Open Access Article
Open Access ArticleLight-driven release of cucurbit[8]uril from a bivalent cage†
Pim J.
de Vink
 ,
Tim
van der Hek
,
Tim
van der Hek
 and
Luc
Brunsveld
and
Luc
Brunsveld
 *
*
Laboratory of Chemical Biology, Department of Biomedical Engineering, Institute for Complex Molecular Systems, Eindhoven University of Technology, P. O. Box 513, 5600 MB Eindhoven, The Netherlands. E-mail: l.brunsveld@tue.nl
First published on 12th April 2021
Abstract
Temporal control over supramolecular systems has great potential for the modulation of binding and assembly events, such as providing orthogonal control over protein activity. Especially light controlled triggering provides unique entries for supramolecular systems to interface in a controlled manner with enzymes. Here we report on the light-induced release of cucurbit[8]uril (CB[8]) from a bivalent cage molecule and its subsequent activation of a proteolytic enzyme, caspase-9, that itself is unresponsive to light. Central to the design is the bivalent binding of the cage with high affinity to CB[8], 100-fold stronger than the UV-inactivated products. The affinity switching occurs in the (sub-)micromolar concentration regime, matching the concentration characteristics required for dimerizing and activating caspase-9 by CB[8]. The light-responsive caged CB[8] concept presented offers a novel platform for tuning and application of switchable cucurbiturils and beyond.
Introduction
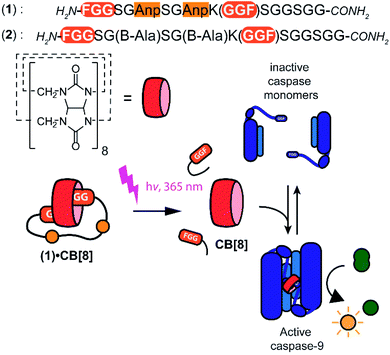
Supramolecular systems have shown great potential for the modulation of biomolecular assemblies, providing orthogonal and additive control to natural recognition and switching events.1–4 Several supramolecular host molecules, such as molecular tweezers, calixarenes, cyclodextrins, and cucurbit[n]urils, are able to selectively recognize specific peptide and protein elements through, for example, combinations of side chain residues.5–10 In particular cucurbit[n]urils have shown to be successful in protein binding, enabling for example protein activity modulation, because of their relatively strong affinity to biomolecular epitopes in water.11–14 Cucurbit[8]uril (CB[8]), specifically, is able to incorporate two N-terminal phenylalanine–glycine–glycine (FGG) motifs simultaneously in its hydrophobic cavity and therewith acts as a supramolecular glue5 allowing to control the dimerization and assembly of biomolecules such as caspases15 and larger complexes.16Light controlled triggering systems offer the possibility to perturb and modulate supramolecular over biological systems with spatio or temporal control.17,18 Beautiful examples of supramolecular light stimulus-responsive systems include: light induced pH jumps,19 photo redox switches20 and photo-caged guests to release drug molecules from CB[8].21 Of particular note are diarylethenes-based switches featuring low to high nanomolar affinity switches between their two states,22 which has been used for example for induction of light-mediated self-sorting.23 Notwithstanding, there is a need for novel switchable CB[8]-based systems for controlling biomolecular assemblies, including those with affinities matching the concentrations and assembly characteristics of the biomolecules involved. Here we report on a supramolecular caged CB[8] capable of undergoing light-triggered release using a photo-cleavable bivalent FGG-cage (Fig. 1). We illustrate its application for subsequent activation in downstream processes, such as fluorescent dye aggregation assembly and control over enzyme activity. For the light triggered CB[8] to work in unison with downstream protein effectors, the affinities were balanced such that the cage completely blocks the CB[8] prior to the stimulus, yet releases the host molecule after the stimulus for efficient binding to the proteins. Central to this design is the usage of a bivalent FGG-cage with a high affinity for CB[8] through an enhanced effective molarity24 and the usage of FGG-tagged enzymes, whose affinity for dimerization is enhanced upon two-fold binding to the CB[8] platform.25 In our design, two FGG-epitopes are bridged by an amino acid linker including two photo-cleavable amino-3-(2-nitrophenyl)propanoic acid (Anp) residues (1).26,27 Upon irradiation with 365 nm light, the amino acid backbone cleaves, resulting in two monovalent FGG-peptide fragments that have significant lower overall affinity, releasing the CB[8] to interact with other guests such as the FGG-tagged caspase-9.
Results and discussion
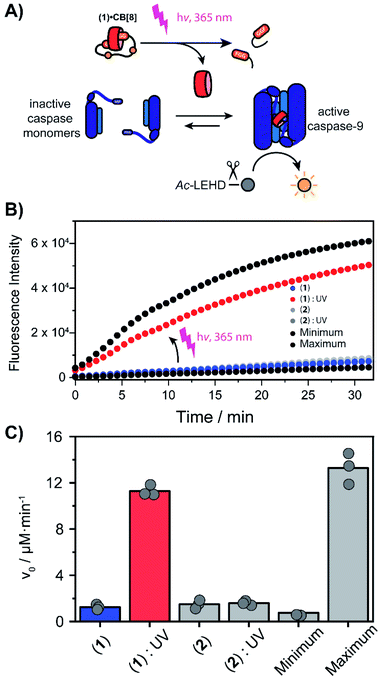
The peptide cages were synthesized using Fmoc-based solid-phase peptide synthesis. The two Anp residues are positioned within the backbone of the supramolecular cage, to ensure a high photo-cleavage yield. The second FGG-tag was introduced from a branching lysine side chain, temporarily protected via a 4-methoxytrityl group. After assembly of the complete backbone, this protecting group was selectively removed, followed by the subsequent installation of the two FGG-tags. The two N-terminal phenylalanines are 11 amino acids apart, giving it sufficient flexibility to encompass the CB[8] and bind the host in a bivalent manner without excessive strain on the peptide backbone (ESI Fig. S1†). As a control, also a non-cleavable variant (2) was made that has β-alanines instead of the Anp-residues.The photo-cleavage of FGG-cage 1 was investigated using LC-MS, UV-Vis and 1H-NMR. The peptide cage was mixed with an internal standard as reference for LC-MS quantification and samples were analyzed at different time points of exposure to irradiation with UV light (365 nm) (Fig. 2A). Within 5 minutes of UV light exposure most of cage 1 was cleaved, with a half-life of around 40 seconds. The control β-alanine-cage 2 remained intact, also after prolonged exposure. Gratifyingly, the presence of CB[8] had no significant influence on the cleavage rate.
 | ||
| Fig. 2 Cage-cleavage by UV-light. (A) Cleavage followed over time by LC-MS of 1 and 2 (control) with an internal standard as reference. The cleavage half-life (τ1/2) ∼ 40 s. The presence of CB[8] has no significant influence on the cleavage rate. (B) UV-Vis absorption spectra of 1 upon UV-radiation at different time points. Inset: absorbance at 230 nm over time. (C) 1H-NMR spectra of peptide cage 1 (500 μM) before (blue) and after (red) UV-radiation in D2O. Spectrum between 5 and 3.5 ppm is omitted for clarity (see ESI Fig. S3† for full spectra). | ||
The photo-cleavage of 1 was also followed by UV-Vis spectra at various time points during irradiation (Fig. 2B). Upon radiation, the characteristic peak at 260 nm of the ortho-nitro group rapidly disappears, while two new peaks at 230 and 300 nm appeared, indicating the successful photo-conversion. The presence of CB[8] had no significant influence on the optical properties of 1 (ESI Fig. S4†). 1H-NMR spectra of 1 before and after UV radiation (Fig. 2C and ESI Fig. S5†) revealed that after 10 minutes of irradiation the signals characteristic of the Anp groups had all disappeared.
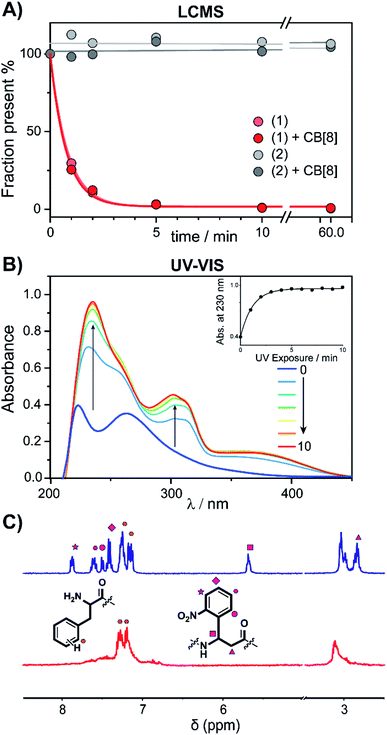
To verify complexation of the bivalent cage 1 with CB[8], 1H-NMR complex formation studies were performed. The aromatic signals belonging to the (N-terminal) phenylalanines, shifted up-field upon addition of CB[8] from 7.2 to 6.4 ppm, indicative of the de-shielding inside the CB[8] cavity for FGG-peptides (ESI Fig. S6†).5 Binding studies were performed to explore whether photo-cleavage of cage 1 would result in differential binding properties between both chemical states. Initial experiments were performed using a fluorometric assay via the displacement of acridine orange (AO) from the cavity of CB[8]. The AO dye is auto-quenched inside the cavity of CB[8], and becomes fluorescent again when another guest displaces it from the cavity (Fig. 3A).28 The FGG-cages 1 and 2 were titrated to a solution of pre-complexed CB[8] and AO. In previous studies,28 a relative high concentration of the FGG-mono peptide was required to displace AO from the CB[8] cavity (IC50 62 μM). In comparison, the bivalent FGG-cages 1 and 2 had a much stronger affinity and displayed AO with IC50's of 400 and 500 nM respectively. Upon UV irradiation of the titration plate, the affinity of the light-sensitive 1 weakened towards the value of the monovalent peptide (Fig. 3B on the left, blue to red). In contrast, the IC50 of non-cleavable 2 was not affected (Fig. 3B on the right).
 | ||
| Fig. 3 (A) Schematic representation of fluorometric assay. (B) Titration of bivalent cage 1 into a solution of acridine orange (1 μM) and CB[8] (1 μM) and titration with bivalent control 2 in HBS (10 mM HEPES, pH 7.4, 150 mM NaCl) at various time points after UV-radiation, performed in triplicate; error bars represent the standard deviation. (C) Apparent KD determined by isothermal titration calorimetry before and after photo cleavage of cage 1 and control 2; error bars represent the confidence interval of the fit. (D) ITC trace of 1 before and after UV-radiation; (see ESI Table S1† for an overview of the isothermal titration calorimetry characteristics). | ||
Isothermal titration calorimetry (ITC) studies were performed to provide insights into the thermodynamic parameters of the supramolecular caging and light induced uncaging (Fig. 3C, D; ESI Table S1; Fig. S7 and S8†). Titration of FGG-cage 1 to CB[8] resulted in a dissociation constant (KD) of 22 nM. Irradiation of 1 resulted in a 100-fold weakening of the apparent KD to 2.4 μM. Interestingly, the ΔH of the binding interactions before and after UV-light induced cleavage remained similar and the lowering of the ΔG results from the change of the ΔS. These results strongly support a binding mode that is molecularly very similar in both states (same molecular interactions), but that has changed from bivalent to monovalent.29 In line with previous results, the affinity of the non-cleavable cage 2 was not affected significantly upon UV irradiation.
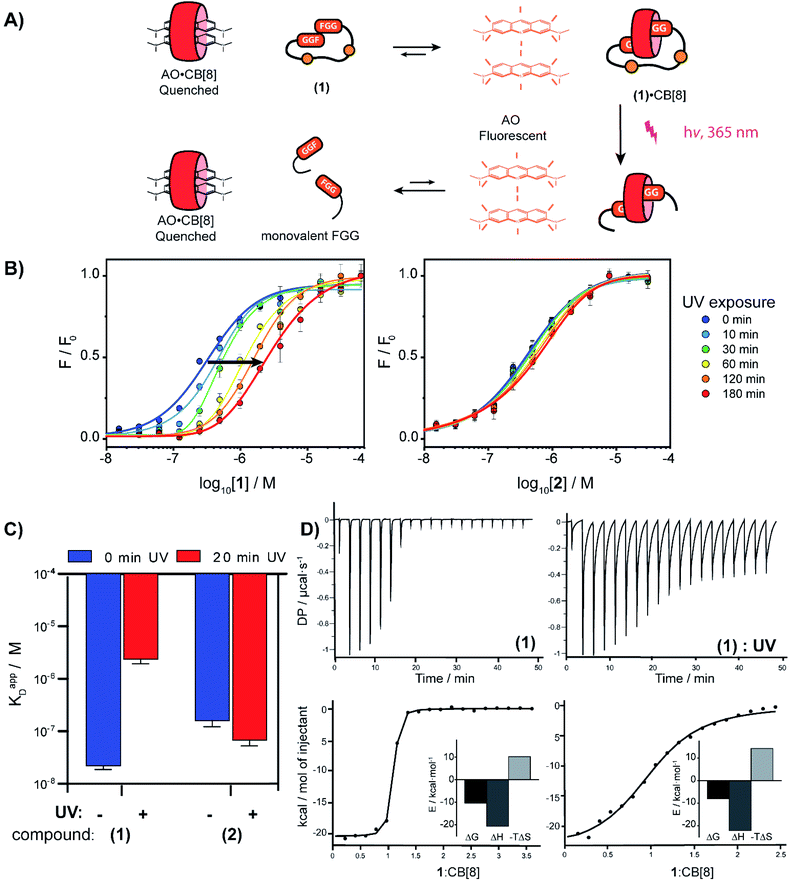
With the light-controllable CB[8] in hand, we investigated its utility as downstream control element on enzyme activity. Protein dimerization is a ubiquitous mechanism to regulate protein activity in a broad range of biological processes including apoptosis. Molecular control over these processes is critical to elucidate and perturb the molecular mechanisms of the proteins involved.30 Caspase-9 (casp-9) is a critical cysteine protease and initiator of the apoptosis pathway and is responsible for cleaving proteins at specific aspartate residues. In the cell, caspase-9 exists primarily in its inactive monomeric form,31 becoming activated only upon induced dimerization by regulatory factors as part of controlled cell death.32 For example extensive UV-damage to the DNA33 leads to mitochondrial release of cytochrome-C which triggers the assembly of the apoptosome that acts a scaffolding platform that recruits multiple copies of caspase-9, initiating apoptosis.34 This scaffolding can be mimicked by bringing two caspase-9 proteins in close proximity by virtue of dual binding of CB[8] to N-terminal FGG-tags on the caspase-9.15 The intrinsic affinity between two caspase monomers (KD ∼ 100 μM)35 causes the caspases to act as a supramolecular bidentate.28 As a consequence, the caspases primarily form active dimers, even when CB[8] is in 10-fold excess over the caspase monomers. We employed a proteolytic activity assay for caspase-9 using the synthetic substrate Ac-LEHD-AFC (where AFC is 7-amino-4-(trifluoromethyl)coumarin), that becomes fluorescent after cleavage by caspase-9 (Fig. 4A). Kinetic traces were recorded by following the fluorescence over time. The optimal concentration of CB[8] for caspase-9 activation, independent of the bivalent cage, was determined to be 10 μM.
We screened for the optimal concentration of cage 1 that had the highest on/off ratio before and after UV cleavage (ESI Fig. S10†). We then investigated the effect of photo-cleavage of cage 1 in comparison with caspase with and without CB[8] as maximum and minimum effect (Fig. 4B). Without UV radiation the caspase-9 activity in the presence of CB[8] and cage 1 is similar to the characteristic caspase-9 background activity, around 1 μM min−1 (Fig. 4C). After irradiation the activity increases dramatically to an initial slope of 11 μM min−1 (the rate decreases over time due to depletion of the AFC substrate). The UV-light activated caspase-9 reaches a comparable level activity as the CB[8]-only positive control (13 μM min−1). As expected, reference inhibitor 2 effectively blocked caspase-9 activity, also after UV-light treatment. Even though the caspase-9 monomers feature a similar FGG-tag as the monovalent fragments of the inhibitor, the intrinsic interaction between two caspase monomers results in self-sorting and hetero complexation in the CB[8] only starts to play a more dominate role when concentrations exceed the effective molarity.36
Conclusions
In conclusion, a versatile photo-triggered release system for CB[8] is presented, as well as its application for subsequent downstream modulation of fluorescent guest binding and binding and activation of caspase-9 enzymes. The UV-sensitive cage exhibits a 100-fold higher affinity in its intact bivalent, inhibitory mode as compared to its cleaved state. The UV-induced switching occurs in the submicromolar regime, highly favourable for supramolecular chemical biology applications, with proteins and alike that typically act in a similar concentration regime.37 Light-activatable supramolecular systems are in high demand, especially those that work at low concentrations and those that can interface with biomolecules.38 The caged CB[8] concept presented here offers many entries for tuning and application to the specific biomolecular system at hand, for example by varying the concentration of cage and CB[8] or by tuning of the affinities by utilizing stronger or weaker binding FGG analogues.39 Similarly, the conceptual approach of a cleavable bivalent inhibitor, as presented here, can be extended to other types of triggers, which are currently being explored.Author contributions
P. J. d. V. and T. v. d. H. performed experiments, P. J. d. V. and L. B. conceptualized the studies, all authors contributed to writing the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by the Netherlands Organization for Scientific Research (NWO) through Gravity program 024.001.035 and VICI grant 016.150.366. We thank Bas Rosier for the discussions about caspases.Notes and references
- L. Zhang, Y. Wu and L. Brunsveld, Angew. Chem., Int. Ed., 2007, 46, 1798–1802 CrossRef CAS PubMed.
- S. Sakamoto and K. Kudo, J. Am. Chem. Soc., 2008, 130, 9574–9582 CrossRef CAS PubMed.
- D. Bier, R. Rose, K. Bravo-Rodriguez, M. Bartel, J. M. Ramirez-Anguita, S. Dutt, C. Wilch, F.-G. Klärner, E. Sanchez-Garcia, T. Schrader and C. Ottmann, Nat. Chem., 2013, 5, 234–239 CrossRef CAS PubMed.
- S. C. Zimmerman, Beilstein J. Org. Chem., 2016, 12, 125–138 CrossRef CAS PubMed.
- L. M. Heitmann, A. B. Taylor, P. J. Hart and A. R. Urbach, J. Am. Chem. Soc., 2006, 128, 12574–12581 CrossRef CAS PubMed.
- H. D. Nguyen, D. T. Dang, J. L. J. van Dongen and L. Brunsveld, Angew. Chem., Int. Ed., 2010, 49, 895–898 CrossRef CAS PubMed.
- D. A. Uhlenheuer, K. Petkau and L. Brunsveld, Chem. Soc. Rev., 2010, 39, 2817–2826 RSC.
- R. E. McGovern, H. Fernandes, A. R. Khan, N. P. Power and P. B. Crowley, Nat. Chem., 2012, 4, 527–533 CrossRef CAS PubMed.
- S. Sonzini, A. Marcozzi, R. J. Gubeli, C. F. van der Walle, P. Ravn, A. Herrmann and O. A. Scherman, Angew. Chem., Int. Ed., 2016, 55, 14000–14004 CrossRef CAS PubMed.
- S. H. Hewitt and A. J. Wilson, Chem. Commun., 2016, 52, 9745–9756 RSC.
- J. M. Chinai, A. B. Taylor, L. M. Ryno, N. D. Hargreaves, C. A. Morris, P. J. Hart and A. R. Urbach, J. Am. Chem. Soc., 2011, 133, 8810–8813 CrossRef CAS PubMed.
- K. L. Kim, G. Sung, J. Sim, J. Murray, M. Li, A. Lee, A. Shrinidhi, K. M. Park and K. Kim, Nat. Commun., 2018, 9, 1712–1721 CrossRef PubMed.
- F. Guagnini, P. M. Antonik, M. L. Rennie, P. O'Byrne, A. R. Khan, R. Pinalli, E. Dalcanale and P. B. Crowley, Angew. Chem., Int. Ed., 2018, 57, 7126–7130 CrossRef CAS PubMed.
- L. A. Logsdon, C. L. Schardon, V. Ramalingam, S. K. Kwee and A. R. Urbach, J. Am. Chem. Soc., 2011, 133, 17087–17092 CrossRef CAS PubMed.
- D. T. Dang, H. D. Nguyen, M. Merkx and L. Brunsveld, Angew. Chem., Int. Ed., 2013, 52, 2915–2919 CrossRef CAS PubMed.
- C. Si, J. Li, Q. Luo, C. Hou, T. Pan, H. Li and J. Liu, Chem. Commun., 2016, 52, 2924–2927 RSC.
- A. Gautier, C. Gauron, M. Volovitch, D. Bensimon, L. Jullien and S. Vriz, Nat. Chem. Biol., 2014, 10, 533–541 CrossRef CAS PubMed.
- M. J. Hansen, W. A. Velema, M. M. Lerch, W. Szymanski and B. L. Feringa, Chem. Soc. Rev., 2015, 44, 3358–3377 RSC.
- C. Parente Carvalho, V. D. Uzunova, J. P. D. Silva, W. M. Nau and U. Pischel, Chem. Commun., 2011, 47, 8793–8795 RSC.
- F. Tian, D. Jiao, F. Biedermann and O. A. Scherman, Nat. Commun., 2012, 3, 1207 CrossRef PubMed.
- M. A. Romero, N. Basílio, A. J. Moro, M. Domingues, J. A. González-Delgado, J. F. Arteaga and U. Pischel, Chem.–Eur. J., 2017, 23, 13105–13111 CrossRef CAS PubMed.
- P. Ferreira, B. Ventura, A. Barbieri, J. P. Da Silva, C. A. T. Laia, A. J. Parola and N. Basílio, Chem.–Eur. J., 2019, 25, 3477–3482 CrossRef CAS PubMed.
- P. Remón, D. González, S. Li, N. Basílio, J. Andréasson and U. Pischel, Chem. Commun., 2019, 55, 4335–4338 RSC.
- M. Ramaekers, S. P. W. Wijnands, J. L. J. van Dongen, L. Brunsveld and P. Y. W. Dankers, Chem. Commun., 2015, 51, 3147–3150 RSC.
- R. P. G. Bosmans, J. M. Briels, L.-G. Milroy, T. F. A. de Greef, M. Merkx and L. Brunsveld, Angew. Chem., Int. Ed., 2016, 55, 8899–8903 CrossRef CAS PubMed.
- E. Profft and F.-J. Becker, J. Prakt. Chem., 1965, 30, 18–38 CrossRef CAS.
- B. B. Brown, D. S. Wagner and H. M. Geysen, Mol. Diversity, 1995, 1, 4–12 CrossRef CAS PubMed.
- P. J. de Vink, J. M. Briels, T. Schrader, L.-G. Milroy, L. Brunsveld and C. Ottmann, Angew. Chem., Int. Ed., 2017, 56, 8998–9002 CrossRef CAS PubMed.
- G. Wu, M. Olesińska, Y. Wu, D. Matak-Vinkovic and O. A. Scherman, J. Am. Chem. Soc., 2017, 139, 3202–3208 CrossRef CAS PubMed.
- J. D. Klemm, S. L. Schreiber and G. R. Crabtree, Annu. Rev. Immunol., 1998, 16, 569–592 CrossRef CAS PubMed.
- S. J. Riedl and G. S. Salvesen, Nat. Rev. Mol. Cell Biol., 2007, 8, 405–413 CrossRef CAS PubMed.
- B. J. H. M. Rosier, A. J. Markvoort, B. Gumí Audenis, J. A. L. Roodhuizen, A. den Hamer, L. Brunsveld and T. F. A. de Greef, Nat. Catal., 2020, 3, 295–306 CrossRef CAS PubMed.
- L. A. Sitailo, S. S. Tibudan and M. F. Denning, J. Biol. Chem., 2002, 277, 19346–19352 CrossRef CAS PubMed.
- P. Li, L. Zhou, T. Zhao, X. Liu, P. Zhang, Y. Liu, X. Zheng and Q. Li, Oncotarget, 2017, 8, 23996–24008 CrossRef PubMed.
- M. Renatus, H. R. Stennicke, F. L. Scott, R. C. Liddington and G. S. Salvesen, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 14250–14255 CrossRef CAS PubMed.
- S. Di Stefano and G. Ercolani, in Advances in Physical Organic Chemistry, ed. I. H. Williams and N. H. Williams, Academic Press, 2016, vol. 50, pp. 1–76 Search PubMed.
- L.-G. Milroy, T. N. Grossmann, S. Hennig, L. Brunsveld and C. Ottmann, Chem. Rev., 2014, 114, 4695–4748 CrossRef CAS PubMed.
- O. Dagliyan and K. M. Hahn, Curr. Opin. Struct. Biol., 2019, 57, 17–22 CrossRef CAS PubMed.
- A. R. Urbach and V. Ramalingam, Isr. J. Chem., 2011, 51, 664–678 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Figures and tables, experimental details of peptide synthesis, protein expression, NMR, fluorometric assays, ITC. See DOI: 10.1039/d1sc01410b |
| This journal is © The Royal Society of Chemistry 2021 |