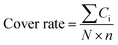Open Access Article
Open Access ArticleDevelopment of a structure-based computational simulation to optimize the blocking efficacy of pro-antibodies†
Bo-Cheng
Huang‡
a,
Yun-Chi
Lu‡
bc,
Jun-Min
Liao
bc,
Hui-Ju
Liu
d,
Shih-Ting
Hong
d,
Yuan-Chin
Hsieh
e,
Chih-Hung
Chuang
cf,
Huei-Jen
Chen
d,
Tzu-Yi
Liao
d,
Kai-Wen
Ho
d,
Yeng-Tseng
Wang
 *h and
Tian-Lu
Cheng
*h and
Tian-Lu
Cheng
 *abcg
*abcg
aInstitute of Biomedical Sciences, National Sun Yat-Sen University, Kaohsiung, Taiwan. E-mail: tlcheng@kmu.edu.tw
bDepartment of Biomedical Science and Environmental Biology, Kaohsiung Medical University, Kaohsiung, Taiwan
cDrug Development and Value Creation Research Center, Kaohsiung Medical University, Kaohsiung, Taiwan
dGraduate Institute of Medicine, College of Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan
eSchool of Medicine for International Students, I-Shou University, Kaohsiung, Taiwan
fDepartment of Medical Laboratory Science and Biotechnology, College of Health Sciences, Kaohsiung Medical University, Kaohsiung, Taiwan
gDepartment of Medical Research, Kaohsiung Medical University Hospital, Kaohsiung, Taiwan
hDepartment of Biochemistry, Kaohsiung Medical University, Kaohsiung, Taiwan. E-mail: c00jsw00@kmu.edu.tw
First published on 14th June 2021
Abstract
The on-target toxicity of monoclonal antibodies (Abs) is mainly due to the fact that Abs cannot distinguish target antigens (Ags) expressed in disease regions from those in normal tissues during systemic administration. In order to overcome this issue, we “copied” an autologous Ab hinge as an “Ab lock” and “pasted” it on the binding site of the Ab by connecting a protease substrate and linker in between to generate a pro-Ab, which can be specifically activated in the disease region to enhance Ab selectivity and reduce side effects. Previously, we reported that 70% of pro-Abs can achieve more than 100-fold blocking ability compared to the parental Abs. However, 30% of pro-Abs do not have such efficient blocking ability. This is because the same Ab lock linker cannot be applied to every Ab due to the differences in the complementarity-determining region (CDR) loops. Here we designed a method which uses structure-based computational simulation (MSCS) to optimize the blocking ability of the Ab lock for all Ab drugs. MSCS can precisely adjust the amino acid composition of the linker between the Ab lock and Ab drug with the assistance of molecular simulation. We selected αPD-1, αIL-1β, αCTLA-4 and αTNFα Ab as models and attached the Ab lock with various linkers (L1 to L7) to form pro-Abs by MSCS, respectively. The resulting cover rates of the Ab lock with various linkers compared to the Ab drug were in the range 28.33–42.33%. The recombinant pro-Abs were generated by MSCS prediction in order to verify the application of molecular simulation for pro-Ab development. The binding kinetics effective concentrations (EC-50) for αPD-1 (200-250-fold), αIL-1β (152-186-fold), αCTLA-4 (68-150-fold) and αTNFα Ab (20-123-fold) were presented as the blocking ability of pro-Ab compared to the Ab drug. Further, there was a positive correlation between cover rate and blocking ability of all pro-Ab candidates. The results suggested that MSCS was able to predict the Ab lock linker most suitable for application to αPD-1, αIL-1β, αCTLA-4 and αTNFα Ab to form pro-Abs efficiently. The success of MSCS in optimizing the pro-Ab can aid the development of next-generation pro-Ab drugs to significantly improve Ab-based therapies and thus patients' quality of life.
Monoclonal antibodies (Abs) have been regarded as potential therapeutics due to their antigen (Ag) specificity that can be applied to multiple diseases, such as malignant cancers,1–4 chronic diseases5 and autoimmune diseases.6,7 However, the targeted Ags are not only expressed in the disease area but also in the healthy region, causing unexpected on-target toxicity during systemic over-activation.8 Neutralization of the targeted Ag can also reduce therapeutic efficacy or even terminate drug treatment. For example, previous studies have reported that the immune checkpoint drug antagonists: Ipilimumab (αCTLA-4 Ab) and Nivolumab (αPD-1 Ab) may systemically target CTLA-4/PD-1 and over-activate immune cells, causing immune-related adverse events such as hepatitis, colitis, thyroid disorders, and even paralysis.9–11 On the other hand, the Ab drugs for rheumatoid arthritis (RA), Infliximab and Adalimumab (αTNFα Ab) have been reported to reduce pathological inflammation and inhibit RA progression through targeting TNFα, which modulates host defense and tumor growth. Systemic neutralization of TNFα may lead to severe infections, reactivation of viral infections (hepatitis or herpes zoster), and raise the risk of malignancy.12–14 Canakinumab (αIL-1β Ab) is an IL-1β blocker for patients who suffer from associated periodic syndromes (CAPS), IL-1β plays a role in resisting inflammation and regulating immune response, which by systemic neutralization causes on-targeted toxicities such as increasing risk of pneumonia, bone and joint infections.15 To sum up, the U.S. Food and Drug Administration (FDA) has indicated that most therapeutic Abs have on-target toxicity issues and therefore limits their application.8,16,17 Hence, enhancement of Ab selectivity and reduction of side effects are necessary if Ab-based therapies are to be effectively used in the clinic.
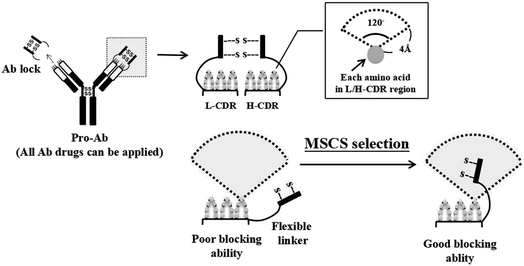
Several technologies have been developed to increase the selectivity of Ab to the disease region and the safety of Ab-based therapies.18 CytomX Therapeutics, Inc. generated an epithelial growth factor receptor (EGFR) pro-Ab by selecting a bacterial display-associated binding peptide to specifically mask the Ag-binding site of an αEGFR Ab and linking it with a substrate peptide of urokinase-type plasminogen activator (uPA).19 The pro-αEGFR Ab exhibited 48-fold weaker Ag binding as compared to the parental Ab and the biological activity could be specifically restored in the tumor microenvironment.19 However, the exogenous binding peptide may not be efficiently released from pro-Ab due to the affinity of the binding peptide to the Ab, and the exogenous nature of the peptide may induce undesirable immune responses. The Beckman Research Institute at the City of Hope also designed a reversible masking technology for two distinct αEGFR single-chain Fv (scFv) by mutual masking with their natural target, mutant domain III of the soluble EGFR and linking with a matrix metalloprotease (MMP) substrate peptide.20 The masked scFvs showed an 8-fold lower association with EGFR as compared with the original αEGFR scFvs after unmasking by MMP treatment.20 Nonetheless, the low masking efficiency of the masked scFvs and the customized masking peptide may limit its broad application to other Abs. In additional, Seattle Genetics, Inc. used leucine-rich and parallel heterodimeric coils with high inter-coil interactions (CC2B) as a spatial hindrance-based masking domain to block the Ag binding site of αCD20 Ab, αHER2 Ab, and mouse αCD3 Ab by using MMP-2 or -9 substrate linkers to develop pro-Ab.21 They proved that the CC2B masking domain can decrease the Ag binding ability of each Ab by at least 80-fold, 470-fold and 1000-fold, respectively. CC2B masking domains are also efficiently removed to restore the Ag binding ability after MMP protease treatment. However, the complex helix structure and customized mask may affect the production efficiency and increase the risk of immune responses. Overall, it's very important to develop a universal and low immunogenic Ab lock with high masking efficiency and release ability. In our previous research, we developed a spatial hindrance-based “Ab lock” to generate pro-Abs which can increase Ab selectivity and safety. We “copied” the autologous Ab hinge as an “Ab lock” and “pasted” it onto the binding site of the Ab by connecting a protease substrate and linker in between to generate a pro-Ab.22 Once the Ab lock is cleaved from the pro-Ab by a protease expressed at the disease region, the Ab is expected to have restored binding ability and neutralize its target Ag. In our previous study, we generated pro-Infliximab which can be selectively activated by MMP-2/9 only at the RA region. The Ab lock significantly inhibits TNFα binding of pro-Infliximab by 395-fold compared with Infliximab, and MMP-2/9 protease treatment to pro-Infliximab can completely restore the neutralization of TNFα. Further, pro-Infliximab not only showed equivalent therapeutic efficacy to Infliximab but also maintained immunity against Listeria infection in the RA mouse model, which led to a 71% survival rate compared to 0% of the Infliximab treatment group, indicating that pro-Abs can enhance Ab selectivity and reduce side effects.22 Our previous results showed that by using the concept of “copy and paste”, we transformed almost 70% of therapeutic Abs into pro-Abs and achieved over 100-fold blocking ability compared to the parental Abs. Hence, this strategy could be further applied to develop customized pro-Abs. However, 30% of pro-Abs did not achieve the blocking ability to even 50-fold compared to parental Abs. The reason for the low blocking ability is that there are distinct differences among the complementarity-determining region (CDR) loop of each Ab,23,24 so the identical linker of the Ab lock cannot be applied to every Ab. In order to overcome this issue, it is essential to design a method to predict and select the Ab lock with a suitable linker for each Ab.
Here we designed a method using structure-based computational simulation (MSCS) to predict the cover rate of the pro-Ab in order to select a suitable linker for the Ab lock applied to different Ab drugs. We precisely extended or shortened the length of the amino acid composition to form linkers (L1, L2, L3, L4, L5, L6 and L7) between the Ab lock and Ab drug. Next, we predicted pro-Abs with each Ab lock linker using Amber and Discovery Studio software to simulate and calculate the cover rate of the Ab lock to each CDR residue. The cover rate was determined by a homemade program which analyzes the trajectories and calculates the frequency if any atom of hinge, linker or substrate above 120° and 4 Å of any atom of CDR amino acids. In order to confirm the accuracy of the MSCS, we selected four Ab drugs: αPD-1, αIL-1β, αCTLA-4, and αTNFα Ab as models to simulate pro-Abs with the following Ab lock linkers: pro-αPD-1 Ab (L2 and L3), pro-αIL-1β Ab (L3 and L4), pro-αCTLA-4 Ab (L1 and L2) and pro-αTNFα Ab (L5, L6, and L7), and calculated their cover rate, respectively. Then, we generated the recombinant pro-Abs and evaluated the blocking ability using biological assays. Finally, we analyzed the correlation between the blocking ability and the cover rate to verify the accuracy of the MSCS. If it is possible to efficiently transform any Ab into a pro-Ab using MSCS, the development of pro-Ab will be facilitated and Ab therapeutic efficacy will be improved. In addition, serious side effects can be avoided, thereby improving patients' quality of life.
Materials and methods
Cells
The Expi293F cells (Thermo Fisher Scientific, Waltham, MA, USA) were cultured in Expi293 Expression Medium (Thermo Fisher Scientific, catalog A1435102) at 37 °C in a humidified atmosphere of 8% CO2.Pro-Ab construction, expression, and purification
The complementary DNA coding for the heavy and light chains of the pro-Abs and Abs were cloned based on the Ab (αCTLA-4, αPD-1, αIL-1β and αTNFα) DNA construct through assembly PCR. Human IgG1 hinge sequences were obtained from the National Center for Biotechnology Information. The hinge-encoding sequences (EPKSCDKTHTCPPCP), linkers (L1-VL: VNGGGGS-GPLGVR-AAQPA/L1-VH: GGRGGGGS-GPLGVR-RS; L2-VL: VN-GPLGVR-AAQPA/L2-VH: GGR-GPLGVR-PGRS; L3-VL: GGGGS-GPLGVR-AAQPA/L3-VH: VNAAAGGGGS-GPLGVR-RS; L4-VL: -GPLGVR-AAQPA/L4-VH: VNAAA-GPLGVR-PRS; L5-VL: GGGGS-GPLGVR-GGGGS/L5-VH: GGGGS-GPLGVR-GGGGS; L6-VL: GGGGS-GPLGVR-GGGFS/L6-VH: GGGGS-GPLGVR-GGGFS; L7-VL: -GPLGVR-GGGDS/L7-VH: -GPLGVR-GGGDS), and MMP-2/9 substrate-encoding sequences (GPLGVR) were introduced upstream of the light chain and heavy chain to generate pro-Abs (pro-αCTLA-4, pro-αPD-1, pro-αIL-1β and pro-αTNFα). All Ab or pro-Ab production was generated through the Expi293 Expression System (Thermo Fisher Scientific, Waltham, MA, USA) and was purified using Protein A-Sepharose (GE Healthcare, Milwaukee, WI, USA).Computational structure predictions for pro-Abs
The structure of the antibodies was extracted from the crystal structure in the protein data bank25 or found by homology modeling if the crystal structure is not solved. Only Fv segments were used in the simulations. The IgG1 hinges were built by homology modeling. These hinges were combined with linkers and MMP-2/9 substrate segments (GPLGVR), and then attached to the N-terminal of the Ab to construct pro-Ab structures. The 3D pro-Ab structures were built using Discovery Studio Software (San Diego, CA, USA). To avoid D-amino acids and cis peptide bonds in pro-Ab, these structures were validated by VMD software.26–29 These initial structures were then simulated using the software package AMBER 18 with the all-hydrogen amino acid AMBER FF14SB force field and generalized Born solvent model (igb = 5).30–32 All MD simulations were performed in the NVT ensembles with a simulation temperature of 310 K, unless otherwise stated, using the Langevin integrator with an integration time step of 2 fs and SHAKE constraints33 for all covalent bonds involving hydrogen atoms. These complex structures were minimized for 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 conjugate gradient steps and then subjected to 20 ns GaMD equilibrium and 1000 ns GaMD production simulations.34 Snapshots of all nine 1000 ns GaMD production simulations were used to calculate the cover rates using our homemade program, in which the cover rate is calculated from the blocking frequencies of the CDR residues. A CDR residue is counted as blocked if any atom of the hinge, linker or substrate reaches above 120° and 4 Å for any atom of the CDR residues. The cover rate is expressed as follows:
000 conjugate gradient steps and then subjected to 20 ns GaMD equilibrium and 1000 ns GaMD production simulations.34 Snapshots of all nine 1000 ns GaMD production simulations were used to calculate the cover rates using our homemade program, in which the cover rate is calculated from the blocking frequencies of the CDR residues. A CDR residue is counted as blocked if any atom of the hinge, linker or substrate reaches above 120° and 4 Å for any atom of the CDR residues. The cover rate is expressed as follows:where the Ci is the counted number of times each CDR residue is blocked over the whole production trajectories. N is the number of snapshots of each trajectory and n is the number of CDR residues.
Comparison of the binding ability of pro-Abs with or without MMP-2/9 treatment
To determine the binding kinetics effective concentration (EC-50) of pro-Abs (pro-αCTLA-4, pro-αPD-1, pro-αIL-1β and pro-αTNFα Ab) and Abs (αCTLA-4, αPD-1, αIL-1β and αTNFα Ab), the recombinant Ag-CTLA-4, PD-1, IL-1β and TNFα were coated onto 96-well plates and blocked with 5% skim milk. The pro-Ab or Ab were added onto the plates at the given concentrations (αCTLA-4 and pro-αCTLA-4 Abs: 107.4–0.0138 nM, 6-fold serial dilutions; αPD-1 and pro-αPD-1 Abs: 4000–0.004 nM, 10-fold serial dilutions; αIL-1β and pro-αIL-1β Abs: 1440–0.0014 nM, 10-fold serial dilutions; αTNFα and pro-αTNFα Ab: 500–0.0005 nM, 10-fold serial dilutions) for 1 h at RT. After washing, the wells were incubated with HRP-goat anti human IgG Fcγ Ab for 1 h at RT, and detection was performed by the addition of ABTS containing 30% H2O2. The binding ability was quantified through absorbance detection at 405 nm.Results
Prediction of the structure of pro-Ab candidates with different linkers to analyze the cover rate of the Ab lock
We previously developed ten pro-Abs, seven of which achieved more than 100-fold blocking ability. However, approximately 30% of the pro-Abs did not show such efficient blocking ability (Table S1†). Here we designed a method using structure-based computational simulation (MSCS) to predict the cover rates of pro-Abs with different linkers, and selected a suitable ones for specific Abs. To construct the structures of the pro-Abs, we first obtained the crystal structure of the Ab Fab fragment from the protein data bank. The IgG1 hinge (Ab lock) was built by homology modeling, and attached to the Ab Fab fragment through protease substrate and linker to generate pro-Ab Fab structure. In order to select a suitable linker to generate a pro-Ab with high blocking efficiency, we designed a method which uses structure-based computational simulation (MSCS) to predict the cover rate of the Ab lock with various linkers. The cover rate of the Ab lock was identified as the frequency of the appearance of the Ab lock above 4 Å within 120 degrees of each amino acid on the CDR loop (Fig. 1). Here, we selected four Abs, αCTLA-4, αPD-1, αIL-1β and αTNFα Ab as candidates with various linkers for which the lengths were extended or shortened between the Ab lock (EPKSCDKTHTCPPCP), MMP-2/9 substrate sequence (GPLGVR) and VL/H region (Table 1). In order to select suitable linkers for each Ab, we designed six linkers based on the patented version of linker, L1. Compared with L1, L2 has a shortened linker 1 to test the masking effect of the shortened spacer (linker 1). Compared with L1, L3 has non-symmetrical linker. Based on L3, L4 has shortened link 1. L5 has symmetrical linker 1 and linker 2. Based on L5, L6 adds a phenylalanine, hydrophobic amino acid in its linker 2, and L7 has aspartic acid, a negative charge amino acid in its linker 2 and without linker 1. Using MSCS to the predict structure of each pro-Ab, these Ab locks with various kinds of linkers were observed to have a cover rate from 18.1% to 42.43% in the four Ab drugs (Table 2). The computer simulated results indicated that variation of the linker length and composition will lead to different cover rates of the Ab lock on the CDR region of the Ab.| Name | IgG1 hinge | Linker 1 | MMP-2/9 substrate | Linker 2 | Feature |
|---|---|---|---|---|---|
| L1 | L: EPKSCDKTHTCPPCP | VNGGGGS | GPLGVR | AAQPA∼ | Patent version |
| H: EPKSCDKTHTCPPCP | GGRGGGGS | GPLGVR | RS∼ | ||
| L2 | L: EPKSCDKTHTCPPCP | VN | GPLGVR | AAQPA∼ | Shortened of L1's linker 1 |
| H: EPKSCDKTHTCPPCP | GGR | GPLGVR | PGRS∼ | ||
| L3 | L: EPKSCDKTHTCPPCP | GGGGS | GPLGVR | AAQPA∼ | Non-symmetrical |
| H: EPKSCDKTHTCPPCP | VNAAAGGGGS | GPLGVR | RS∼ | ||
| L4 | L: EPKSCDKTHTCPPCP | GPLGVR | AAQPA∼ | Shortened of L3's linker 1 | |
| H: EPKSCDKTHTCPPCP | VNAAA | GPLGVR | PRS∼ | ||
| L5 | L: EPKSCDKTHTCPPCP | GGGGS | GPLGVR | GGGGS∼ | Symmetrical |
| H: EPKSCDKTHTCPPCP | GGGGS | GPLGVR | GGGGS∼ | ||
| L6 | L: EPKSCDKTHTCPPCP | GGGGS | GPLGVR | GGGFS∼ | Hydrophobic of L5 |
| H: EPKSCDKTHTCPPCP | GGGGS | GPLGVR | GGGFS∼ | ||
| L7 | L: EPKSCDKTHTCPPCP | GPLGVR | GGGDS∼ | No link 1 and negative charge of L5 | |
| H: EPKSCDKTHTCPPCP | GPLGVR | GGGDS∼ |
| Types of linkers | αCTLA-4 | αPD-1 | αIL-1β | αTNFα | |||||
|---|---|---|---|---|---|---|---|---|---|
| L1 | L2 | L2 | L3 | L3 | L4 | L5 | L6 | L7 | |
| Cover rate% | 29.96 | 42.43 | 41.19 | 28.33 | 37.18 | 38 | 18.1 | 22.28 | 23 |
| Blocking fold | 68 | 150 | 250 | 200 | 152 | 186 | 31 | 38 | 39 |
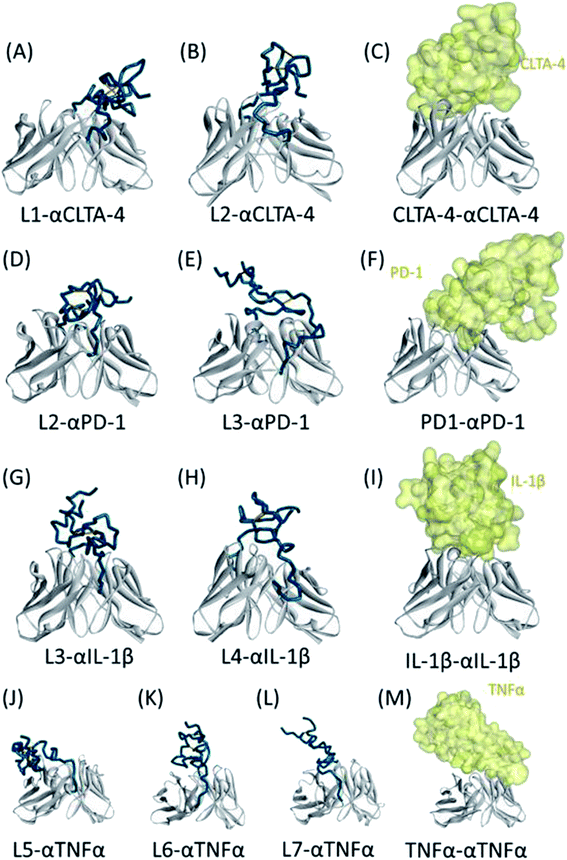
For clarity, the final simulated structures of pro-Abs and the corresponding X-ray crystal structures of the Ab–Ag complexes are shown in Fig. 3. A comparison of the 3D structures of the pro-αCTLA-4 Abs with the X-ray crystal complex structures showed that the CDR region can be effectively locked with the L2 linker (Fig. 3A–C). The predicted cover rates of L1 and L2 were 29.96 and 42.43%, respectively. A comparison of the 3D structures of the pro-αPD-1 Abs with the X-ray crystal complex structures showed that the CDR region can be effectively blocked with the L2 linker (Fig. 3D–F). The predicted cover rates of L2 and L3 were 41.19 and 28.33%, respectively. A comparison of the 3D structures of the pro-αIL-1β Abs with the X-ray crystal complex structures showed that the L3 and L4 linkers are similarly placed on the middle of the CDR3 loop regions (Fig. 3G–I). The predicted cover rates of L3 and L4 were 37.18 and 38.00%, respectively. A comparison of the 3D structures of the pro-αTNFα Abs with the X-ray crystal complex structures showed that the L5, L6 and L7 linkers are similarly bending toward the heavy chains with TNFα binding to the light chain (Fig. 3J–M). The predicted cover rates of the L5, L6 and L7 were 18.10, 22.28 and 23.00%, respectively.
Generation of the recombinant pro-Abs predicted by MSCS to confirm their blocking ability
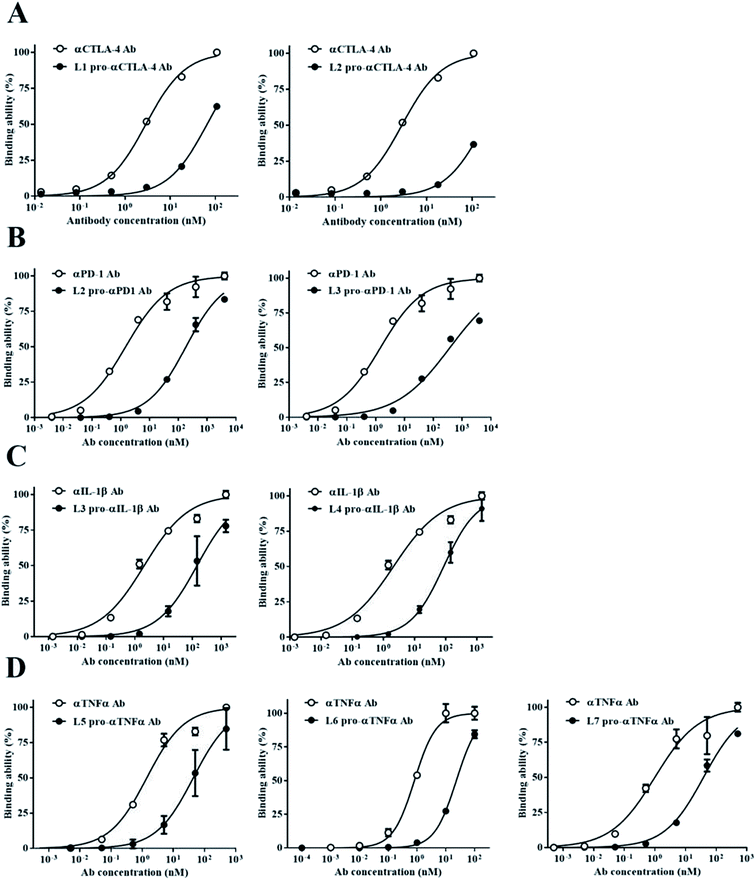
We wanted to confirm the blocking ability of each pro-Ab with different Ab lock linker candidates, so we generated the pro-Abs and detected their binding kinetic by ELISA. We used gene engineering to generate the computer simulation selected-pro-Abs candidates of αCTLA-4 (L1 and L2), αPD-1 (L2 and L3), αIL-1β (L3 and L4) and αTNFα (L5, L6 and L7), respectively, and analyzed the blocking ability of the pro-Abs via Ag-base ELISA. The results of pro-αCTLA-4 Abs showed that the EC-50 of L1 Ab lock and L2 Ab lock had a 68-fold and 150-fold masking effect, respectively, compared to αCTLA-4 Ab alone (Fig. 2A). As shown in Fig. 2B, the EC-50 of the L2 Ab lock had a 250-fold, and L3 had a 200-fold masking effect compared to αPD-1 Ab alone. The αIL-1β revealed that the EC-50 of the L3 Ab lock had a 152-fold, and L4 had a 186-fold masking effect compared to αIL-1β Ab alone (Fig. 2C). And αTNFα showed that the EC-50 of the L5 Ab lock had a 31-fold, L6 Ab lock had a 38-fold, and L7 had a 39-fold masking effect compared to αTNFα Ab alone (Fig. 2D). These data indicated that among various linker candidates, the higher the cover rate the Ab lock had, the greater its blocking ability was. Also each Ab lock with different Ab lock linkers led to a distinct blocking ability (Table 2). We successfully used this system to construct four kinds of pro-Abs, and the Ab locks with the computer simulation-selected linkers showed binding ability reduced by 31- to 200-fold compared to parental Ab drugs.Analysis of the correlation between the cover rate and blocking ability of pro-Abs
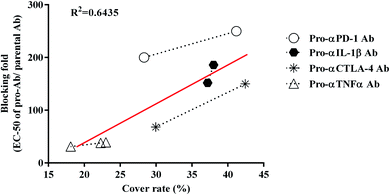
To verify that the pro-Abs simulated by computer do not differ from the biological results, we analyzed the correlation between the computer simulated results-cover rate and blocking ability (Fig. 4). The candidates, which included αCTLA-4, αPD-1, αIL-1β and αTNFα Ab, showed positive correlations between the cover rate and blocking fold. The R2 value of each pro-Ab we analyzed was 0.6435. These results indicated that the MSCS could predict the cover rate of the pro-Ab via computer modeling. By simulating the Ab lock and predicting the blocking ability of the pro-Ab by MSCS before generating it, we efficiently and accurately selected the optimal Ab lock candidate; therefore, customizing each Ab lock to different Ab drugs.Discussion
We successfully designed a method which used structure-based computational simulation (MSCS) to efficiently select the most suitable linker for enhancing the blocking ability of various pro-Abs. We applied the concept of “copy and paste” to develop an autologous Ab lock which is connected by various linkers. Here we compared the cover rate of different Abs applied with different linkers, L1, L2, L3, L4, L5, L6 and L7, and connected them to αCTLA-4, αPD-1, αIL-1β and αTNFα Ab, respectively, as pro-Abs. We found that pro-αCTLA-4 with L2 linker can achieve higher cover rate (42.43%) compared to L1 linker (29.96%). Producing the recombinant pro-Abs mentioned above biologically to evaluate their blocking ability confirmed that the linker with a higher cover rate had better masking ability. Likewise, pro-αPD-1 with the L2 linker (41.19% of cover rate) had better masking ability (250-fold) compared to L3 linker (28.33% of cover rate, 200-fold). During the analysis we found that there was a positive correlation between the cover rate and masking ability, indicating our MSCS can select the Ab lock with the appropriate linker depending on different Ab drug characteristics to efficiently optimize pro-Abs. This way, scientists can easily apply the “copy and paste” concept to generate an efficient pro-Ab according to the MSCS. Using this method, pro-Abs can be more widely developed to enhance drug selectivity and reduce on-toxicity of Abs.GaMD approach can enhance conformational sampling for biomolecules, and the GaMD simulations were previously applied in the pro-Abs.35 In the pro-αCTLA-4 Ab GaMD simulations, our final simulated structures showed that L1 linker bent toward the light chain of the Ab, leading to a lower cover rate (29.96%). L2 linker was in the middle of the CDR3 loops and showed higher cover rate (42.43%) (Fig. 3A and B). The X-ray crystal complex structures of CTLA-4 (Fig. 3C) shows that the Ag majorly binds on the heavy chain, and thus the pro-Ab with the L2 linker could supply the better blocking fold to the pro-Ab (Fig. 3C and Table 2). Likewise, in the pro-αPD-1Ab GaMD simulated results, the L2 linker was majorly positioned in the middle, but the L3 linker was bent toward the light chain (Fig. 3D and E). The X-ray complex crystal structure showed that the binding site of PD-1 involves both the middle of CDR3 loops and the light chain regions of the Ab (Fig. 3F), and therefore the L2 linker could supply a better blocking fold but L3 still kept the blocking function. In these two cases, the short linker L2 provided better cover rates than longer linkers L1 and L3. We propose that the shorter linkers might possibly enhance the coverage by limiting the Ab lock in the middle of CDR loops, and therefore provide greater blocking fold. Overall, the in vitro blocking fold test results were close to our GaMD simulations and structural analysis. We contend that our predictions yield valuable insight into making effective blocking fold predictions for pro-Abs with various linkers.
It is important to develop pro-Abs depending on the diversity of the CDR loop of each Ab. The structure motif of the CDR loop is a key to determining the difference in bio-activity of each Ab. For example, Regep and colleagues screened 1779 structures and calculated that over 30% of CDR-H3 loops play a key role in the ability of the Ab to bind to the diverse spaces of potential Ags.36 Avnir and colleagues developed an influenza hemagglutinin neutralizing Ab (anti-influenza A virus Ab) for which the major structure was achieved through two CDR-H2 loop anchor residues with the Phe54 residue having the dominant position.37 In addition to the CDR loop of the heavy chain, in some cases the CDR loop of light chain has also been reported to play the dominant role in Ag recognition. Van den Beucken and colleagues reported that they generated the affinity maturation of anti-streptavidin Ab fragments, R2H10 (3.2 nM) and R3B1 (5.5 nM), by yeast-display, then identified that mutation in CDR-L1 and CDR-L3 could improve the affinity 10.7-fold and 6.3-fold, respectively, compared to the starting Ab.38 These examples indicate that the diversity of the CDR loop affects the characteristics of the Ab. Therefore, it is essential to customize the Ab lock for any Ab. Here we precisely adjusted the amino acid composition of the linker between the Ab lock and the Ab drug and predicted the most suitable linker via MSCS depending on variable CDRs. As a result, we confirmed that there is positive correlation between the cover rate simulated by MSCS and the blocking ability measured through biological assay. Our results not only showed the cover rate of the Ab lock with each linker but also the individual cover rate of each CDR loop. With MSCS, the linker could be adjusted in order to coordinate with the CDR loop of an Ab, which is the key dominant region. We thus designed a universal method-MSCS that can efficiently overcome the issue of poor pro-Ab blocking ability due to CDR diversity and generate better pro-Abs accurately.
Furthermore, blocking the binding of anti-idiotypic Ab (anti-Id Ab) and prolonging the serum half-life of Ab drug is desirable, because generation of anti-Id Abs have been reported to neutralize the activity of Ab drugs, thereby accelerating the clearance rate, reducing therapeutic effect of Ab drug, and even increasing the frequency of immune-mediated adverse effects.39 Previous studies suggested that 28% Adalimumab-treated RA patients induced anti-Adalimumab Ab (>97% are anti-Id Ab) during a 3 year treatment period. Anti-Adalimumab Ab reduces the serum concentration of Adalimumab and causes a 2-fold increase in treatment failure rate as compared with the anti-Adalimumab-negative Ab.40 Ruiz Garcia et al also reported that Certolizumab pegol (α-TNFα Fab' fragment bound to polyethylene glycol) induces anti-Id Ab generation to Certolizumab pegol in 8.1% RA patients at 24 weeks, thus, reducing the drug response (ACR20).41 In addition, Weinblatt et al. indicated that a high incidence of anti-Id Ab (79%) was found following treatment with Alemtuzumab (anti-CD52 Ab) in patients with multiple sclerosis, and with reduced therapeutic efficacy.4 These clinical findings indicated that preventing the interference effect of anti-Id Ab can solve many of the clinical problems of Ab drugs. Our team have demonstrated that the spatial hindrance-based Ab lock can prevent the response of anti-Id Ab to Infliximab (anti-I-Id Ab); the binding of the anti-I-Id Ab to pro-Infliximab was 108-fold weaker than that to Infliximab,19 and the Ab lock maintained the TNFα binding ability of pro-Infliximab which was pre-incubated with anti-Id Ab. In the future, we will be able to efficiently transform Abs into optimized pro-Abs to prevent the binding of anti-Id Ab using MSCS. The custom-designed linker can enhance the masking ability of Ab lock to cover the CDR region of each Ab, thereby restricting the generation of anti-Id Abs. We believe MSCS can generate safer pro-Abs and provide better therapeutic efficacy.
Last but not least, it is important to develop pro-Abs with changeable protease substrates depending on different protease-expressing diseases. Proteases are disease-specific diagnostic markers which offer a therapeutic index. Qu and colleagues reported high levels of expression of matrix metalloproteinase-9 (MMP-9) in breast cancer, which plays the role of tumor migration and invasion. MMP-9 belongs to the family of Ca2+ and Zn2+ ion-dependent endopeptidases and has the ability to degrade extracellular matrix components, which indicates that MMP-9 is involved in tumorigenesis of breast cancer.42 Choi and colleagues have reported proteases that are overexpressed in ovarian cancer, such as cathepsin D, which can promote tumor progression. Cathepsin D is localized in lysosomes, and is also detected at a high level in the area surrounding tumors which causes the invasion.43 Duong and colleagues reported that osteoclasts express high levels of cathepsin K that can cause osteoporosis. Cathepsin K is a cysteine protease and has the ability to degrade the bone matrix and decrease bone mass, meaning cathepsin K may have the potential to be a new treatment target.44 Many cases show that different diseases express several proteases. If we generate pro-Abs with different protease substrates, the distinct substrate will change the composition of the linker and the efficacy of the spatial-hindrance of the Ab lock, even affecting the blocking ability of the pro-Ab. In order to overcome this issue, we can utilize MSCS to calculate the cover rate of the pro-Ab with different protease substrates in the pro-Ab. MSCS will adjust the amino acid composition of the linker between the Ab lock and Ab drug with the assistance of molecular simulation, so that we can efficiently generate protease-independent pro-Abs, and thereby replace any protease as the substrate peptide of the pro-Ab depending on the disease. By MSCS, we can accelerate pro-Ab development which is dependent on different protease-expressing diseases.
In conclusion, we provide MSCS that can predict the cover rate of Ab locks to optimize and customize pro-Abs. MSCS has the following advantages: (1) it can adjust the cover rate of an Ab lock to the Ab binding site depending on the diversity of each CDR loop; (2) it can be utilized to predict the pro-Ab that may prevent anti-Id Ab neutralizing the Ab in the blood; (3) it can predict the pro-Abs that need to change the protease substrate for different protease-expressing diseases; (4) any scientist could easily apply MSCS to select the Ab lock with a most suitable linker to generate a pro-Ab. We expect that by using the MSCS strategy to develop pro-Abs it will be possible to accelerate the development of pro-Abs. In addition, it is possible to apply this system which make all Abs transform pro-Abs and simulate the cover rate of the inhibited domain. The development of pro-Abs can be expected to change the behavior of next-generation Abs and significantly improve Ab therapy.
Funding
This work was supported by grants from the Ministry of Science and Technology, Taipei, Taiwan (MOST 107-2320-B-037-024-MY3) (MOST 109-2320-B-037-010-MY3); the National Health Research Institutes, Taiwan (NHRI-EX108-10729EI); Academia Sinica, Taiwan (AS-TP-107-L11); the Ministry of Education, Taiwan (108RSB0029); and the KMU-KMUH Co-Project of Key Research (KMU-DK109001), Research Foundation (KMU-DK109004) from Kaohsiung Medical University, Taiwan. This study is also supported partially by Kaohsiung Medical University Research Center Grant (Drug Development and Value Creation Research Center) (KMU-TC108A03).Author contributions
B.-C. H. and Y.-C. L. contributed equally to this work. T.-L. C., B.-C. H., Y.-C. L., J.-M. L. and S.-T. H. designed the research, B.-C. H., Y.-C. L., S.-T. H., H.-J. C., T.-Y. L. and K.-W. H. performed the research and analyzed the data, B.-C. H., Y.-C. L. and H.-J. L. wrote the paper. Y.-C. H., C.-H. C., Y.-T. W. and T.-L. C assisted in eliminating problems during the experiments and provided suggestions for the manuscript.Conflicts of interest
T.-L. C., C.-H. C. and Y.-C. L. are listed as inventors on patents [U.S. application number: 14/893,509] related to the technology described in this work. The other authors declare that they have no competing interests.References
- J. A. Seidel, A. Otsuka and K. Kabashima, Anti-PD-1 and Anti-CTLA-4 Therapies in Cancer: Mechanisms of Action, Efficacy, and Limitations, Front. Oncol., 2018, 8, 86 CrossRef PubMed.
- X. Qi, F. Li, Y. Wu, C. Cheng, P. Han, J. Wang and X. Yang, Optimization of 4-1BB antibody for cancer immunotherapy by balancing agonistic strength with FcγR affinity, Nat. Commun., 2019, 10(1), 2141 CrossRef PubMed.
- M. Fakih and R. Wong, Efficacy of the monoclonal antibody EGFR inhibitors for the treatment of metastatic colorectal cancer, Curr. Oncol., 2010, 17(suppl. 1), S3–S17 CrossRef.
- M. E. Weinblatt, P. J. Maddison, K. J. Bulpitt, B. L. Hazleman, M. B. Urowitz, R. D. Sturrock, J. S. Coblyn, A. L. Maier, W. R. Spreen and V. K. Manna, et al., CAMPATH-1H, a humanized monoclonal antibody, in refractory rheumatoid arthritis. An intravenous dose-escalation study, Arthritis Rheumatol., 1995, 38(11), 1589–1594 CrossRef CAS.
- J. Nixon, P. Newbold, T. Mustelin, G. P. Anderson and R. Kolbeck, Monoclonal antibody therapy for the treatment of asthma and chronic obstructive pulmonary disease with eosinophilic inflammation, Pharmacol. Ther., 2017, 169, 57–77 CrossRef CAS PubMed.
- I. Cludts, F. R. Spinelli, F. Morello, J. Hockley, G. Valesini and M. Wadhwa, Anti-therapeutic antibodies and their clinical impact in patients treated with the TNF antagonist Adalimumab, Cytokines, 2017, 96, 16–23 CrossRef CAS PubMed.
- S. Vermeire, M. Noman, G. Van Assche, F. Baert, K. Van Steen, N. Esters, S. Joossens, X. Bossuyt and P. Rutgeerts, Autoimmunity associated with anti-tumor necrosis factor α treatment in Crohn's disease: a prospective cohort study, Gastroenterology, 2003, 125(1), 32–39 CrossRef CAS.
- T. T. Hansel, H. Kropshofer, T. Singer, J. A. Mitchell and A. J. T. George, The safety and side effects of monoclonal antibodies, Nat. Rev. Drug Discovery, 2010, 9(4), 325–338 CrossRef CAS PubMed.
- K. C. Kähler, J. C. Hassel, L. Heinzerling, C. Loquai, R. Mössner, S. Ugurel, L. Zimmer and R. Gutzmer, Management of side effects of immune checkpoint blockade by anti-CTLA-4 and anti-PD-1 antibodies in metastatic melanoma, J. Dtsch. Dermatol. Ges., 2016, 14(7), 662–681 Search PubMed.
- J. Pol and G. Kroemer, Anti-CTLA-4 immunotherapy: uncoupling toxicity and efficacy, Cell Res., 2018, 28(5), 501–502 CrossRef CAS PubMed.
- R. Bajwa, A. Cheema, T. Khan, A. Amirpour, A. Paul, S. Chaughtai, S. Patel, T. Patel, J. Bramson, V. Gupta, M. Levitt, A. Asif and M. A. Hossain, Adverse Effects of Immune Checkpoint Inhibitors (Programmed Death-1 Inhibitors and Cytotoxic T-Lymphocyte-Associated Protein-4 Inhibitors): Results of a Retrospective Study, J. Clin. Med. Res., 2019, 11(4), 225–236 CrossRef CAS PubMed.
- I. Ferreira and D. Isenberg, Vaccines and biologics, Ann. Rheum. Dis., 2014, 73(8), 1446 CrossRef CAS PubMed.
- L. K. Mercer, J. Askling, P. Raaschou, W. G. Dixon, L. Dreyer, M. L. Hetland, A. Strangfeld, A. Zink, X. Mariette, A. Finckh, H. Canhao, F. Iannone, J. Zavada, J. Morel, J.-E. Gottenberg, K. L. Hyrich and J. Listing, Risk of invasive melanoma in patients with rheumatoid arthritis treated with biologics: results from a collaborative project of 11 European biologic registers, Ann. Rheum. Dis., 2017, 76(2), 386 CrossRef PubMed.
- S. Mori and S. Fujiyama, Hepatitis B virus reactivation associated with antirheumatic therapy: risk and prophylaxis recommendations, World J. Gastroenterol., 2015, 21(36), 10274–10289 CrossRef CAS PubMed.
- A. Bettiol, G. Lopalco, G. Emmi, L. Cantarini, M. L. Urban, A. Vitale, N. Denora, A. Lopalco, A. Cutrignelli, A. Lopedota, V. Venerito, M. Fornaro, A. Vannacci, D. Rigante, R. Cimaz and F. Iannone, Unveiling the Efficacy, Safety, and Tolerability of Anti-Interleukin-1 Treatment in Monogenic and Multifactorial Autoinflammatory Diseases, Int. J. Mol. Sci., 2019, 20(8), 1898 CrossRef CAS PubMed.
- D. E. Johnson, Biotherapeutics: Challenges and Opportunities for Predictive Toxicology of Monoclonal Antibodies, Int. J. Mol. Sci., 2018, 19(11), 3685 CrossRef PubMed.
- P. Chames, M. Van Regenmortel, E. Weiss and D. Baty, Therapeutic antibodies: successes, limitations and hopes for the future, Br. J. Pharmacol., 2009, 157(2), 220–233 CrossRef CAS.
- W. W. Lin, Y. C. Lu, C. H. Chuang and T. L. Cheng, Ab locks for improving the selectivity and safety of antibody drugs, J. Biomed. Sci., 2020, 27(1), 76 CrossRef CAS.
- L. Desnoyers, O. Vasiljeva, J. Richardson, A. Yang, E. Menendez, T. Liang, C. Wong, P. Bessette, K. Kamath, S. Moore, J. Sagert, D. Hostetter, F. Han, J. Gee, J. Flandez, K. Markham, M. Nguyen, M. Krimm, K. Wong and H. Lowman, Tumor-Specific Activation of an EGFR-Targeting Probody Enhances Therapeutic Index, Sci. Transl. Med., 2013, 5, 207ra144 Search PubMed.
- J. M. Donaldson, C. Kari, R. C. Fragoso, U. Rodeck and J. C. Williams, Design and development of masked therapeutic antibodies to limit off-target effects: application to anti-EGFR antibodies, Cancer Biol. Ther., 2009, 8(22), 2147–2152 CrossRef CAS PubMed.
- V. H. Trang, X. Zhang, R. C. Yumul, W. Zeng, I. J. Stone, S. W. Wo, M. M. Dominguez, J. H. Cochran, J. K. Simmons, M. C. Ryan, R. P. Lyon, P. D. Senter and M. R. Levengood, A coiled-coil masking domain for selective activation of therapeutic antibodies, Nat. Biotechnol., 2019, 37(7), 761–765 CrossRef CAS PubMed.
- Y. C. Lu, C. H. Chuang, K. H. Chuang, I. J. Chen, B. C. Huang, W. H. Lee, H. E. Wang, J. J. Li, Y. A. Cheng, K. W. Cheng, J. Y. Wang, Y. C. Hsieh, W. W. Lin and T. L. Cheng, Specific activation of pro-Infliximab enhances selectivity and safety of rheumatoid arthritis therapy, PLoS Biol., 2019, 17(6), e3000286 CrossRef PubMed.
- W. K. Wong, J. Leem and C. M. Deane, Comparative Analysis of the CDR Loops of Antigen Receptors, Front. Immunol., 2019, 10, 2454 CrossRef CAS PubMed.
- I. Sela-Culang, V. Kunik and Y. Ofran, The structural basis of antibody-antigen recognition, Front. Immunol., 2013, 4, 302 Search PubMed.
- R. J. Read, P. D. Adams, W. B. Arendall 3rd, A. T. Brunger, P. Emsley, R. P. Joosten, G. J. Kleywegt, E. B. Krissinel, T. Lütteke, Z. Otwinowski, A. Perrakis, J. S. Richardson, W. H. Sheffler, J. L. Smith, I. J. Tickle, G. Vriend and P. H. Zwart, A new generation of crystallographic validation tools for the protein data bank, Structure, 2011, 19(10), 1395–1412 CrossRef CAS PubMed.
- S. Das, S. Ramakumar and D. Pal, Identifying functionally important cis-peptide containing segments in proteins and their utility in molecular function annotation, FEBS J., 2014, 281(24), 5602–5621 CrossRef CAS PubMed.
- A. P. Joseph, N. Srinivasan and A. G. de Brevern, Cis-trans peptide variations in structurally similar proteins, Amino Acids, 2012, 43(3), 1369–1381 CrossRef CAS PubMed.
- A. Jabs, M. S. Weiss and R. Hilgenfeld, Non-proline cis peptide bonds in proteins, J. Mol. Biol., 1999, 286(1), 291–304 CrossRef CAS.
- W. Humphrey, A. Dalke and K. Schulten, VMD: visual molecular dynamics, J. Mol. Graphics Modell., 1996, 14(1), 33–38 CrossRef CAS.
- D. A. Case, et al., The Amber biomolecular simulation programs, J. Comput. Chem., 2005, 26(16), 1668–1688 CrossRef CAS.
- A. V. Onufriev and D. A. Case, Generalized Born Implicit Solvent Models for Biomolecules, Annu. Rev. Biophys., 2019, 48(1), 275–296 CrossRef CAS PubMed.
- J. A. Maier, et al., ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB, J. Chem. Theory Comput., 2015, 11(8), 3696–3713 CrossRef CAS PubMed.
- J.-P. Ryckaert, G. Ciccotti and H. J. C. Berendsen, Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes, J. Comput. Phys., 1977, 23(3), 327–341 CrossRef CAS.
- A. Bhattarai and Y. Miao, Gaussian accelerated molecular dynamics for elucidation of drug pathways, Expert Opin. Drug Discovery, 2018, 13(11), 1055–1065 CrossRef CAS PubMed.
- Y. Miao, V. A. Feher and J. A. McCammon, Gaussian Accelerated Molecular Dynamics: Unconstrained Enhanced Sampling and Free Energy Calculation, J. Chem. Theory Comput., 2015, 11(8), 3584–3595 CrossRef CAS PubMed.
- C. Regep, G. Georges, J. Shi, B. Popovic and C. M. Deane, The H3 loop of antibodies shows unique structural characteristics, Proteins, 2017, 85(7), 1311–1318 CrossRef CAS PubMed.
- Y. Avnir, A. S. Tallarico, Q. Zhu, A. S. Bennett, G. Connelly, J. Sheehan, J. Sui, A. Fahmy, C. Y. Huang, G. Cadwell, L. A. Bankston, A. T. McGuire, L. Stamatatos, G. Wagner, R. C. Liddington and W. A. Marasco, Molecular Signatures of Hemagglutinin Stem-Directed Heterosubtypic Human Neutralizing Antibodies against Influenza A Viruses., PLoS Pathog., 2014, 10(5), e1004103 CrossRef.
- T. van den Beucken, H. Pieters, M. Steukers, M. van der Vaart, R. C. Ladner, H. R. Hoogenboom and S. E. Hufton, Affinity maturation of Fab antibody fragments by fluorescent-activated cell sorting of yeast-displayed libraries, FEBS Lett., 2003, 546(2–3), 288–294 CrossRef CAS.
- M. Godar, V. Morello, A. Sadi, A. Hultberg, N. De Jonge, C. Basilico, V. Hanssens, M. Saunders, B. N. Lambrecht, M. El Khattabi, H. de Haard, P. Michieli and C. Blanchetot, Dual anti-idiotypic purification of a novel, native-format biparatopic anti-MET antibody with improved in vitro and in vivo efficacy, Sci. Rep., 2016, 6(1), 31621 CrossRef CAS.
- G. M. Bartelds, C. L. M. Krieckaert, M. T. Nurmohamed, P. A. van Schouwenburg, W. F. Lems, J. W. R. Twisk, B. A. C. Dijkmans, L. Aarden and G. J. Wolbink, Development of Antidrug Antibodies Against Adalimumab and Association With Disease Activity and Treatment Failure During Long-term Follow-up, J. Am. Med. Assoc., 2011, 305(14), 1460–1468 CrossRef CAS PubMed.
- V. Ruiz Garcia, A. Burls, J. B. Cabello, P. Vela Casasempere, S. Bort-Marti and J. A. Bernal, Certolizumab pegol (CDP870) for rheumatoid arthritis in adults, Cochrane Database Syst. Rev., 2017, 9(9), CD007649 Search PubMed.
- J. Qu, X. Zhao, X. Liu, Y. Sun, J. Wang, L. Liu, J. Wang and J. Zhang, Natriuretic peptide receptor a promotes breast cancer development by upregulating MMP9, Am. J. Cancer Res., 2019, 9(7), 1415–1428 CAS.
- K. Y. Choi, M. Swierczewska, S. Lee and X. Chen, Protease-Activated Drug Development, Theranostics, 2012, 2(2), 156–178 CrossRef CAS PubMed.
- T. Duong le, A. T. Leung and B. Langdahl, Cathepsin K Inhibition: A New Mechanism for the Treatment of Osteoporosis, Calcif. Tissue Int., 2016, 98(4), 381–397 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Table S1: development of various pro-Abs with different blocking fold. We identified good candidate pro-Abs that achieved ≥100-fold blocking with targeted Ags including pro-Infliximab, anti-hu-CD19, PD-1, IL-1β, CTLA-4, mouse PD-1 and mouse CD3 Ab. In contrast, the bad pro-Abs did not reach 100-fold such as anti-mouse CTLA-4, human EGFR and CD52 Ab. See DOI: 10.1039/d1sc01748a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |