Open Access Article
Open Access ArticleTurning waste into wealth: facile and green synthesis of carbon nanodots from pollutants and applications to bioimaging†
Weifeng
Chen‡
a,
Jialu
Shen‡
a,
Zuo
Wang‡
b,
Xiang
Liu
 *a,
Yanyi
Xu
b,
Haiyu
Zhao
*a,
Yanyi
Xu
b,
Haiyu
Zhao
 *b and
Didier
Astruc
*b and
Didier
Astruc
 *c
*c
aCollege of Materials and Chemical Engineering, Engineering Research Center of Eco-environment in Three Gorges Reservoir Region, Ministry of Education, Key Laboratory of Inorganic Nonmetallic Crystalline and Energy Conversion Materials, China Three Gorges University, Yichang, Hubei 443002, P. R. China. E-mail: xiang.liu@ctgu.edu.cn
bGansu Key Laboratory of Biomonitoring and Bioremediation for Environmental Pollution, School of Life Sciences, Lanzhou University, Lanzhou, Gansu 730000, P. R. China. E-mail: zhaohy@lzu.edu.cn
cISM, UMR CNRS No. 5255, Univ. Bordeaux, 351 Cours de la Libération, 33405 Talence Cedex, France. E-mail: didier.astruc@u-bordeaux.fr
First published on 2nd August 2021
Abstract
In an effort to turn waste into wealth, Reactive Red 2 (RR2), a common and refractory organic pollutant in industrial wastewater, has been employed for the first time as a precursor to synthesize carbon nanodots (CNDs) by a facile, green and low-cost route, without utilization of any strong acids or other oxidizers. The detailed characterizations have confirmed that the synthesized CNDs exhibit good water dispersibility, with a mean particle size of 2.43 nm and thickness of 1–3 layers. Importantly, the excellent fluorescence properties and much reduced biotoxicity of the CNDs confer its potential applications in further biological imaging, which has been successfully verified in both in vitro (cell culture) and in vivo (zebrafish) model systems. Thus, it is demonstrated that the synthesized CNDs exhibit nice biocompatibility and fluorescence properties for bioimaging. This work not only provides a novel economical and environmentally friendly approach in recycling a chemical pollutant, but also greatly promotes the potential application of CNDs in biological imaging.
1. Introduction
Carbon nanodots (CNDs) have gained tremendous research attention worldwide due to their preeminent water solubility, highly tunable band gaps, superior photo and chemical stability, low cytotoxicity, excellent biocompatibility, easy functionalization, brilliant catalytic properties and high photoluminescence (PL) intensity.1 These nanomaterials that usually possess heavily functionalized graphene-like structures2 have been extensively applied in many areas including biomedical optical imaging, catalysis, heterojunction solar cells, drug delivery, environmental monitoring, supercapacitors and biosensing.3 The synthesis of CNDs has been typically classified into top-down and bottom-up strategies. The top-down strategies involve the synthesis of CNDs from graphite, graphene nanosheets, carbon nanotubes, candle soot, coal and graphite rods by physical or chemical cutting treatment;4 on the other hand, the bottom-up strategies target the fabrication of CNDs from conventional organic molecules or biomass by plasma treatment, ultrasonic/microwave irradiation, hydrothermal treatment or thermal decomposition.5 However, many of these methods are associated with some demerits, such as high cost, low yields, time-consuming processes, or the utilization of strong acids.6 Therefore, it is still a considerable challenge to develop more facile and green approaches for the synthesis of CNDs from abundant and low-cost precursors.The rapidly increasing usage of organic dyes (e.g., reactive red 2) in industrial manufacture, such as textile, papermaking, plastics, pigments and paint, has rendered them among the most globally hazardous pollutants of water.7 Approximately 10% of organic dyes containing aromatic groups and azo groups (–N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–) have been discharged into natural water.8 The mass discharge of these azo dyes represents a great threat for aquatic organisms and ecological environments since most of azo dyes are highly toxic, carcinogenic and teratogenetic.9 A large number of methods, such as ozonation, biological treatment, ionic exchange, photocatalysis, oxidative process and adsorption have been developed for the removal of organic dyes from water.10 However, these efforts are always associated with high chemical and energy consumption, as well as costly equipment investment. Therefore, developing more eco-friendly and efficient tactics for the removal of the organic dyes and turning these chemical pollutants into wealth is still a very meaningful and challenging research.11
N–) have been discharged into natural water.8 The mass discharge of these azo dyes represents a great threat for aquatic organisms and ecological environments since most of azo dyes are highly toxic, carcinogenic and teratogenetic.9 A large number of methods, such as ozonation, biological treatment, ionic exchange, photocatalysis, oxidative process and adsorption have been developed for the removal of organic dyes from water.10 However, these efforts are always associated with high chemical and energy consumption, as well as costly equipment investment. Therefore, developing more eco-friendly and efficient tactics for the removal of the organic dyes and turning these chemical pollutants into wealth is still a very meaningful and challenging research.11
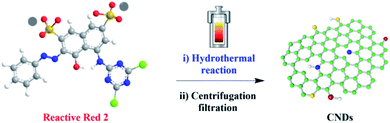
For this purpose, our laboratories have a long-term interest in the synthesis of carbon nanodots from abundant and low-cost biomass and their applications in catalysis, sensor and bioimaging.12 Herein, we report a facile, green and turning wastes into wealth route to synthesize the typical material carbon nanodots (CNDs) by using chemical pollutant reactive red 2 (RR2) as the precursor for the first time, via simple hydrothermal treatment (Fig. 1). Thereafter, the morphology structure, fluorescence properties and chemical composition of the as-synthesized CNDs have been comprehensively investigated. Importantly, this work provides some insights into the biotoxicity of the synthesized CNDs in comparison with the RR2 precursor, at the cellular and organismal level. Our results verified that the biotoxicity was dramatically reduced when the pollutant was fully converted into CNDs. In addition, the synthesized CNDs with excellent fluorescent properties have been successfully applied in the bioimaging of both living cell and the popular zebrafish model system, demonstrating practically biological applications in the future.
2. Results and discussion
2.1 Structural characterization of the CNDs
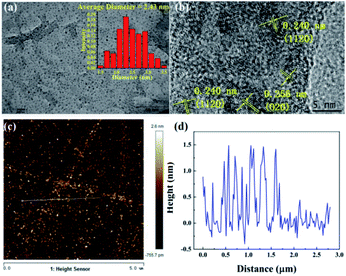
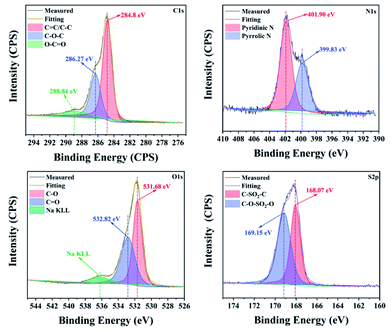
First, the CNDs were synthesized from only RR2 and water under green hydrothermal conditions. Indeed, RR2 was successfully fully converted into CNDs, because the adsorption peak at approximately 500 nm and the characteristic peaks of RR2 completely disappeared in the UV-vis spectra (Fig. 2). Meanwhile, the peaks at 234 nm, 269 nm and 358 nm (the characteristic peaks of CNDs) appeared in the UV-vis spectra. Among them, the peaks at 234 nm and 269 nm are assigned to π–π* transitions of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C–C bonds in the sp2 hybridization region, while the peak of 358 nm is due to the n–π* transition.13 The CNDs solution exhibited an intense blue fluorescence under 365 nm ultraviolet lamp (Fig. 2). The transmission electron microscope (TEM) image shows that the CNDs are well dispersed with an average diameter of 2.43 nm (Fig. 3a). Besides, the interplanar spacings of 0.240 nm and 0.256 nm were observed by high resolution TEM (HRTEM, Fig. 3b), and assigned to the (1120) and (020) plane of graphitic carbon, respectively (JCPDS 26-1076).14 The atomic force microscope (AFM) image of CNDs (Fig. 3c) indicates that the height is less than 1.5 nm (Fig. 3d), suggesting that the thickness of CNDs is 1–3 layers.15 Then, X-ray photoelectron spectroscopy (XPS) was utilized to study the valence states of the different atoms of CNDs, i.e., C, N, O, and S (Fig. 4). The spectrum of C 1s (Fig. 4a) shows the typical peaks at 284.8 eV, 286.27 eV and 288.84 eV corresponding to C
C/C–C bonds in the sp2 hybridization region, while the peak of 358 nm is due to the n–π* transition.13 The CNDs solution exhibited an intense blue fluorescence under 365 nm ultraviolet lamp (Fig. 2). The transmission electron microscope (TEM) image shows that the CNDs are well dispersed with an average diameter of 2.43 nm (Fig. 3a). Besides, the interplanar spacings of 0.240 nm and 0.256 nm were observed by high resolution TEM (HRTEM, Fig. 3b), and assigned to the (1120) and (020) plane of graphitic carbon, respectively (JCPDS 26-1076).14 The atomic force microscope (AFM) image of CNDs (Fig. 3c) indicates that the height is less than 1.5 nm (Fig. 3d), suggesting that the thickness of CNDs is 1–3 layers.15 Then, X-ray photoelectron spectroscopy (XPS) was utilized to study the valence states of the different atoms of CNDs, i.e., C, N, O, and S (Fig. 4). The spectrum of C 1s (Fig. 4a) shows the typical peaks at 284.8 eV, 286.27 eV and 288.84 eV corresponding to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C–C, C–O–C and O–C
C/C–C, C–O–C and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, respectively.16 Moreover, the XPS of N 1s (Fig. 4b) exhibited two peaks at 399.83 eV and 401.90 eV belonging to the pyrrolic N and pyridinic N, respectively. As shown in Fig. 4c, two characteristic peaks at 531.68 eV and 532.82 eV are assigned to C–O and C
O, respectively.16 Moreover, the XPS of N 1s (Fig. 4b) exhibited two peaks at 399.83 eV and 401.90 eV belonging to the pyrrolic N and pyridinic N, respectively. As shown in Fig. 4c, two characteristic peaks at 531.68 eV and 532.82 eV are assigned to C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, respectively. Besides, the XPS spectrum of S 2p (Fig. 4d) shows that the S 2p spectra is decomposed into two peaks, C–SO2–C at 168.07 eV and C–O–SO2–O at 169.15 eV, respectively.
O, respectively. Besides, the XPS spectrum of S 2p (Fig. 4d) shows that the S 2p spectra is decomposed into two peaks, C–SO2–C at 168.07 eV and C–O–SO2–O at 169.15 eV, respectively.
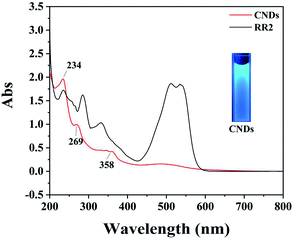 | ||
| Fig. 2 UV-vis spectra of RR2 and CNDs; inset image: CNDs solution exhibits intense blue fluorescence under 365 nm ultraviolet lamp. | ||
The structure of the produced CNDs was further investigated by Raman spectroscopy (Fig. 5a), revealing that the peaks of the D-band, G-band and 2D-band of the CNDs are located at 1300 cm−1, 1584 cm−1 and 2778 cm−1, respectively. The D-band and 2D-band are due to the disordered carbon, while the G-band is assigned to the graphite carbon. The value of ID/IG is 1.7, indicating the CNDs have some defects in the crystal lattices.17 Fourier transform infrared spectroscopy (FT-IR) was also been used to investigate the chemical groups of CNDs, as shown in Fig. 5b. The peak at 3418 cm−1 is corresponding to the stretching vibration of O–H/N–H, whereas the characteristic peaks of 3166 cm−1, 1660 cm−1, 1405 cm−1, 1055 cm−1 and 1624 cm−1 are attributed to the stretching vibrations of C–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–N and C–O, and bending vibration of N–H, respectively. Fig. 5c also exhibits a typical profile with a broad peak at approximately 24°, the characteristic peak of (002) graphene appears in the X-ray diffraction (XRD) spectrum (JCPDS card no. 75-0444). In addition, it indicates that the synthesized CNDs solution shows excitation-dependent PL property, as shown in Fig. 5d. When the excitation wavelength is set from 300 nm to 580 nm, the emission wavelength is improved from 423 nm to 619 nm. The maximum emission is at the position of 428 nm when being excited by 360 nm. In this work, rhodamine B in ethanol (QY = 69.0%) was used as a standard for CNDs. The quantum yield (QY) of CNDs was calculated to be 15.7%.18 The fluorescence of CNDs is mainly attributed to emission from oxygen-rich surface defects.19 To sum up, these results confirmed that the graphene-like structure containing sp2 hybridization of these fluorescent CNDs has been formed from RR2 by hydrothermal reaction.
O, C–N and C–O, and bending vibration of N–H, respectively. Fig. 5c also exhibits a typical profile with a broad peak at approximately 24°, the characteristic peak of (002) graphene appears in the X-ray diffraction (XRD) spectrum (JCPDS card no. 75-0444). In addition, it indicates that the synthesized CNDs solution shows excitation-dependent PL property, as shown in Fig. 5d. When the excitation wavelength is set from 300 nm to 580 nm, the emission wavelength is improved from 423 nm to 619 nm. The maximum emission is at the position of 428 nm when being excited by 360 nm. In this work, rhodamine B in ethanol (QY = 69.0%) was used as a standard for CNDs. The quantum yield (QY) of CNDs was calculated to be 15.7%.18 The fluorescence of CNDs is mainly attributed to emission from oxygen-rich surface defects.19 To sum up, these results confirmed that the graphene-like structure containing sp2 hybridization of these fluorescent CNDs has been formed from RR2 by hydrothermal reaction.
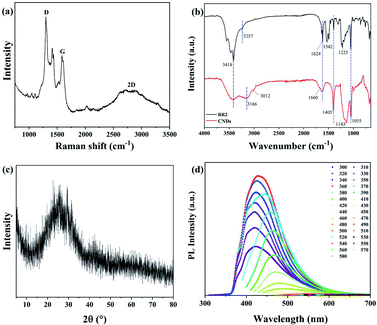 | ||
| Fig. 5 (a) Raman spectrum, (b) FT-IR spectra, (c) XRD and (d) corresponding PL spectra at different excitation wavelengths from 300 nm to 580 nm for CNDs. | ||
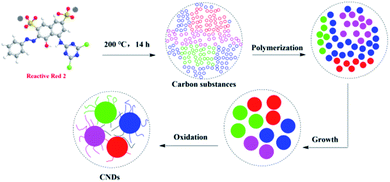
The formation mechanism of CNDs from RR2 involves two steps (Fig. 6); first RR2 was decomposed into small carbon substances under hydrothermal conditions. Then the small carbon substances were polymerized, grown and oxidized into these fluorescent CNDs with abundant surface O, N and S-containing functional groups. Finally, the fluorescence of CNDs is mainly attributed to these highly functional surface groups.19,20
2.2 Cell viability
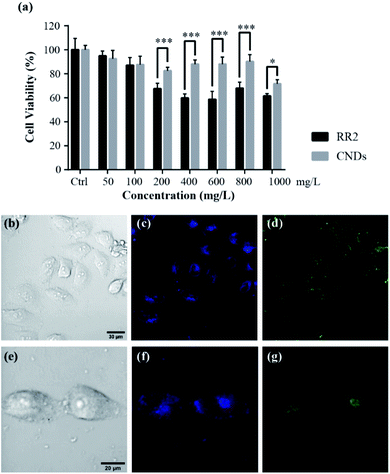
To test whether the cytotoxicity of the synthesized CNDs has been significantly reduced in comparison with the RR2, as well as its suitability for biological applications, we evaluated their cytotoxicity on HeLa cells by using a MTT assay. The HeLa cells were treated with RR2 and CNDs respectively for 24 h, and the MTT assay was performed to analyze their effect on cell viability. In comparison with the control (Ctrl) group, RR2 induced decreases in the viability of the HeLa cells in a dose-dependent manner, it reduced the cell viability by nearly 60% at the concentration of 1000 mg L−1. However, the CNDs showed significantly reduced cytotoxicity (p < 0.05 or p < 0.001). It was so especially at high doses (above 200 mg L−1), the average cell viability being greater than 70% even when they were treated with CNDs at a concentration of up to 1000 mg L−1 (Fig. 7a). This reveals the great bio-compatibility of the synthesized CNDs.2.3 Living cells fluorescence imaging
Living cell fluorescence imaging was performed to demonstrate their practicability in biological applications.21 The zeta potential value of CNDs is −24.1 mV (Fig. S2†), suggesting that the as-synthesized CNDs are relatively stable.22 After incubating the HeLa cells with CNDs (100 mg L−1) for 24 h at 37 °C, a significant fluorescence in the HeLa cells was observed (Fig. 7b–i). The CNDs were mostly localized at the cytoplasm of HeLa cells, since the cytoplasmic area exhibited a strong photoluminescence. Importantly, no photobleaching was observed within one hour for the cell imaging, suggesting that they are suitable for long-term cellular imaging. In addition, distinctive colors were available for the observation of the cells incubated with CNDs when they were excited with 405 nm and 488 nm laser beams, respectively. This intrinsic excitation separation property is particularly ideal for multichannel imaging, suggesting that CNDs are an excellent counterstain in cooperation with other dyes to obtain cellular images. This robust photostability as well as the multichannel tunability of our synthesized CNDs will certainly be beneficial for bioimaging applications.2.4 Biotoxicity assay with the zebrafish model
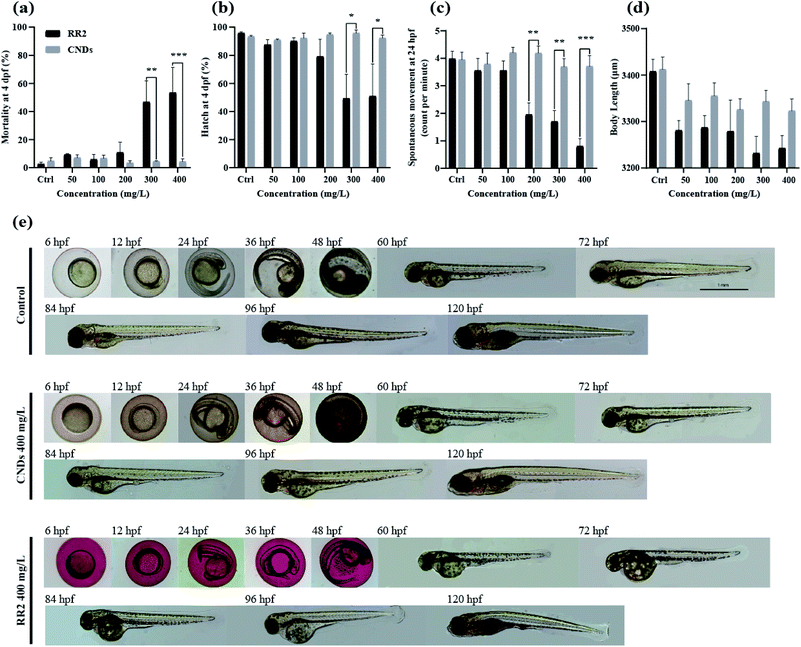
The wide range applications of carbon nanodots in biomedical studies have attracted significant attention of the potential hazards to biological systems.23 Zebrafish has been widely reported as a valuable vertebrate model and “gold standard” for assessing genotoxicity and developmental toxicity of chemicals and pollutants due to its high fecundity, cost-effectiveness, well-characterized developmental stages, and optical transparency.24 Therefore, in this study the biotoxicity of RR2 and CNDs was systematically examined by exposing zebrafish embryos at 4 hpf to different concentrations of each regents for at least 120 hours. As shown in Fig. 8a, RR2 significantly induced mortality of zebrafish embryos at higher doses (above 300 mg L−1). The hatching rate (at 4 dpf, Fig. 8b), spontaneous movement (at 24 hpf, Fig. 8c) and body length (at 4 dpf, Fig. 8d) of zebrafish embryos also declined significantly after exposure to RR2, in a dose dependent manner. However, within 120 hours, zebrafish embryos exposed to the CNDs (from 50 to 400 mg L−1) were observed to develop normally without any sign of lethality and delay of hatching, no significant difference was observed between the CNDs treatment group and control group (Fig. 8a–d, grey panels). In addition, there were no noticeable influence on the morphology of zebrafish after exposure to different concentrations of CNDs, in sharp contrast with the RR2 treatment group, in which the body length of zebrafish was significantly reduced, and the yolk sac edema was obvious (Fig. 8e). All these notable morphological differences implied that in comparison with the RR2, the biotoxicity of CNDs was significantly reduced, and the CNDs (up to 400 mg L−1) had no potential toxicity hazard to zebrafish development. CNDs have been reported to be unable to infiltrate the cell nucleus, which suggested that they did not disrupt the genetic structure of the organisms and caused cellular toxicity. This supports the hypotoxicity of our CNDs and its bio-compatibility for further utilization.2.5 Neurobehavioral alteration induced by RR2 and CNDs
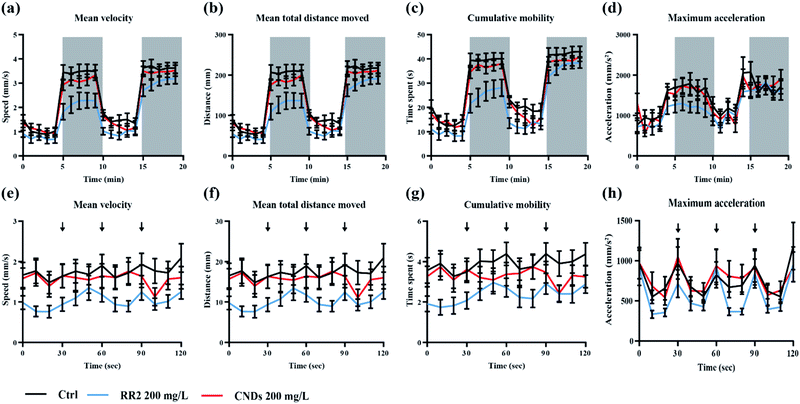
To further evaluate the developmental toxicity of RR2 and our synthesized CNDs, the neurobehavior of the zebrafish larvae exposed chronically was tested.25 Specifically, we detected their locomotor activity as well as the response to environmental signals, for example light and vibration stimulations. The swimming speed, distance moved, cumulative mobility as well as maximum acceleration of zebrafish larvae was quantified in different assays. In our results, the developmental exposure to RR2 (200 mg L−1) caused behavioral impairment including hypoactivity in locomotor activity as well as reduced reactivity to light–dark cycles. The swimming behaviors was significantly repressed by RR2 exposure especially during the dark period (Fig. 9a–d, black and blue traces, p < 0.05). Similarly, in the experiment testing the zebrafish response to vibration stimulations, a significant hypoactivity in swimming activity was observed in the RR2 exposure group during the whole process (Fig. 8e–h, black and blue traces, p < 0.05). However, in contrast to the RR2 that dramatically decreased the activity as well as the reactivity, no significant difference was observed between the Ctrl group and the CNDs exposure group (Fig. 9a–h, black and red traces, p > 0.05). These results are consistent with the previous morphological observations showing the hypotoxicity of these synthesized CNDs, suggesting that they significantly elevated bio-compatibility (Table 1).| Toxicological criteria | Timepoints | Exposure concentrations (mg L−1) | Results |
|---|---|---|---|
| Survival rates (mortality) | 4 hpf to 5 dpf | 0, 50, 100, 200, 300, 400 | Fig. 8a |
| Hatching rates | 2 dpf to 5 dpf | 0, 50, 100, 200, 300, 400 | Fig. 8b |
| Spontaneous movement | 24 hpf | 0, 50, 100, 200, 300, 400 | Fig. 8c |
| Body length | 96 hpf | 0, 50, 100, 200, 300, 400 | Fig. 8d |
| Morphological feature | 6 hpf to 120 hpf | 400 | Fig. 8e |
| Behavioral test | 24 hpf, 6 dpf | 0, 50, 100, 200, 300, 400 | Fig. 9 |
2.6 Fluorescence imaging of CNDs in zebrafish model
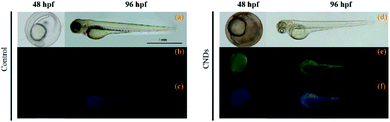
The rapid development and transparency of zebrafish embryos facilitate the application of CNDs in the biological imaging.26 The imaging was implemented by acute exposing zebrafish embryos in CNDs solution (400 mg L−1). Multicolor fluorescence imaging of zebrafish at different developmental stages (48 hpf and 96 hpf) was shown in Fig. 10. The small size of CNDs (2.4 nm), much smaller than the nanoscale pores of the zebrafish chorion (approximately 0.17 μm2), enables their rapid entrance into the pores of the chorion upon simple immersion. In addition, the zebrafish embryo or larvae exhibited green and blue fluorescence under different excitation fields, and the multicolor fluorescent CNDs enabled the in vivo observation of their distribution in zebrafish embryos and larvae. This phenomenon indicates that the CNDs present high biocompatibility during zebrafish development and might be used as an excellent candidate for in vivo fluorescence imaging in biological studies.3. Conclusions
In summary, a facile, green and low-cost synthesis of carbon nanodots from the chemical pollutant reactive red 2 has allowed various full-scale biological assays demonstrating that the biotoxicity of the CNDs was significantly reduced in the cellular as well as organismal levels in comparison to its azo dye precursor, in which azo groups are the main cause of cytotoxicity.27 The fluorescence properties and low cytotoxicity of the synthesized CNDs confer its very favorable potential applications in biological imaging, as verified in the in vitro (cell culture) and in vivo (zebrafish embryo) model systems. In an effort to turn waste into wealth, the discovery of CNDs from RR2 provides new economically and environmentally friendly opportunities in both bioimaging and waste-water remediation.Ethical statement
All zebrafish husbandry and experimental procedures were performed in accordance with the Guiding Principles for the Care and Use of Laboratory Animals and were reviewed and approved by the Animal Care Committee of Lanzhou University (ethic approval ID no. EAF2020007).Data availability
All data, models, or code generated or used during the study are available from the corresponding author by request.Author contributions
X. L., D. A. and H. Z. conceived this project; W. C., J. S., Z. W. and Y. X. performed the experiments; X. L., D. A. and H. Z. analyzed the data and wrote the paper. All authors provided scientific input, edited, and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Funding by the National Natural Science Foundation of China (21805166), the 111 Project (D20015) and the Engineering Research Center of Eco-environment in Three Gorges Reservoir Region, Ministry of Education, China Three Gorges University (KF2019-05), the outstanding young and middle-aged science and technology innovation teams, Ministry of Education, Hubei province, China (T2020004), Foundation of Science and Technology Bureau of Yichang City (A21-3-012), the Fundamental Research Funds for the Central Universities (lzujbky-2019-74), the “Double First-Class” Research Start-up Funds of Lanzhou University (561119203), the University of Bordeaux and the Centre National de la Recherche Scientifique (CNRS) is gratefully acknowledged.Notes and references
- (a) M. J. H. Ku, T. X. Zhou, Q. Li, Y. J. Shin, J. K. Shi, C. Burch, L. E. Anderson, A. T. Pierce, Y. Xie, A. Hamo, U. Vool, H. Zhang, F. Casola, T. Taniguchi, K. Watanabe, M. M. Fogler, P. Kim, A. Yacoby and R. L. Walsworth, Nature, 2020, 583, 537–541 CrossRef CAS; (b) A. El Fatimy, R. L. Myers-Ward, A. K. Boyd, K. M. Daniels, D. K. Gaskill and P. Barbara, Nat. Nanotechnol., 2016, 11, 335–338 CrossRef CAS PubMed; (c) M. Kaur, M. Kaur and V. K. Sharma, Adv. Colloid Interface Sci., 2018, 259, 44–64 CrossRef CAS; (d) J. Zhu, Y. Tang, G. Wang, J. Mao, Z. Liu, T. Sun, M. Wang, D. Chen, Y. Yang, J. Li, Y. Deng and S. Yang, ACS Appl. Mater. Interfaces, 2017, 9, 14470–14477 CrossRef CAS PubMed.
- (a) A. Cadranel, J. T. Margraf, V. Strauss, T. Clark and D. M. Guldi, Acc. Chem. Res., 2019, 52, 955–963 CrossRef CAS PubMed; (b) X. Xu, R. Ray, Y. Gu, H. J. Ploehn, L. Gearheart, K. Raker and W. A. Scrivens, J. Am. Chem. Soc., 2018, 126, 12736–12737 CrossRef; (c) X. Xu, R. Ray, Y. Gu, H. J. Ploehn, L. Gearheart, K. Raker and W. A. Scrivens, J. Am. Chem. Soc., 2004, 126, 12736–12737 CrossRef CAS PubMed; (d) Y.-P. Sun, B. Zhou, Y. Lin, W. Wang, K. A. Fernando, P. Pathak, M. J. Meziani, B. A. Harruff, X. Wang, H. Wang, P. G. Luo, H. Yang, M. E. Kose, B. Chen, L. M. Veca and S. F. Xie, J. Am. Chem. Soc., 2006, 128, 7756–7757 CrossRef CAS PubMed; (e) M. Zhang, X. Zhai, M. Sun, T. Ma, Y. Huang, B. Huang, Y. Du and C. Yan, Chem. Soc. Rev., 2020, 49, 9220–9248 RSC; (f) F. Arcudi, L. Đorđević and M. Prato, Acc. Chem. Res., 2019, 52, 2070–2079 CrossRef CAS PubMed; (g) V. S. Sivasankarapillai, A. V. Kirthi, M. Akksadha, S. Indu, U. D. Dharshini, J. Pushpamalar and L. Karthik, Nanoscale Adv., 2020, 2, 1760–1773 RSC; (h) Z. Hassanvand, F. Jalali, M. Nazari, F. Parnianchi and C. Santoro, ChemElectroChem, 2021, 8, 15–35 CrossRef CAS; (i) C. Liu, F. Zhang, J. Hu, W. Gao and M. Zhang, Front. Chem., 2021, 8, 605028 CrossRef PubMed.
- (a) Y.-T. Long and T. J. Meade, Chem. Sci., 2020, 11, 6940–6941 RSC; (b) J. Wang, X. Wu, J. Pan, T. Feng, D. Wu, X. Zhang, B. Yang, X. Zhang and J. Jie, Adv. Mater., 2020, 32, 2003315 CrossRef CAS; (c) Y. Uemura, K. Yamato, R. Sekiya and T. Haino, Angew. Chem., Int. Ed., 2018, 57, 4960–4964 CrossRef CAS; (d) W. Xu, D. Wang and B. Z. Tang, Angew. Chem., Int. Ed., 2021, 60, 7476–7487 CrossRef CAS PubMed; (e) J. H. Zhang, T. Sun, A. Niu, Y. M. Tang, S. Deng, W. Luo, Q. Xu, D. Wei and D. S. Pei, Biomaterials, 2017, 133, 49–59 CrossRef CAS PubMed; (f) S. Deng, P. P. Jia, J. H. Zhang, M. Junaid, A. Niu, Y. B. Ma, A. Fu and D. S. Pei, J. Hazard. Mater., 2018, 357, 146–158 CrossRef CAS PubMed.
- (a) Y. Yan, J. Gong, J. Chen, Z. Zeng, W. Huang, K. Pu, J. Liu and P. Chen, Adv. Mater., 2019, 31, 1808283 CrossRef; (b) G. Gollavelli and Y. C. Ling, Biomaterials, 2012, 33, 2532–2545 CrossRef CAS PubMed.
- (a) S. Deng, A. Fu, M. Junaid, Y. Wang, Q. Yin, C. Fu, L. Liu, D. S. Su, W. P. Bian and D. S. Pei, Biomaterials, 2019, 206, 61–72 CrossRef CAS PubMed; (b) Y. F. Huang, X. Zhou, R. Zhou, H. Zhang, K. B. Kang, M. Zhao, Y. Peng, Q. Wang, H. L. Zhang and W. Y. Qiu, Chem.–Eur. J., 2014, 20, 5640–5648 CrossRef CAS; (c) X. Wei, L. Li, J. Liu, L. Yu, H. Li, F. Cheng, X. Yi, J. He and B. Li, ACS Appl. Mater. Interfaces., 2019, 11, 9832–9840 CrossRef CAS PubMed; (d) S. Li, Z. Peng, J. Dallman, J. Baker, A. M. Othman, P. L. Blackwelder and R. M. Leblanc, Colloids Surf., B, 2016, 145, 251–256 CrossRef CAS; (e) S. Li, I. Skromne, Z. Peng, J. Dallman, A. O. Al-Youbi, A. S. Bashammakh, M. S. El-Shahawi and R. M. Leblanc, J. Mater. Chem. B, 2016, 4, 7398–7405 RSC; (f) F. Huo, W. Liang, Y. Tang, W. Zhang, X. Liu, D. Pei, H. Wang, W. Jia, P. Jia and F. Yang, J. Mater. Sci., 2019, 54, 6815–6825 CrossRef CAS; (g) S. Y. Park, C. Y. Lee, H. R. An, H. Kim, Y. C. Lee, E. C. Park, H. S. Chun, H. Y. Yang, S. H. Choi, H. S. Kim, K. S. Kang, H. G. Park, J. P. Kim, Y. Choi, J. Lee and H. U. Lee, Nanoscale, 2017, 9, 9210–9217 RSC; (h) Z. Zhu, J. Qian, X. Zhao, W. Qin, R. Hu, H. Zhang, D. Li, Z. Xu, B. Z. Tang and S. He, ACS Nano, 2016, 10, 588–597 CrossRef CAS PubMed.
- (a) C.-L. Huang, C.-C. Huang, F.-D. Mai, C.-L. Yen, S.-H. Tzing, H.-T. Hsieh, Y.-C. Ling and J.-Y. Chang, J. Mater. Chem. B, 2015, 3, 651–664 RSC; (b) P. Roy, A. P. Periasamy, C. Y. Lin, G. M. Her, W. J. Chiu, C. L. Li, C. L. Shu, C. C. Huang, C. T. Liang and H. T. Chang, Nanoscale, 2017, 7, 2504–2510 RSC; (c) L. Ma, Y. Gao and X. Hu, Biomaterials, 2015, 52, 301–311 CrossRef; (d) S. H. Chiu, G. Gedda, W. M. Girma, J. K. Chen, Y. C. Ling, A. V. Ghule, K. L. Ou and J. Y. Chang, Acta Biomater., 2016, 46, 151–164 CrossRef CAS; (e) F. Du, Z. Cheng, M. Kremer, Y. Liu, X. Wang, S. Shuang and C. Dong, J. Mater. Chem. B, 2020, 8, 5089–5095 RSC.
- (a) F. Lu and D. Astruc, Coord. Chem. Rev., 2018, 356, 147–164 CrossRef CAS; (b) F. Lu and D. Astruc, Coord. Chem. Rev., 2020, 408, 213180 CrossRef CAS.
- M. de la Luz-Asunción, E. E. Pérez-Ramírez, A. L. Martínez-Hernández, P. E. García-Casillas, J. G. Luna-Bárcenas and C. Velasco-Santos, Diamond Relat. Mater., 2020, 109, 108002 CrossRef.
- (a) Z. Oruç, M. Ergüt, D. Uzunoğlu and A. Özer, J. Environ. Chem. Eng., 2019, 7, 103231 CrossRef; (b) Z. Wang, Q. Yin, M. Gu, K. He and G. Wu, J. Hazard. Mater., 2018, 357, 226–234 CrossRef CAS.
- (a) X. Liu, Y. Huang, P. Zhao, X. Meng and D. Astruc, ChemCatChem, 2020, 12, 175–180 CrossRef CAS; (b) Y. Huang, K. Zheng, X. Liu, X. Meng and D. Astruc, Inorg. Chem. Front., 2020, 7, 939–945 RSC; (c) J. Ma, X. Yang, X. Jiang, J. Wen, J. Li, Y. Zhong, L. Chi and Y. Wang, Chem. Eng. J., 2020, 389, 123422 CrossRef CAS; (d) S. Ye, G. Zeng, X. Tan, H. Wu, J. Liang, B. Song, N. Tang, P. Zhang, Y. Yang, Q. Chen and X. Li, Appl. Catal., B, 2020, 269, 118850 CrossRef CAS; (e) L. Peng, Y. Shang, B. Gao and X. Xu, Appl. Catal., B, 2021, 282, 119484 CrossRef CAS; (f) S. Zhang, B. Li, X. Wang, G. Zhao, B. Hu, Z. Lu, T. Wen, J. Chen and X. Wnag, Chem. Eng. J., 2020, 390, 124642 CrossRef CAS; (g) A. Tkaczyk, K. Mitrowska and A. Posyniak, Sci. Total Environ., 2020, 717, 37222 CrossRef; (h) W. Hou, S. Wang, X. Bi, X. Meng, P. Zhao and X. Liu, Chin. Chem. Lett., 2021 DOI:10.1016/j.cclet.2021.01.023; (i) L. Zhang, X. Bi, M. Gou, M. Sun, L. Tao, G. Chen, X. Liu, X. Meng and P. Zhao, Sep. Purif. Technol., 2021, 264, 118397 CrossRef; (j) X. Bi, Y. Huang, X. Liu, N. Yao, P. Zhao, X. Meng and D. Astruc, Sep. Purif. Technol., 2021, 275, 119141 CrossRef CAS; (k) L. Peng, X. Duan, Y. Shang, B. Gao and X. Xu, Appl. Catal., B, 2021, 287, 118863 CrossRef; (l) Y. Shang, X. Xu, B. Gao, S. Wang and X. Duan, Chem. Soc. Rev., 2021, 50, 5281–5322 RSC.
- (a) J. Hu, X. Yuan, C. Wng, X. Shao, B. Yang, A. A. Razzaq, X. Zhao, Y. Lian, Z. Zhao, M. Chen and Y. Peng, Small, 2020, 16, 2000755 CrossRef CAS; (b) H. Xu, Y. Liu, Q. Bai and R. Wu, J. Mater. Chem. A, 2019, 7, 3558–3562 RSC; (c) G. Ding, Y. Li, Y. Zhang, C. Huang, X. Yao, K. Lin, K. Shen, W. Yan, F. Sun and L. Zhou, ACS Appl. Mater. Interfaces, 2019, 11, 19096–19103 CrossRef CAS PubMed; (d) Y. Yao and F. Wu, ACS Appl. Mater. Interfaces, 2017, 9, 31907–31912 CrossRef CAS PubMed; (e) W. Yuan, A. Jie, S. Li, F. Huang, P. Zhang and Y. Shen, Energy, 2016, 115, 397–403 CrossRef CAS; (f) T. K. Marella, I. Y. López-Pacheco, R. Parra-Saldívar, S. Dixit and A. Tiwari, Sci. Total Environ., 2020, 724, 137960 CrossRef CAS.
- (a) W. Chen, D. Li, L. Tian, W. Xiang, T. Wang, W. Hu, Y. Hu, S. Chen, J. Chen and Z. Dai, Green Chem., 2018, 20, 4438–4442 RSC; (b) W. Chen, J. Shen, Y. Huang, X. Liu and D. Astruc, ACS Sustainable Chem. Eng., 2020, 8, 7513–7522 CrossRef CAS; (c) J. Shen, W. Chen, G. Lv, Z. Yang, J. Yan, X. Liu and Z. Dai, Int. J. Hydrogen Energy, 2021, 46, 796–805 CrossRef CAS; (d) W. Chen, G. Lv, Q. Zhou, J. Shen, J. Cao, X. Liu and Z. Dai, Nano Res., 2021, 14, 1228–1231 CrossRef CAS.
- (a) C.-L. Shen, Q. Lou, J.-H. Zang, K.-K. Liu, S.-N. Qu, L. Dong and C.-X. Shan, Adv. Sci., 2020, 7, 1903525 CrossRef CAS; (b) P. Roy, P.-C. Chen, A. P. Periasamy, Y.-N. Chen and H.-T. Chang, Mater. Today, 2015, 18, 447–458 CrossRef CAS.
- (a) B. Kong, J. Tang, Y. Zhang, T. Jiang, X. Gong, C. Peng, J. Wei, J. Yang, Y. Wang, X. Wang, G. Zheng, C. Selomulya and D. Zhao, Nat. Chem., 2016, 8, 171–178 CrossRef CAS; (b) C. Kgtahya, Y. Zhai, S. Li, S. Liu, J. Li, V. Strehmel, Z. Chen and B. Strehmel, Angew. Chem., Int. Ed., 2021, 60, 10983–10991 CrossRef; (c) N. Soni, S. Singh, S. Sharma, G. Batra, K. Kaushik, C. Rao, N. C. Verma, B. Mondal, A. Yadav and C. K. Nandi, Chem. Sci., 2021, 12, 3615–3626 RSC.
- J. Liu, M. Zheng, X. Shi, H. Zeng and H. Xia, Adv. Funct. Mater., 2016, 26, 919–930 CrossRef CAS.
- S. Zhang, J. Zhu, Y. Qing, L. Wang, J. Zhao, J. Li, W. Tian, D. Jia and Z. Fan, Adv. Funct. Mater., 2018, 28, 1805898 CrossRef.
- (a) Y.-C. Liang, K.-K. Liu, X.-Y. Wu, Q. Lou, L.-Z. Sui, L. Dong, K.-J. Yuan and C.-X. Shan, Adv. Sci., 2021, 8, 2003433 CrossRef CAS; (b) Y. Gao, W. Xu, F. He, P. Nie, Q. Yang, Z. Si, H. Meng and G. Wei, Nano Res., 2021, 14, 2294–2300 CrossRef CAS; (c) S.-Y. Song, L.-Z. Sui, K.-K. Liu, Q. Cao, W.-B. Zhao, Y.-C. Liang, C.-F. Lv, J.-H. Zang, Y. Shang, Q. Lou, X.-G. Yang, L. Dong, K. J. Yuan and C.-X. Shan, Nano Res., 2021, 14, 2231–2240 CrossRef CAS; (d) S. Zhao, D. Zhang, S. Jiang, Y. Cui, H. Li, J. Dong, Z. Xie, D.-W. Wang, R. Amal, Z. Xia and L. Dai, Nano Res., 2021 DOI:10.1007/s12274-021-3358-3.
- S. J. Zhu, Q. N. Meng, L. Wang, J. H. Zhang, Y. B. Song, H. Jin, K. Zhang, H. C. Sun, H. Y. Wang and B. Yang, Angew. Chem., Int. Ed., 2013, 52, 3953–3957 CrossRef CAS.
- (a) H. A. Nguyen, I. Srivastava, D. Pan and M. Gruebele, ACS Nano, 2020, 14, 6127–6137 CrossRef CAS PubMed; (b) N. C. Verma, A. Yadav, C. Rao, P. M. Mishra and C. K. Nandi, J. Phys. Chem. C, 2021, 125, 1637–1653 CrossRef CAS.
- (a) S. Zhu, Y. Song, X. Zhao, J. Shao, J. Zhang and B. Yang, Nano Res., 2015, 8, 355–381 CrossRef CAS; (b) B.-P. Qi, L. Bao, Z.-L. Zhang and D.-W. Pang, ACS Appl. Mater. Interfaces, 2016, 8, 28372–28382 CrossRef CAS; (c) H. Li, H. Ming, Y. Liu, H. Yu, X. He, H. Huang, K. Pan, Z. Kang and S.-T. Lee, New J. Chem., 2011, 35, 2666–2670 RSC; (d) L. Ai, Y. Yang, B. Wang, J. Chang, Z. Tang, B. Yang and S. Lu, Sci. Bull., 2021, 66, 839–856 CrossRef.
- (a) S. N. Baker and G. A. Baker, Angew. Chem., Int. Ed., 2010, 49, 6726–6744 CrossRef CAS; (b) P. G. Luo, S. Sahu, S.-T. Yang, S. K. Sonkar, J. Wang, H. Wang, G. E. LeCroy, L. Cao and Y.-P. Sun, J. Mater. Chem. B, 2013, 1, 2116–2127 RSC; (c) K. Hola, Y. Zhang, Y. Wang, E. P. Giannelis, R. Zboril and A. L. Rogach, Nano Today, 2014, 9, 590–603 CrossRef CAS; (d) S. Zhu, J. Zhang, S. Tang, C. Qiao, L. Wang, H. Wang, X. Liu, B. Li, Y. Li, W. Yu, X. Wang, H. Sun and B. Yang, Adv. Funct. Mater., 2012, 22, 4732–4740 CrossRef CAS; (e) S. Zhu, J. Zhang, C. Qiao, S. Tang, Y. Li, W. Yuan, B. Li, L. Tian, F. Liu, R. Hu, H. Gao, H. Wei, H. Zhang, H. Sun and B. Yang, Chem. Commun., 2011, 47, 6858–6860 RSC; (f) J. Shen, Y. Zhu, X. Yang and C. Li, Chem. Commun., 2012, 48, 3686–3699 RSC; (g) S. Zhu, Q. Meng, L. Wang, J. Zhang, Y. Song, H. Jin, K. Zhang, H. Sun, H. Wang and B. Yang, Angew. Chem., Int. Ed., 2013, 52, 3953–3957 CrossRef CAS PubMed.
- (a) K. Tak, R. Sharma, V. Dave, S. Jain and S. Sharma, ACS Chem. Neurosci., 2020, 11, 3741–3748 CrossRef CAS; (b) A. Kumar, A. Kumari, P. Mukherjee, T. Saikia, K. Pal and S. K. Sahu, Microchem. J., 2020, 159, 105590 CrossRef CAS.
- R. S. A. Sonthanasamy, W. Y. W. Ahmad, S. Fazry, N. I. Hassan and A. M. Lazim, Carbohydr. Polym., 2016, 137, 488–496 CrossRef CAS.
- (a) E. Garcia-Calvo, P. Cabezas-Sanchez and J. L. Luque-Garcia, Chemosphere, 2021, 263, 128170 CrossRef CAS; (b) K. A. Horzmann and J. L. Freeman, Toxicol. Sci., 2018, 163, 5–12 CrossRef CAS; (c) Y.-J. Dai, Y.-F. Jia, N. Chen, W.-P. Bian, Q.-K. Li, Y.-B. Ma, Y.-L. Chen and D.-S. Pei, Environ. Toxicol. Chem., 2014, 33, 11–17 CrossRef CAS PubMed; (d) S. Cassar, I. Adatto, J. L. Freeman, J. T. Gamse, I. Iturria, C. Lawrence, A. Muriana, R. T. Peterson, S. Van Cruchten and L. I. Zon, Chem. Res. Toxicol., 2020, 33, 95–118 Search PubMed.
- G. S. Schorr, E. A. Falcone, D. J. Moretti and R. D. Andrews, PLoS One, 2014, 9, e92633 CrossRef.
- (a) D. Li, D. Wang, X. Zhao, W. Xi, A. Zebibula, N. Alifu, J.-F. Chen and J. Qian, Mater. Chem. Front., 2018, 2, 1343–1350 RSC; (b) Z. Peng, E. H. Miyanji, Y. Zhou, J. Pardo, S. D. Hettiarachchi, S. Li, P. L. Blackwelder, I. Skromne and R. M. Leblanc, Nanoscale, 2017, 16, 17533–17543 RSC; (c) W. Liu, G. Huang, X. Su, S. Li, Q. Wang, Y. Zhao, Y. Liu, J. Luo, Y. Li, C. Li, D. Yuan, H. Hong, X. Chen and T. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 49012–49020 CrossRef CAS.
- (a) T. Deb, D. Choudhury, P. S. Guin, M. B. Saha, G. Chakrabarti and S. Das, Chem. Biol. Interact., 2011, 189, 206–214 CrossRef CAS; (b) L. Isaacs, Acc. Chem. Res., 2014, 47, 2052–2062 CrossRef CAS; (c) F. Zhang, Y.-H. Li, J.-Y. Li, Z.-R. Tang and Y.-J. Xu, Environ. Pollut., 2019, 253, 365–376 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc02837e |
| ‡ These authors have equally contributed to this work and should be considered as co-first authors. |
| This journal is © The Royal Society of Chemistry 2021 |