. Nishiyama and D. Citterio, Anal. Chem., 2020, 92, 4235–4243 CrossRef CAS PubMed.
- G. R. Reddy, W. C. Thompson and S. C. Miller, J. Am. Chem. Soc., 2010, 132, 13586–13587 CrossRef CAS PubMed.
- J. C. Anderson, C.-H. Chang, A. P. Jathoul and A. J. Syed, Tetrahedron, 2019, 75, 347–356 CrossRef CAS.
- S. Iwano, M. Sugiyama, H. Hama, A. Watakabe, N. Hasegawa, T. Kuchimaru, K. Z. Tanaka, M. Takahashi, Y. Ishida, J. Hata, S. Shimozono, K. Namiki, T. Fukano, M. Kiyama, H. Okano, S. Kizaka-Kondoh, T. J. McHugh, T. Yamamori, H. Hioki, S. Maki and A. Miyawaki, Science, 2018, 359, 935–939 CrossRef CAS PubMed.
- D. Cao, Z. Liu, P. Verwilst, S. Koo, P. Jangjili, J. S. Kim and W. Lin, Chem. Rev., 2019, 119, 10403–10519 CrossRef CAS.
- M. V. Sednev, V. N. Belov and S. W. Hell, Methods Appl. Fluoresc., 2015, 3, 042004 CrossRef PubMed.
- A. Gandioso, S. Contreras, I. Melnyk, J. Oliva, S. Nonell, D. Velasco, J. García-Amorós and V. Marchán, J. Org. Chem., 2017, 82, 5398–5408 CrossRef CAS PubMed.
- A. Gandioso, R. Bresolí-Obach, A. Nin-Hill, M. Bosch, M. Palau, A. Galindo, S. Contreras, A. Rovira, C. Rovira, S. Nonell and V. Marchán, J. Org. Chem., 2018, 83, 1185–1195 CrossRef CAS.
- S. S. Matikonda, J. Ivanic, M. Gomez, G. Hammersley and M. J. Schnermann, Chem. Sci., 2020, 11, 7302–7307 RSC.
- Z. Lei, C. Sun, P. Pei, S. Wang, D. Li, X. Zhang and F. Zhang, Angew. Chem., Int. Ed., 2019, 58, 8166–8171 CrossRef CAS PubMed.
- H. Chen, B. Dong, Y. Tang and W. Lin, Acc. Chem. Res., 2017, 50, 1410–1422 CrossRef CAS PubMed.
- W.-C. Sun, K. R. Gee and R. P. Haugland, Bioorg. Med. Chem. Lett., 1998, 8, 3107–3110 CrossRef CAS.
- T. Kuchimaru, S. Iwano, M. Kiyama, S. Mitsumata, T. Kadonosono, H. Niwa, S. Maki and S. Kizaka-Kondoh, Nat. Commun., 2016, 7, 11856 CrossRef CAS PubMed.
- M. P. Hall, C. C. Woodroofe, M. G. Wood, I. Que, M. van't Root, Y. Ridwan, C. Shi, T. A. Kirkland, L. P. Encell, K. V. Wood, C. Löwik and L. Mezzanotte, Nat. Commun., 2018, 9, 132 CrossRef PubMed.
- A. Abdel-Fattah Mostafa, C. SathishKumar, A. A. Al-Askar, S. R. M. Sayed, R. SurendraKumar and A. Idhayadhulla, RSC Adv., 2019, 9, 25533–25543 RSC.
- G. Bianchi, M. Feroci and L. Rossi, Eur. J. Org. Chem., 2009, 2009, 3863–3866 CrossRef.
- S. J. Williams, C. S. Hwang and J. A. Prescher, Biochemistry, 2021, 60, 563–572 CrossRef CAS PubMed.
- H. Zhao, T. C. Doyle, O. Coquoz, F. Kalish, B. W. Rice and C. H. Contag, J. Biomed. Opt., 2005, 10, 41210 CrossRef.
- T. Vreven and S. C. Miller, J. Comput. Chem., 2019, 40, 527–531 CrossRef CAS PubMed.
- D. S. Liu, L. G. Nivón, F. Richter, P. J. Goldman, T. J. Deerinck, J. Z. Yao, D. Richardson, W. S. Phipps, A. Z. Ye, M. H. Ellisman, C. L. Drennan, D. Baker and A. Y. Ting, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E4551–E4559 CrossRef CAS PubMed.
- A. D. Pearson, J. H. Mills, Y. Song, F. Nasertorabi, G. W. Han, D. Baker, R. C. Stevens and P. G. Schultz, Science, 2015, 347, 863–867 CrossRef CAS PubMed.
- C. E. Tinberg, S. D. Khare, J. Dou, L. Doyle, J. W. Nelson, A. Schena, W. Jankowski, C. G. Kalodimos, K. Johnsson, B. L. Stoddard and D. Baker, Nature, 2013, 501, 212–216 CrossRef CAS PubMed.
- A. Zanghellini, L. Jiang, A. M. Wollacott, G. Cheng, J. Meiler, E. A. Althoff, D. Röthlisberger and D. Baker, Protein Sci., 2006, 15, 2785–2794 CrossRef CAS.
- J. A. Sundlov, D. M. Fontaine, T. L. Southworth, B. R. Branchini and A. M. Gulick, Biochemistry, 2012, 51, 6493–6495 CrossRef CAS PubMed.
- V. R. Viviani, A. Simões, V. R. Bevilaqua, G. V. M. Gabriel, F. G. C. Arnoldi and T. Hirano, Biochemistry, 2016, 55, 4764–4776 CrossRef CAS.
- B. R. Branchini, R. A. Magyar, M. H. Murtiashaw, S. M. Anderson, L. C. Helgerson and M. Zimmer, Biochemistry, 1999, 38, 13223–13230 CrossRef CAS PubMed.
- A. K. Choudhury, S. Y. Golovine, L. M. Dedkova and S. M. Hecht, Biochemistry, 2007, 46, 4066–4076 CrossRef CAS PubMed.
- B. R. Branchini, T. L. Southworth, M. H. Murtiashaw, H. Boije and S. E. Fleet, Biochemistry, 2003, 42, 10429–10436 CrossRef CAS PubMed.
- K. R. Harwood, D. M. Mofford, G. R. Reddy and S. C. Miller, Chem. Biol., 2011, 18, 1649–1657 CrossRef CAS PubMed.
- A. Sarrion-Perdigones, L. Chang, Y. Gonzalez, T. Gallego-Flores, D. W. Young and K. J. T. Venken, Nat. Commun., 2019, 10, 5710 CrossRef CAS PubMed.
- J. Chu, Y. Oh, A. Sens, N. Ataie, H. Dana, J. J. Macklin, T. Laviv, E. S. Welf, K. M. Dean, F. Zhang, B. B. Kim, C. T. Tang, M. Hu, M. A. Baird, M. W. Davidson, M. A. Kay, R. Fiolka, R. Yasuda, D. S. Kim, H.-L. Ng and M. Z. Lin, Nat. Biotechnol., 2016, 34, 760–767 CrossRef CAS.
- S. T. Gammon, W. M. Leevy, S. Gross, G. W. Gokel and D. Piwnica-Worms, Anal. Chem., 2006, 78, 1520–1527 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc03114g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |