Open Access Article
Open Access ArticleA facile combinatorial approach to construct a ratiometric fluorescent sensor: application for the real-time sensing of cellular pH changes†
Eiji
Nakata
 a,
Hisaaki
Hirose
a,
Hisaaki
Hirose
 b,
Khongorzul
Gerelbaatar
a,
Jan Vincent V.
Arafiles
b,
Khongorzul
Gerelbaatar
a,
Jan Vincent V.
Arafiles
 b,
Zhengxiao
Zhang
a,
Shiroh
Futaki
b,
Zhengxiao
Zhang
a,
Shiroh
Futaki
 b and
Takashi
Morii
b and
Takashi
Morii
 *a
*a
aInstitute of Advanced Energy, Kyoto University, Kyoto, Japan. E-mail: t-morii@iae.kyoto-u.ac.jp
bInstitute for Chemical Research, Kyoto University, Kyoto, Japan
First published on 8th May 2021
Abstract
Realtime monitoring of the cellular environment, such as the intracellular pH, in a defined cellular space provides a comprehensive understanding of the dynamics processes in a living cell. Considering the limitation of spatial resolution in conventional microscopy measurements, multiple types of fluorophores assembled within that space would behave as a single fluorescent probe molecule. Such a character of microscopic measurements enables a much more flexible combinatorial design strategy in developing fluorescent probes for given targets. Nanomaterials with sizes smaller than the microscopy spatial resolution provide a scaffold to assemble several types of fluorophores with a variety of optical characteristics, therefore providing a convenient strategy for designing fluorescent pH sensors. In this study, fluorescein (CF) and tetramethylrhodamine (CR) were assembled on a DNA nanostructure with controlling the number of each type of fluorophore. By taking advantage of the different responses of CF and CR emissions to the pH environment, an appropriate assembly of both CF and CR on DNA origami enabled a controlled intensity of fluorescence emission and ratiometric pH monitoring within the space defined by DNA origami. The CF and CR-assembled DNA origami was successfully applied for monitoring the intracellular pH changes.
Introduction
Spatial and temporal dynamics of biologically important molecules and ions play central roles in controlling the specific activities inside the cell. Such specific activities are also regulated by local variations1,2 such as pH3–5 and temperature.6 Therefore, real time assessment of such physical parameters is critical to characterize the specific biological activity inside the cell.7 To assess the real time dynamics of these physical parameters in a single cell, target-specific fluorescent probes in combination with fluorescence microscopy have proven to be one of the most powerful techniques.8–14 Application of spectrally distinguishable fluorescent probes further enables simultaneous detection and identification of various factors at the same time. The fluorescent probe changes its optical properties in response to binding with the target molecule, a chemical reaction, and changes of environmental parameters. Various kinds of fluorescent probes, such as fluorescent protein-based probes, organic molecular probes, or inorganic fluorescent nanoparticular probes, were developed. While these fluorescent probes have been successfully applied to visualize various types of biological processes,7–14 including pH change,15–19 there is still room for improvement for their application in the fluorescence microscopy measurements. In particular, a fluorescent probe with high emission intensity at a given monitoring wavelength and emission properties suitable for ratiometric monitoring are in demand for fluorescence microscopy measurements.20–28Considering the limitation of spatial resolution in conventional microscopy measurements, a combinatorial design strategy of assembling multiple types of fluorophores within a defined space provides much more flexibility in developing fluorescent probes for given targets. Assembling an appropriate number of the same type of fluorophore within a space of 100 × 100 nm would increase the emission intensity at the resolution limit.29–31 Likewise, a combination of two types of fluorophores within a defined space would enable the ratiometric monitoring of the target within the limited space. Thus, a combinatorial assembly of fluorophores on nanomaterials with a size below the spatial resolution of microscopy potentially provides fluorescent probes with optical characteristics that are tunable on demand by selecting appropriate fluorescent probes. However, most of the nanomaterials used to assemble multiple organic fluorescent probes to date27,32–34 are not suited for precise control of the number and the spatial arrangement of the fluorophores. The distance and polarity of the fluorophores should be restricted to prevent undesirable steric and/or optical interactions between them. The 2D and 3D DNA nanostructures with precisely controlled geometry, such as the ones constructed by DNA origami,35 are ideal scaffolds for spatially organizing a defined number of functional molecules including fluorophores with nanometer precision by taking advantage of their sequence-driven programmability.36–39 While the brightness and characteristics of the multiple fluorophores assembled on a single DNA scaffold are tuned by controlling the number and types of the fluorophores,40,41 application of such fluorophore assemblies for the fluorescent probe has not been well explored. For this purpose, optical responses of the assembled fluorophores to a given target should be characterized and evaluated quantitatively by comparing with the responses by corresponding free fluorophores.
Here, we report a ratiometric fluorescent pH probe prepared by assembling two types of fluorophores on a DNA nanostructure. Based on the optical properties of each fluorophore, optical responses of assembled fluorophores were verified for the ratiometric pH probe. As a proof of principle, the constructed ratiometric fluorescent pH probe, termed as a pH endoscope, was applied to monitor its internalization process to the cell by fluorescence microscope measurements. We identified that the pH endoscope was internalized through macropinocytosis. Furthermore, real time pH changes inside macropinosome,42 a pinosome formed during the macropinocytosis, were monitored by using the pH endoscope.
Results
Design of fluorophore assembled DNA scaffolds
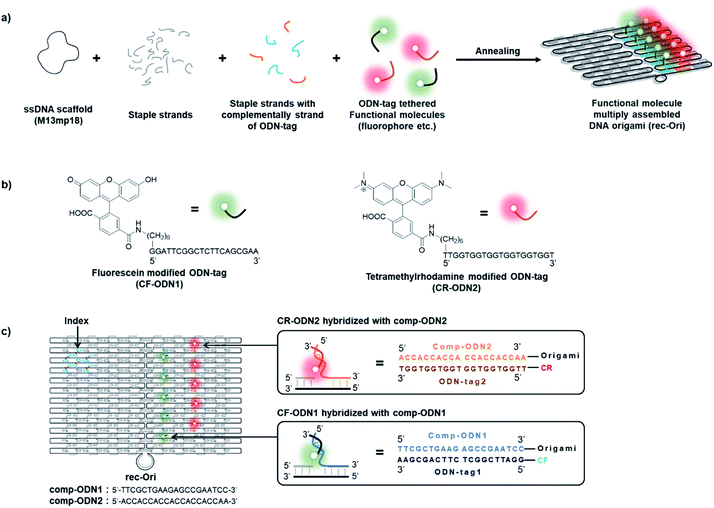
A two-dimensional (2D) rectangular DNA origami consisting of 24 parallel DNA double helices with dimensions of 90 × 60 nm (rec-Ori) was adapted as a scaffold to assemble fluorophores (Fig. 1b and S1†).35,43 In DNA origami, a long single-strand DNA (ssDNA scaffold) is folded with over 200 short single-strand DNA (staple strands) into programmable shapes (Fig. 1a). Each staple strand has a defined sequence to hybridize at a unique location of the ssDNA scaffold to fold it to a designed shape. A fluorophore was tethered to a specific single stranded DNA (ODN-tag) that hybridizes to its complementary sequence of ODN-tag (comp-tag) (Fig. 1b). To a staple strand for a given position to locate the fluorophore, comp-tag was appended either in the 5′- or 3′- direction. Alternatively, the fluorophore was directly modified at the 5′-end of the staple strand for the given location (see Note S1†). Among the methods, comp-tag appended to the 5′-end of the staple strand (Fig. 1c) was adapted to assemble a fluorophore on the DNA scaffold as described in Note S1.† The position of each fluorophore was designed to ensure that there was enough distance (more than 11 nm) between each fluorophore to prevent possible steric or optical interferences between them (Fig. 1c and S1†).35 A pH sensitive fluorophore 6-carboxyfluorescein (CF) with pKa of 6.5 (ref. 15) and 6-carboxytetramethylrhodamine (CR), a pH insensitive fluorophore under the physiological conditions, were chosen to construct a ratiometric pH probe on the DNA scaffold (rec-Ori). CF and CR were labelled at the 5′-ends of orthogonal 20-nt ODN-tags, termed ODN-tag1 and ODN-tag2, respectively (Fig. 1b and S2†). The fluorophore-modified rec-Ori was synthesized, purified and characterized as described in the Experimental procedures section.Tuning the number of fluorophore molecules on the DNA origami scaffold
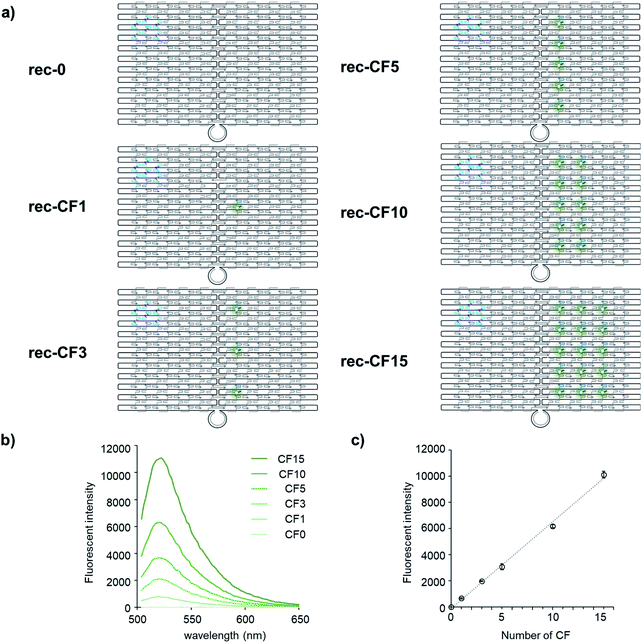
Optical properties of each of the fluorophores CF and CR on rec-Ori in response to the assembled number was evaluated. A series of rec-Ori assembled with various numbers of CF, in the range from 0 to 15 molecules on a single scaffold, were prepared as illustrated in Fig. 2a and S3.† Upon the assembly and purification (Fig. S3†), the fluorescence intensity of CF for each rec-Ori at the same concentration of rec-Ori was measured (Fig. 2b) and plotted against the number of CF on rec-Ori. A linear relationship between the concentration and the emission intensity of CF for rec-CF1, rec-CF3 rec-CF5, rec-CF10 and rec-CF15 (Fig. 2c) indicates that the brightness of the confined space of 90 × 60 nm is tunable for CF loaded rec-Ori by changing the number of CF.41 The same results were confirmed in the case of a series of rec-Ori assembled with various numbers of CR (Fig. S4†).Ratiometric detection of pH changes by rec-Ori assembled with two types of fluorophores
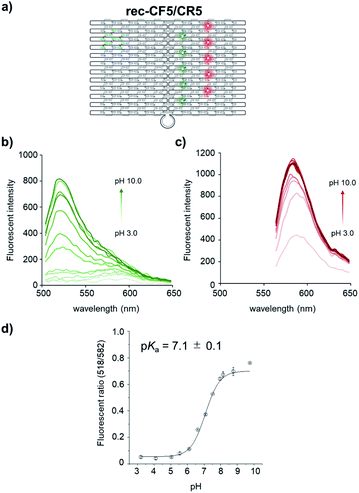
Next, the fluorescence emission properties of CF assembled rec-Ori were investigated at various pH. The pH insensitive 6-carboxytetramethylrhodamine (CR) was co-assembled on rec-Ori as an internal standard for the fluorescence intensity of CF, thus enabling evaluation of the ratio of fluorescence intensities of CF and CR within the confined space. Five molecules of each of CF and CR were assembled on rec-Ori, termed as rec-CF5/CR5 (Fig. 3a and S5†), in a similar manner to that described for rec-CF5 (Fig. S3†). Fluorescence emission intensities of CF and CR on rec-CF5/CR5 were measured at various pH in the range from pH 3 to 10 (Fig. 3b and c). The fluorescence emission of CF showed a gradual increase in intensity at the emission maxima as the pH increased from 3 to 10 (Fig. 3b). On the other hand, that of CR did not change in the pH range of 5 to 10 with a slight decrease in the pH range of 3 to 5 (Fig. 3c). A plot for the ratio of emission intensity of rec-CF5/CR5 at 518 nm excited at 470 nm, corresponding to CF, to that at 582 nm excited at 530 nm, corresponding to CR, at each pH is shown in Fig. 3d. The plot gave an estimation of pKa for CF on rec-CF5/CR5 to be 7.1 ± 0.1. The estimated value was consistent with the pKa of CF modified single strand DNA (CF-ODN1) (pKa of 6.9 ± 0.1) and that of the duplex form of CF-ODN1 with comp-ODN1 (pKa of 7.1 ± 0.1), though these values shifted to the basic side as compared with the 2-aminoethanol modified fluorescein derivative (CF-AE, pKa of 6.3 ± 0.1) (Fig. S6†). The observed pKa shift likely resulted from destabilization of the dianionic form of CF by the negative charge of the DNA phosphate back bone. Stability of rec-CF5/CR5 within the pH range tested was confirmed by gel electrophoreses and AFM images of the sample treated at each pH (Fig. S7†). The well-formed structure of rec-Ori was identified to the same extent within the pH range of 4 to 10 in AFM images and gel electrophoresis. Mobility of the band corresponding to rec-Ori in gel electrophoresis remained constant for all the samples treated at the pH in the range from 4 to 10. On the other hand, the band corresponding to rec-Ori disappeared in gel electrophoresis for the samples treated at pH 3 (Fig. S7a and b†). Likewise, little well-formed rec-Ori was observed in AFM images for the samples treated at pH 3 (Fig. S7c†). These results indicated that the rec-Ori coassembled with CF and CR maintained its structure and responded to pH changes in the ratiometric manner from pH 5 to 10.Ratiometric detection of the pH change by rec-CF5/CR5 in the presence of a macropinocytosis inducible peptide
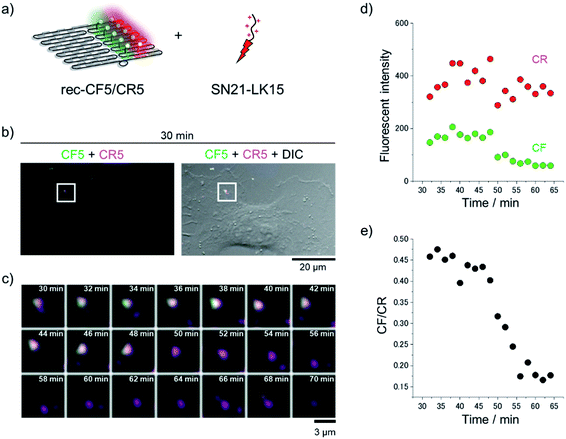
The ratiometric pH response of rec-CF5/CR5 was further verified in the presence of a fusion peptide of a macropinocytosis-inducing peptide SN21 and a cationic amphiphilic peptide LK15 (SN21-LK15),44 which has been shown to enhance the cellular uptake of DNA. Macropinocytosis is a form of endocytosis accompanied by a massive uptake of extracellular fluid and solutes.42 Considering the contribution of macropinocytosis in the cellular uptake of various nanomaterials, viruses, and peptides/proteins, this pathway would provide a promising route for delivery of bioactive molecules into cells.45 To understand the conditions within endosomes of this pathway (i.e., macropinosomes), rec-CF5/CR5 was applied to evaluate the pH change in macropinosomes. While LK15 is considered to perturb endosomal membranes in the original report,44 this peptide segment is expected to decorate SN21 on rec-CF5/CR5 to facilitate its endosomal encapsulation. In the presence of SN21-LK15 at the charge ratio 3, an optimized condition for cellular application as described later and in Fig. S11,† the fluorescence emission of rec-CF5/CR5 originating from CF was gradually increased from pH 3 to 10 in a similar manner to the condition in the absence of SN21-LK15 (Fig. S8†). Little or no change was observed for the fluorescence emission corresponding to CR under the same conditions. From the plot for fluorescence emission ratio (518/582) against each monitored pH (Fig. S8d†), the pKa value of rec-CF5/CR5 in the presence of SN21-LK15 was estimated to be 5.9 ± 0.1 at ambient temperature. Compared to the estimated pKa in the absence of SN21-LK15 (7.1 ± 0.1) (Fig. 3d), the pKa value of rec-CF5/CR5 was shifted to the acidic side in the presence of SN21-LK15, possibly because of neutralization of the negatively charged surface of DNA origami scaffold through the interaction with positively charged SN21-LK15. These results indicated that the changes of pH from the neutral pH around 7.0 to the acidic pH around 5.0 were successfully monitored by rec-CF5/CR5 in a ratiometric manner either in its free form or in the complex with SN21-LK15.Cellular uptake of DNA scaffold rec-CR5
A DNA scaffold assembled with five molecules of CR (rec-CR5, Fig. S9†), which revealed little or no change in the emission within the pH range of 5 to 10 (Fig. S10†), was first used as a marker for tracing the cellular internalization process of the fluorophore modified DNA scaffold. In order to enhance the cellular uptake of DNA origami scaffold, SN21-LK15 was mixed with rec-CR5 at various charge ratios between rec-CR5 and SN21-LK15. Internalization of rec-CR5 was also investigated in the absence of SN21-LK15 and in the presence of lipofectamine 2000 (LF2000), a widely-used gene delivery reagent. HeLa cells treated with rec-CR5 were analyzed by confocal microscopy after washing the cells (Fig. S11†). The cells treated with rec-CR5 alone revealed little or no punctates with fluorescence, indicating that rec-CR5 by itself was not taken up by the cell efficiently. On the other hand, endosome-like punctates emitting characteristic fluorescence of CR were observed in the cells treated with the complex of rec-CR5 and SN21-LK15 at all the charge ratios. Among them, the cells treated by the complex with a charge ratio of 3 revealed confocal microscopy images with strong fluorescence emission and well distributed punctates. Cells treated with the complex in the charge ratio of 1 showed large fluorescent punctates that could have resulted from the aggregation of negatively charged rec-CR5 and positively charged SN21-LK15. Treatment of cells by the complex with a charge ratio of 5 resulted in weak fluorescence emission in dispersed punctates with certain toxicity to the cells. These results indicated that the complex of SN21-LK15 and rec-CR5 with the charge ratio of 3 was suitable for monitoring the fluorescence emission of endosome-like punctates in the cell.The cellular uptake process of rec-CR5 was investigated by confocal microscopy (Fig. 4). The cells were treated with the rec-CR5/SN21-LK15 complex with the charge ratio of 3 (Fig. 4a), FITC conjugated dextran (FITC-dextran, 70 kDa) and LysoTracker™ Deep Red (LysoDR). FITC-dextran is known as a representative macropinosome marker.46 LysoDR is a deep red fluorescent dye for labelling and tracking acidic organelles such as lysosomes in live cells. After incubation for 40 min, the fluorescence localization corresponding to each dye in the cells was observed by confocal microscopy (Fig. 4b and c). The internalized rec-CR5 colocalized well with FITC-dextran and occasionally with LysoDR (Fig. 4c). Images from time lapse monitoring of rec-CR5 and LysoDR (Fig. 4d and S12†) provided further insights on the internal localization of rec-CR5. The fluorescent punctates corresponding to rec-CR5 marked in Fig. 4d migrated alone until 66 min. At 74 min, the fluorescent punctates colocalized with LysoDR and after that they migrated together within the same area, suggesting that the macropinosome including rec-CR5 was fused to lysosome labeled by LysoDR. These results indicated that the complex of rec-CR5 and macropinocytosis-inducible peptide SN21-LK15 was taken up by the cell through macropinocytosis, and then trapped into macropinosome, and subsequently the macropinosome was fused to lysosome. If this is the case, pH changes from the pH inside macropinosome to the one inside lysosome would be monitored by the pH endoscope rec-CF5/CR5.
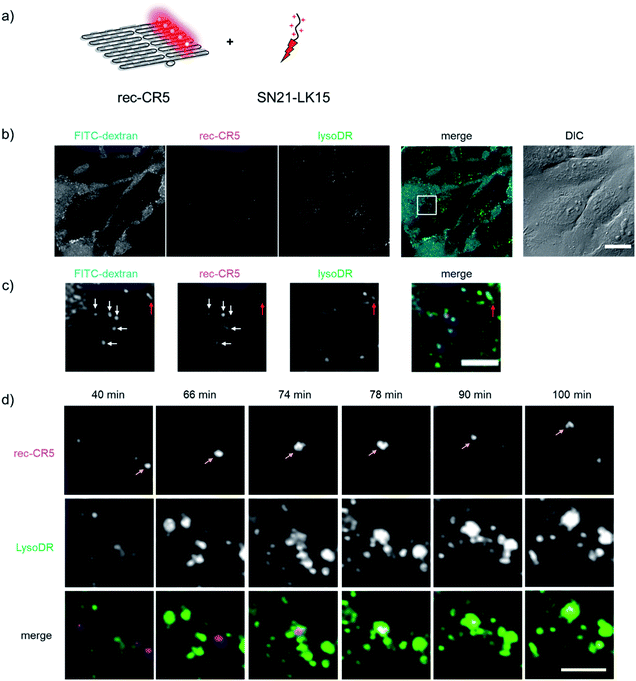 | ||
| Fig. 4 Identification of the internalized rec-CR5 treated with SN21-LK15 (rec-CR5/SN21-LK15). (a) An illustration of rec-CR5 treated with SN21-LK15. (b) Confocal microscopy images of HeLa cells treated with rec-CR5/SN21-LK15 (magenta), FITC-dextran (cyan), and LysoDR (green). The DIC image includes a scale bar (20 μm). (c) Expanded image of the square in the merged image of (b). The arrows indicate FITC-dextran, rec-CR5, or LysoDR colocalized in the same area (white arrows: FITC-dextran and rec-CR5, red arrows: FITC-dextran, rec-CR5 and LysoDR). The merged image (green: FITC-dextran, red: rec-CR5, blue: LysoDR) includes a scale bar (5 μm). (d) Time lapse images of rec-CR5 (magenta) and LysoDR (green) from 40 min to 100 min after addition of rec-CR5 complexed with SN21-LK15. The merged image (magenta: rec-CR5, green: LysoDR) includes a scale bar (5 μm). The full image is shown in Fig. S12.† | ||
Ratiometric detection of pH changes during the internalization process of the DNA scaffold
Finally, dynamic monitoring of the pH change during macropinocytosis was investigated. A complex of rec-CF5/CR5 and SN21-LK15 with the charge ratio of 3 was incubated with HeLa cells for 30 min, and then time-lapse imaging was carried out by confocal microscopy. In the time-lapse images (Fig. 5b and c), fluorescent endosome-like punctates revealed emissions corresponding to both CF and CR. While the fluorescence emission originating from CR remained constant during the observation, the fluorescence emission corresponding to CF was drastically reduced at 50 min (Fig. 5c). The time-course change of emission intensity originating from CF for an endosome (Fig. 5c) was plotted with that from CR (Fig. 5d). These data for time-course of the emission intensity for CF and CR were utilized to estimate the time-course changes in the emission intensity ratios (518/582) as shown in Fig. 5e. By comparing with the calibration plot (Fig. S13a†), the ratiometric changes (518/582) corresponded to the pH changes from 7.0 to 5.6 (Fig. S13b†).Discussion
The conventional fluorescence microscopy has a limitation in resolution known as the diffraction limit of light,47 which could be overcome by applying the super-resolution microscopy system. Due to the limitation, individual molecules existing within the space below the spatial resolution of microscopy, e.g. excitation wavelength at 500 nm limits at most 250 nm of spatial resolution, could not be distinguished by means of conventional fluorescence microscopy.29 Due to this limitation in resolution, an individual fluorescent probe on a nanomaterial with the size of 100 nm will not be spatially distinguished in the conventional fluorescence microscopy measurements. Therefore, a combinatorial assembly of multiple fluorophores within a defined space provides a flexible strategy in developing fluorescent probes for given targets with desired properties, such as emission intensity, color, size, and affinity to the target in combination with the fluorescence microscopy analyses.41 While several reports have pointed out the possible advantage of tunable optical properties using the DNA origami scaffold,40,41,43 to our knowledge, this is the first report to evaluate the emission properties of multiple fluorophores assembled on a DNA origami scaffold.Though DNA origami is one of the ideal scaffolds for assembling a defined number of various functional molecules, only a few applications of fluorophore-assembled DNA origami scaffolds were reported for monitoring intracellular events.48,49 One of the possible reasons for the limited application is the low cell permeability of the DNA scaffold.50 Recently, several examples reported that the DNA origami scaffold could be internalized into the cell and localized in lysosomes after incubation for a long time (e.g. 12 h),48–52 however, these events were highly dependent on the structure, size and compactness of DNA nanostructures.52 As shown in Fig. 4b, the cells treated by CR assembled DNA origami alone did not show a significant fluorescence emission inside the cell. An efficient cellular uptake of DNA origami alone was not observed at least in 1 h incubation.
The macropinocytosis-inducible peptide SN21-LK15 was adapted to improve the cellular uptake efficiency of DNA origami (Fig. S11†). As reported previously,44 it is known that the efficiency of cellular uptake of DNA can be improved by the assistance of various transfection reagents, such as lipofectamine,53 PEI48 and cationic polymer54 and/or targeting ligands or aptamer to interact with the proteins or protein receptors on the cell surface. SN21-LK15 also contains several positively charged amino acid residues that strongly interact with the DNA origami scaffold and neutralize its highly negative charge. The SN21-LK15 assisted efficient cellular uptake of the DNA origami scaffold was observed with a charge ratio of 3, where the complex was positively charged to accelerate the electrostatic interaction with the negatively charged cell surface. Such considerations were strongly supported by the pKa shift of CF as described in Fig. 3, S6 and S8.† The pKa of CF on the DNA scaffold alone (7.1 ± 0.1) was shifted to the basic side compared with CF-AE (6.3 ± 0.1), which represented the standard pKa of CF. On the other hand, the pKa of CF on the DNA scaffold with SN21-LK15 was shifted to the acidic side (5.9 ± 0.1). These results suggested a positively charged environment for CF on the DNA origami scaffold complexed with SN21-LK15 at the charge ratio 3.
Interestingly, not only the efficiency of the cellular uptake, but also the stability of DNA origami scaffold at acidic pH was enhanced in the presence of SN21-LK15. CR is known as the pH insensitive fluorophore. Actually, the emission spectra of CR-TE remain unchanged from pH 3 to pH 10 (Fig. S10a†). The emission spectra of CR modified with a single strand DNA (CR-ODN2) are almost constant at the same pH range (Fig. S10b†), whereas those of CR modified with a double strand DNA (CR-dODN2) and with the DNA origami scaffold (rec-CR) show a decrease in the emission intensity at the pH range of 3 to 5 in the absence of SN21-LK15 (Fig. S10†). The reduction in emission intensity likely resulted from the dissociation of duplex and the instability of the DNA origami scaffold at pH 4 as reported previously.55–57 On the other hand, in the presence of SN21-LK15, such decrements of emission intensity for CR on rec-CR5 were not observed above pH 3 (Fig. S8d†). The local pH shift on rec-Ori in the complex of rec-CR5 and SN21-LK15, as evident from the observed pKa shift mentioned above, is the likely factor for the possible stabilization of the DNA origami scaffold at pH below 5.
The internalized rec-CR5 treated with SN21-LK15 co-localized well with FITC-dextran (70 kDa) in the initial stage and later co-localized with LysoDR. Our observation suggests that the complex of rec-CR5 and SN21-LK15 was taken up by the cell through macropinocytosis, trapped into macropinosome, and fused to lysosome. Therefore, the observed ratiometric fluorescence emission changes of rec-CF5/CR5 with SN21-LK15 during its cellular internalization likely reflect the pH gradient of the endocytosis pathway from early endosome to lysosome. It is known that the pH gradient follows the endocytosis pathway as ∼6.3 in early endosomes, ∼5.5 in late endosomes, and ∼4.6 lysosomes.4,58,59 In this study, the estimated pH change of macropinosome is in the range from pH 7.0 to pH 5.6, which is consistent with the previous reports.4,58,59
Conclusion
We have investigated a combinatorial strategy to design a ratiometric fluorescent pH probe, termed as a pH endoscope, by assembling two types of fluorophores on a DNA nanostructure. Assembly of appropriate numbers of CF and CR on a rectangular DNA origami scaffold provided a fluorescent pH probe with increased emission intensity and enabled the ratiometric detection of pH. When the pH was changed, CF showed the expected fluorescence intensity change in response to the changes in pH, while CR emitted with almost constant fluorescence intensity within the pH ranging from 5 to 10. The efficient cellular uptake of the pH endoscope was observed by complexation with the macropinocytosis inducible peptide SN21-LK15 to successfully monitor the intracellular pH changes. By taking advantage of the characteristics of the DNA origami scaffold design, such as the flexibility in loading given fluorophores at defined positions, it is possible to design various pH endoscopes with different detection wavelengths by using appropriate fluorophores and with high sensitivity by increasing the number of fluorophore molecules to be assembled. Furthermore, not only pH changes, but also several other intracellular events can be detected by applying appropriate fluorophores and/or fluorescent probes on the DNA origami scaffold. Work in this direction is ongoing in our laboratory.Experimental procedures
Materials
Single-stranded M13mp18 viral DNA (7249) was purchased from Guild Biosciences. Purified DNA origami staple strands and all other oligonucleotides including fluorophore modified oligonucleotides were obtained from Sigma-Aldrich (St. Louis, MO), Japan Bio Services Co., LTD (Saitama, Japan) or Thermal Fisher Scientific (Tokyo, Japan). Sephacryl S-400 was purchased from GE Healthcare Japan Inc. (Tokyo, Japan). Ultrafree-MC-DV column was obtained from Merck Millipore (Darmstadt, Germany). Low-binding microtube (BT-150L, 1.5 ml, nonpyrogenic & RNase-/DNase-free) was purchased from Ina OPTIKA CO., LTD (Osaka, Japan).Preparation of the fluorophore assembled DNA origami scaffold (rec-Ori)
A solution (50 μl) containing M13mp18 single-stranded DNA (New England Biolabs, 20 nM) and staple DNA strands (5 equiv. for scaffold ssDNA, 100 nM) with fluorophore modified ODN-tag (1 equiv. for each comp-ODN appended staple DNA strands, e.g. rec-CF5/CR5: CF-ODN1 500 nM and CR-ODN2 500 nM) in 1× DNA origami buffer (40 mM Tris–HCl, 20 mM acetic acid, 12.5 mM MgCl2, pH 7.6) was heated at 95 °C for 1 min, annealed at 53 °C for 30 min, and held at 4 °C in a C1000 Thermal cycler by Bio-Rad. Excess staple strands in the samples were removed by size-exclusion chromatography (500 μl Sephacryl S-400 (GE Healthcare) and equilibrated with a 1× DNA origami buffer (pH 7.6) containing 40 mM Tris–HCl, 20 mM acetic acid, and 12.5 mM MgCl2 in an Ultrafree-MC-DV centrifugal unit (Millipore)). The designed rec-Ori was prepared and purified as described and the characterization of the purified rec-Ori was conducted by AFM imaging and agarose gel electrophoresis.Absorption at 260 nm was measured with a Nanodrop 2000 spectrophotometer (Thermo Scientific) to estimate the concentration of any oligonucleotide or rec-Ori. Theoretical molar extinction coefficients used for the estimation were calculated by an OligoAnalyzer (Integrated DNA Technologies, https://sg.idtdna.com/calc/analyzer). For example, a theoretical molar extinction coefficient of 1.169 × 108 M−1 cm−1 was used for calculating the concentration of the DNA scaffolds.
Agarose gel electrophoresis analysis
All of the samples were mixed with a 6× Orange G DNA loading dye in 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. Then, 6 μl 1 kb DNA ladder and 8 μl samples were loaded into the 1.0% agarose gel (in 1× TAE buffer) and run at 50 V for 1.5 hours inside fridge at 4 °C. Before staining for visualization, the gel was scanned in fluorophore channels (CF: excitation wavelength at 488 nm, emission wavelength at 530 nm; CR: excitation wavelength at 555 nm, emission wavelength at 585 nm). Then, the gel was scanned at EtBr channel likewise using a Molecular Imager Pharox FX (Bio-Rad) after stained in 0.5 μg ml−1 ethidium bromide for 30 min with mild shaking. The images were analyzed using software Image Lab by Bio-Rad Laboratories.
1 ratio. Then, 6 μl 1 kb DNA ladder and 8 μl samples were loaded into the 1.0% agarose gel (in 1× TAE buffer) and run at 50 V for 1.5 hours inside fridge at 4 °C. Before staining for visualization, the gel was scanned in fluorophore channels (CF: excitation wavelength at 488 nm, emission wavelength at 530 nm; CR: excitation wavelength at 555 nm, emission wavelength at 585 nm). Then, the gel was scanned at EtBr channel likewise using a Molecular Imager Pharox FX (Bio-Rad) after stained in 0.5 μg ml−1 ethidium bromide for 30 min with mild shaking. The images were analyzed using software Image Lab by Bio-Rad Laboratories.
AFM imaging
The purified sample (2 μl) was dropped on freshly cleaved mica (1.5 mm in diameter) surface and adsorbed for 5 min at room temperature. Then, the surface was gently washed three times with the 1× DNA buffer (pH 7.6, 40 mM Tris–HCl, 20 mM acetic acid, 12.5 mM MgCl2). The sample was scanned in solution with the tapping mode using a High-Speed Atomic Force Microscope (HS-AFM) system (Nano Live Vision, RIBM Co. Ltd.) with a silicon nitride cantilever (Olympus BL-AC10DS-A2).Fluorescence spectra measurement and pH titration
Fluorescence spectra were measured by TECAN Infinite M Nano+ with excitation at 470 nm and 530 nm in the 40 mM citrate (pH 3.0 to 5.5), MES (pH 6.0 to 6.5), HEPES (pH 7.0 to 8.0) or CHES (pH 8.5 to 10.0) buffer containing 12.5 mM MgCl2. Fluorescence measurements were performed in the 96-plate (Greiner Microplate, 655906, 96-well, PS, F-bottom (chimney well) μCLEAR®, black, non-binding).Peptide synthesis
SN21-LK15 (H-KPVSLSYRCPCRFFESHVARAGGKLLKLLLKLLLKLLK-NH2)44 was synthesized via Fmoc-solid-phase peptide synthesis using a Rink amide resin and purified by reverse-phase high-performance liquid chromatography using a C18 column as previously described.44 The mass of the peptide was confirmed by MALDI-TOF mass spectroscopy.Cell culture
HeLa cells (human epithelial carcinoma cell line) (Riken BRC Cell Bank) were cultured in minimum essential medium alpha (α-MEM) (FUJIFILM Wako Pure Chemical Corporation) containing 10% (v/v) heat-inactivated bovine serum (BS) [α-MEM(+)] in a humidified 5% CO2 atmosphere at 37 °C and a subculture was conducted every 2–3 days.Optimization of the mixing ratio of rec-Ori with SN21-LK15
To optimize the mixing ratio of rec-Ori and SN21-LK15, the ratio of positively-chargeable amine groups to negatively-charged nucleic acid phosphate groups (referred to NP ratio or charge ratio) was considered as previously described.60 To prepare rec-Ori with SN21-LK15 at a 1, 3 or 5 NP ratio, 50 μl of Opti-MEM (Gibco) containing 1 μg of rec-Ori were mixed with 50 μl of Opti-MEM containing 333 pmol, 999 pmol or 1665 pmol of SN21-LK15, respectively, for 15 min at room temperature (∼23 °C). For a control experiment using lipofectamine2000 (Invitrogen), 50 μl of Opti-MEM containing 2.5 μl of lipofectamine2000 was mixed with 1 μg of rec-Ori. HeLa cells were washed with α-MEM without serum [α-MEM(−)] twice and 1 ml of α-MEM(−) was added into the dish. Then the above respective mixture of rec-Ori with SN21-LK15 was added to the medium dropwise and mixed evenly by gentle shaking. After incubation for 1 h at 37 °C, the cells were washed with α-MEM(−) three times and observed using an FV3000 Olympus confocal laser scanning microscope (CLSM) equipped with a 60× objective lens (UPlanSApo, NA 1.35, oil immersion). The images were analyzed using ImageJ software (NIH).Observation of localization of rec-Ori with SN21-LK15, FITC-dextran and lysosomes
HeLa cells (2 × 105 cells) were seeded onto a 35 mm glass-bottom dish (IWAKI) and incubated overnight. For staining lysosomes, the cells were washed with α-MEM(−) twice and incubated with 75 nM of LysoTracker Deep Red (LysoDR) (Invitrogen) in α-MEM(−) for 30 min at 37 °C before incubating with rec-Ori (rec-CR5 or rec-CF5/CR5) and SN21-LK15. After preparing rec-Ori and SN21-LK15 with the charge ratio of 3, the cells were washed with α-MEM(−) twice and 900 μl of α-MEM(−) containing FITC-dextran (70 kDa) (final concentration, 0.5 mg ml−1) (Invitrogen) was added to the dish. Then 100 μl of the mixture of rec-Ori and SN21-LK15 was added dropwise and mixed evenly by gentle shaking. After incubation for 40 min, fluorescence localization was observed using an FV3000 Olympus confocal laser scanning microscope (CLSM) equipped with a 60× objective lens (UPlanSApo, NA 1.35, oil immersion). For time-lapse imaging to observe cellular uptake and cellular trafficking of rec-Ori with or without LysoDR, the dish was mounted onto a stage top microchamber (Tokai Hit) (in a humidified 5% CO2 atmosphere) attached on the FV3000 inverted microscope. Time-lapse images were acquired every 2 min. The images and fluorescence intensities to quantitate CF5 and CR5 to get the CF/CR ratio were analyzed and measured using ImageJ software (NIH).Author contributions
T. M. conceived the original idea and supervised the project. E. N., H. H., S. F., and T. M. designed the experiments. K. G. performed the majority of in vitro experiments and analyzed data in collaboration with E. N. H. H. performed confocal microscopy observation and analyzed the data. J. V. V. A. performed peptide synthesis. Z. Z. synthesized fluorophores. All authors discussed the results and contributed to the final manuscript.Conflicts of interest
The authors have no conflicts of interest directly relevant to the content of this article.Acknowledgements
This work was supported by JST CREST Grant Number JPMJCR18H5 (S. F. and T. M.).Notes and references
- A. L. Lehninger, D. L. Nelson and M. M. Cox, Lehninger principles of biochemistry, Worth Publishers, New York, 2000 Search PubMed.
- Proteomic and metabolomic approaches to biomarker discovery, ed. H. Issaq, Academic Press, London, 2013 Search PubMed.
- M. C. Brahimi-Horn and J. Pouysségur, FEBS Lett., 2007, 581, 3582–3591 CrossRef CAS PubMed.
- J. R. Casey, S. Grinstein and J. Orlowski, Nat. Rev. Mol. Cell Biol., 2010, 11, 50–61 CrossRef CAS PubMed.
- X. Zhang, Y. Lin and R. J. Gillies, J. Nucl. Med., 2010, 51, 1167–1170 CrossRef CAS PubMed.
- M. Karnebogen, D. Singer, M. Kallerhoff and R. H. Ringert, Thermochim. Acta, 1993, 229, 147–155 CrossRef.
- A. Sigaeva, Y. Ong, V. G. Damle, A. Morita, K. J. van der Laan and R. Schirhagl, Acc. Chem. Res., 2019, 52, 1739–1749 CrossRef CAS PubMed.
- L. D. Lavis and R. T. Raines, ACS Chem. Biol., 2008, 3, 142–155 CrossRef CAS PubMed.
- H. Kobayashi, M. Ogawa, R. Alford, P. L. Choyke and Y. Urano, Chem. Rev., 2010, 110, 2620–2640 CrossRef CAS PubMed.
- T. Ueno and T. Nagano, Nat. Methods, 2011, 8, 642–645 CrossRef CAS PubMed.
- J. Chan, S. C. Dodani and C. J. Chang, Nat. Chem., 2012, 4, 973–984 CrossRef CAS PubMed.
- T. Terai and T. Nagano, Pflügers Archiv: European Journal of Physiology, 2013, 465, 347–359 CrossRef CAS PubMed.
- A. T. Aron, K. M. Ramos-Torres, J. A. Cotruvo and C. J. Chang, Acc. Chem. Res., 2015, 48, 2434–2442 CrossRef CAS PubMed.
- J. V. Jun, D. M. Chenoweth and E. J. Petersson, Org. Biomol. Chem., 2020, 18, 5747–5763 RSC.
- J. Han and K. Burgess, Chem. Rev., 2010, 110, 2709–2728 CrossRef CAS PubMed.
- Y. Yue, F. Huo, S. Lee, C. Yin and J. Yoon, Analyst, 2017, 142, 30–41 RSC.
- J. T. Hou, W. X. Ren, K. Li, J. Seo, A. Sharma, X. Q. Yu and J. S. Kim, Chem. Soc. Rev., 2017, 46, 2076–2090 RSC.
- M. Shamsipur, A. Barati and Z. Nematifar, J. Photochem. Photobiol., C, 2019, 39, 76–141 CrossRef CAS.
- A. Steinegger, O. S. Wolfbeis and S. M. Borisov, Chem. Rev., 2020, 120, 12357–12489 CrossRef CAS PubMed.
- K. Kikuchi, H. Takakusa and T. Nagano, TrAC, Trends Anal. Chem., 2004, 23, 407–415 CrossRef CAS.
- A. P. Demchenko, Anal. Biochem., 2005, 343, 1–22 CrossRef CAS PubMed.
- J. Young, Work. U. S. A., 2003, 7, 100–102 Search PubMed.
- H. Wang, E. Nakata and I. Hamachi, ChemBioChem, 2009, 10, 2560–2577 CrossRef CAS PubMed.
- T. Doussineau, A. Schulz, A. Lapresta-Fernandez, A. Moro, S. Körsten, S. Trupp and G. J. Mohr, Chem.–Eur. J., 2010, 16, 10290–10299 CrossRef CAS PubMed.
- A. P. Demchenko, J. Fluoresc., 2010, 20, 1099–1128 CrossRef PubMed.
- L. Yuan, W. Lin, K. Zheng and S. Zhu, Acc. Chem. Res., 2013, 46, 1462–1473 CrossRef CAS PubMed.
- X. Huang, J. Song, B. C. Yung, X. Huang, Y. Xiong and X. Chen, Chem. Soc. Rev., 2018, 47, 2873–2920 RSC.
- L. D. Lavis, Annu. Rev. Biochem., 2017, 86, 825–843 CrossRef CAS PubMed.
- E. Abbe, Beiträge zur Theor. des Mikroskops und der mikroskopischen Wahrnehmung, Arch. Mikrosk. Anat., 1873, 9, 413–420 CrossRef.
- S. W. Hell, Nat. Biotechnol., 2003, 21, 1347–1355 CrossRef CAS PubMed.
- S. W. Hell, Nat. Methods, 2009, 6, 24–32 CrossRef CAS PubMed.
- O. S. Wolfbeis, Chem. Soc. Rev., 2015, 44, 4743–4768 RSC.
- J. L. Kolanowski, F. Liu and E. J. New, Chem. Soc. Rev., 2018, 47, 195–208 RSC.
- P. Reineck and B. C. Gibson, Adv. Opt. Mater., 2017, 5, 1600446 CrossRef.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- V. Linko, S. Nummelin, L. Aarnos, K. Tapio, J. Toppari and M. Kostiainen, Nanomaterials, 2016, 6, 139 CrossRef PubMed.
- A. Rajendran, M. Endo and H. Sugiyama, Angew. Chem., Int. Ed., 2012, 51, 874–890 CrossRef CAS PubMed.
- A. Rajendran, E. Nakata, S. Nakano and T. Morii, ChemBioChem, 2017, 18, 696–716 CrossRef CAS PubMed.
- T. A. Ngo, H. Dinh, T. M. Nguyen, F. F. Liew, E. Nakata and T. Morii, Chem. Commun., 2019, 55, 12428–12466 RSC.
- C. Lin, R. Jungmann, A. M. Leifer, C. Li, D. Levner, G. M. Church, W. M. Shih and P. Yin, Nat. Chem., 2012, 4, 832–839 CrossRef CAS PubMed.
- J. B. Woehrstein, M. T. Strauss, L. L. Ong, B. Wei, D. Y. Zhang, R. Jungmann and P. Yin, Sci. Adv., 2017, 3, 18–26 Search PubMed.
- S. L. Schmid and S. D. Conner, Nature, 2003, 422, 37–44 CrossRef PubMed.
- T. Tørring, N. V Voigt, J. Nangreave, H. Yan and K. V Gothelf, Chem. Soc. Rev., 2011, 40, 5636–5646 RSC.
- J. V. V. Arafiles, H. Hirose, M. Akishiba, S. Tsuji, M. Imanishi and S. Futaki, Bioconjugate Chem., 2020, 31, 547–553 CrossRef CAS PubMed.
- A. S. Desai, M. R. Hunter and A. N. Kapustin, Philos. Trans. R. Soc., B, 2019, 374, 20180156 CrossRef CAS PubMed.
- L. Li, T. Wan, M. Wan, B. Liu, R. Cheng and R. Zhang, Cell Biol. Int., 2015, 39, 531–539 CrossRef CAS PubMed.
- S. Weiss, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 8747–8749 CrossRef CAS PubMed.
- A. Ora, E. Järvihaavisto, H. Zhang, H. Auvinen, H. A. Santos, M. A. Kostiainen and V. Linko, Chem. Commun., 2016, 52, 14161–14164 RSC.
- X. Shen, Q. Jiang, J. Wang, L. Dai, G. Zou, Z. G. Wang, W. Q. Chen, W. Jiang and B. Ding, Chem. Commun., 2012, 48, 11301–11303 RSC.
- D. Balakrishnan, G. D. Wilkens and J. G. Heddle, Nanomedicine, 2019, 14, 911–925 CrossRef CAS PubMed.
- P. Wang, T. A. Meyer, V. Pan, P. K. Dutta and Y. Ke, Chem, 2017, 2, 359–382 CAS.
- A. Udomprasert and T. Kangsamaksin, Cancer Sci., 2017, 108, 1535–1543 CrossRef CAS PubMed.
- A. H. Okholm, J. S. Nielsen, M. Vinther, R. S. Sørensen, D. Schaffert and J. Kjems, Methods, 2014, 67, 193–197 CrossRef CAS PubMed.
- J. K. Kiviaho, V. Linko, A. Ora, T. Tiainen, E. Järvihaavisto, J. Mikkilä, H. Tenhu, Nonappa and M. A. Kostiainen, Nanoscale, 2016, 8, 11674–11680 RSC.
- S. Ramakrishnan, H. Ijäs, V. Linko and A. Keller, Comput. Struct. Biotechnol. J., 2018, 16, 342–349 CrossRef CAS PubMed.
- D. Wang, Z. Da, B. Zhang, M. A. Isbell, Y. Dong, X. Zhou, H. Liu, J. Y. Y. Heng and Z. Yang, RSC Adv., 2015, 5, 58734–58737 RSC.
- N. Wu and I. Willner, Nano Lett., 2016, 16, 6650–6655 CrossRef CAS PubMed.
- S. Modi, M. G. Swetha, D. Goswami, G. D. Gupta, S. Mayor and Y. Krishnan, Nat. Nanotechnol., 2009, 4, 325–330 CrossRef CAS PubMed.
- I. Canton and G. Battaglia, Chem. Soc. Rev., 2012, 41, 2718–2739 RSC.
- V. Weissig, Liposomes: methods and protocols. Volume 1, Humana Press, Totowa, N.J., 2010, vol. 1 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Design of a DNA scaffold (Fig. S1), absorption and fluorescence emission spectra (Fig. S2, S4 and S6), characterization of rec-Ori (Fig. S3, S5, S7 and S9), fluorescent pH titration (Fig. S8 and S10), comparison of the difference of internalization efficiency (Fig. S11), identification of the internalized rec-CR5 treated with SN21-LK15 (Fig. S12), fluorescent pH titration by using confocal microscopy (Fig. S13), and the comparison of CF properties on rec-Ori with different assembling methods (Note S1, Fig. S14 and 15). See DOI: 10.1039/d1sc01575c |
| This journal is © The Royal Society of Chemistry 2021 |