Open Access Article
Open Access ArticleClickable amino acid tuned self-assembly of a nucleus-selective multi-component nanoplatform for synergistic cancer therapy†
Lan
Yang
a,
Xiao
He
a,
Zhiying
Zeng
a,
Jiakun
Tang
a,
Dongmei
Qi
a,
Huijie
Ma
a,
Hui
Chen
*a,
Xinghai
Ning
 *b and
Xuli
Feng
*b and
Xuli
Feng
 *a
*a
aChongqing Key Laboratory of Natural Product Synthesis and Drug Research, School of Pharmaceutical Sciences, Chongqing University, Chongqing 401331, China. E-mail: chenhui0429@cqu.edu.cn; fengxuli@cqu.edu.cn
bNational Laboratory of Solid State Microstructures, Chemistry and Biomedicine Innovation Center, College of Engineering and Applied Sciences, Jiangsu Key Laboratory of Artificial Functional Materials, Nanjing University, Nanjing 210093, China. E-mail: xning@nju.edu.cn
First published on 14th May 2021
Abstract
Nucleus-targeted therapy holds great promise in cancer treatment; however, a lack of effective nucleus-specific delivery significantly limits its application potential. Here, we report a nucleus-targeted synergistic chemo-photodynamic therapy based on the self-assembly of chlorin e6 (Ce6) and doxorubicin (DOX) tuned by clickable dibenzocyclooctyne (DIBO) functionalized lysine (D-K) and subsequent reaction with crosslinkers. The assembled nanodrugs with high loading efficiency and long-term stability show enhanced cellular uptake and accumulation in the nucleus, resulting in greatly improved in vitro and in vivo chemo-photodynamic efficacy. Notably, D-K can promote the rapid self-assembly of Ce6 and DOX in aqueous solution, avoiding the introduction of organic solvents or tedious preparations. In addition, the introduction of the DIBO group can effectively expand the types of self-assembly material and enhance the self-assembly behaviour through a copper-free click reaction. Therefore, we present an effective nucleus-targeted combination drug delivery strategy, which has great potential in the treatment of many diseases.
Introduction
Over the past decade, organelle-targeted therapy has been emerging as a promising anticancer approach. In comparison to other apoptosis-associated cellular components, such as mitochondria and lysosomes, the nucleus plays a critical role in most biological cellular processes, and exhibits high sensitivity to various types of DNA damage, making it a promising target for antitumor therapy. Chemotherapy is one of the most commonly used treatments for various cancers.1,2 In recent years, chemotherapy has been combined with other treatment strategies such as photodynamic therapy (PDT) to overcome the limitations of traditional single treatments through different modes of action.3–8 PDT has been recognized as a potent strategy for advanced combination treatment.9–12 However, the inherent drawbacks of concurrent PDT, including low bioavailability and poor systemic circulation, are still major obstacles to its clinical applications.13 To improve the PDT efficiency, various carriers have been developed, such as liposomes, and polymeric and inorganic nanoparticles.14–17 Unfortunately, most of these carriers suffer from technical problems of inconvenient preparation, low drug loading efficiency and poor stability.18,19 In addition, the efficacy of PDT largely depends on the subcellular localization of photosensitizers (PS) due to the short transient lifetime and diffusion range of reactive oxygen species (ROS).20 Thus, selective generation of ROS in specific organelles, such as the nucleus, that are more susceptible to oxidative damage may significantly improve therapeutic efficacy. Furthermore, the nucleus-targeting delivery strategy makes it efficient to transport DOX into the nucleus, which can induce direct inhibition of the DNA or RNA synthesis process, while avoiding the P-glycoprotein associated efflux pumping of DOX. Therefore, an effective strategy that directly transports PS and DOX into cancer cell nuclei is greatly needed for improving the combination treatment of PDT and chemotherapy.Molecular self-assembly is a versatile process that involves an organized arrangement of molecules through non-covalent interactions.21–23 Various biomolecules including proteins, peptides, and even amino acids have been employed as building blocks for the construction of functional nanomaterials.24–28 In particular, amino acids and their derivatives (e.g., fluorenylmethyloxycarbonyl (Fmoc) modified amino acids) offer appreciable advantages over proteins and peptides in the construction of nanoarchitectured functional systems, due to their molecular simplicity, biostability, and inherent biocompatibility.29,30 Recent studies show that amino acids can spontaneously assemble to form ordered nanomaterials with various morphologies by simply manipulating the coordination and noncovalent interactions.31–35 Although amino acid self-assembly exhibits benefits for integrating drugs into therapeutic nanoparticles,36,37 no studies have actually managed to develop nucleus-targeted nanodrugs. To accomplish this task, there is an urgent need for a new amino acid self-assembly method to achieve selective delivery of therapeutics not only to the same cells, but also to therapeutically active sites in the nucleus.
Here, we present a sequential self-click-assembly strategy, which utilized amphiphilic amino acid D-K mediated self-assembly to construct multi-component nanoplatforms for nucleus-targeted delivery of PS and anticancer drugs. As shown in Scheme 1a, the traditional amino acid protecting group Fmoc was changed to the dibenzocyclooctyne (DIBO) functional group in D-K mainly for copper-free click chemistry, which can effectively expand the types of self-assembly material and enhance the self-assembly behaviour under mild conditions. Ce6 and DOX molecules can be integrated into the amphiphilic networks of D-K through multiple weak intermolecular interactions, including π–π stacking, hydrophobic and electrostatic interactions, and then assembled with azide crosslinkers (4-A) through a copper-free click reaction to generate a D-KCD/A multi-component nanoplatform. D-KCD/A could not only improve the drug loading efficacy, but also enhance the accumulation of Ce6 and DOX in the nucleus, which greatly enhanced the synergistic effects of in vitro and in vivo PDT and chemotherapy (Scheme 1B). Thus, D-KCD/A represents a promising nucleus-targeted platform for combination therapy, and may have potential for improving the management of malignant diseases.
Results and discussion
Synthesis of D-K and 4-A
The synthesis of D-K and 4-A is illustrated in Fig. 1. Initially, Boc-lysine-NHS was reacted with ethylenediamine to afford compound 1, which was reacted with 4-nitrophenyl chloroformate-substituted DIBO to get compound 2. The final D-K was obtained by deprotecting the Boc groups with hydrochloric acid. 3-Azido-1-propanol was reacted with triflic anhydride to get compound 3, which was further reacted with tetraazacyclododecane to afford 4-A. All intermediates and final compounds were carefully characterized and confirmed by nuclear magnetic resonance (NMR) and mass spectroscopy (MS).Preparation and characterization of D-K tuned self-assembly
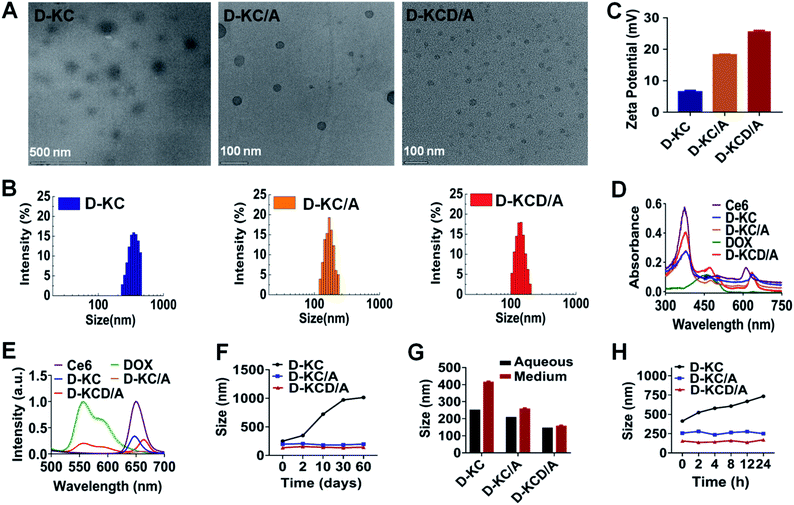
We next investigated the capability of D-K to regulate the self-assembly of nanoparticles loaded with Ce6 and DOX. The morphology and size of nanoparticles containing different components were identified using transmission electron microscopy (TEM) and dynamic light scattering (DLS). As shown in Fig. 2A and B, loose nanoparticle agglomerates (D-KC) were formed upon mixing D-K (0.233 mg mL−1) and Ce6 (0.08 mg mL−1), and had an average diameter of 250 nm and zeta potential of about 8 mV (Fig. 2C). However, D-KC was transformed into a compact nanostructure after crosslinking with 4-A (0.067 mg mL−1), generating D-KC/A with the decrease of particle sizes and the increase of zeta potentials. Importantly, we found that the integration of DOX (0.08 mg mL−1) into D-KC/A had no effects on the morphology of the nanoparticles (D-KCD/A), but a smaller size and higher zeta potential were observed, indicating that DOX further enhances the hydrophobic and electrostatic interactions within nanoparticles. It is worth noting that the particle size of both D-KC/A and D-KCD/A measured by DLS was larger than that observed by TEM, which may be due to the fact that DLS provided the hydrodynamic diameter of nanoparticles with a hydride corona, while the TEM image depicted the real size of dry state samples. In addition, UV-vis absorption and fluorescence spectroscopy were applied to investigate the intermolecular interactions to gain insight into the molecular configuration and arrangement of the self-assembled nanostructures (Fig. 2D and E). In comparison to monomeric Ce6, D-KC showed a weak red-shifted absorbance in both the Soret and Q bands, due to Ce6 aggregation. The crosslinking of 4-A enhanced the Q-band absorption of Ce6 in D-KC nanoparticles, indicating the formation of a stable Ce6 complex. The integration of DOX into D-KC/A could enhance the absorption of Ce6. Moreover, we found that the fluorescence of D-KC and D-KC/A dramatically decreased due to the aggregation of assembled Ce6 (λex = 400 nm), compared with monomeric Ce6. However, the fluorescence of Ce6 was partially restored after integrating DOX; meanwhile, the fluorescence of DOX (λex = 488 nm) was greatly quenched. All these data indicate that D-K mediated self-assembly can effectively integrate Ce6 into nanoplatforms and DOX could coordinate with Ce6 and D-K to further increase the intermolecular hydrophobic and electrostatic interactions.38 The loading efficiency of both Ce6 and DOX was 17%, respectively. Notably, the assembled D-KC/A and D-KCD/A nanoparticles with cross-linking agents were stable both in aqueous solution and culture medium (Fig. 2F–H), thus facilitating further biological applications.Cellular uptake and nuclear accumulation of various self-assembled nanoparticles regulated by D-K
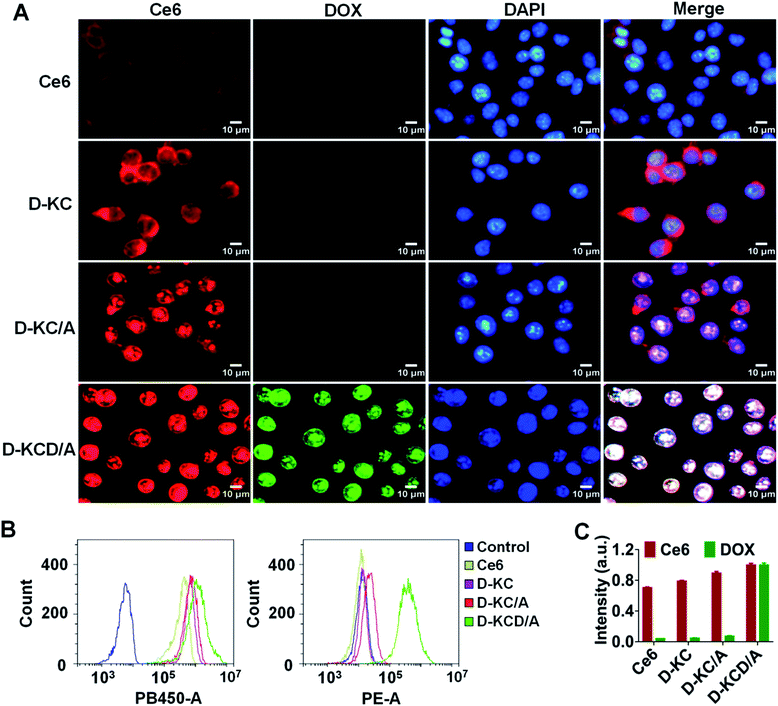
To investigate the capability of self-assembled nanoparticles to improve the cellular uptake and nuclear localization of Ce6 and DOX, breast cancer 4T1 cells were incubated with free Ce6, D-KC, D-KC/A or D-KCD/A for 2 h, followed by imaging with a fluorescence microscope. The cell nucleus was stained with 4,6-diamidino-2-phenylindole (DAPI) for identifying nuclear accumulation. As shown in Fig. 3A, all self-assembled nanoformulations were effectively transported into the cells, while free Ce6 exhibited weak intracellular uptake. Importantly, 4-A mediated click crosslinking in the nanoparticles facilitated nuclear translocation, causing a high accumulation of Ce6 and DOX in the nucleus (Fig. 3B and C). These data demonstrate that D-K coordinated self-assembly provides a potent strategy to improve cellular uptake and nuclear targeting of drugs.Cellular transport mechanism and in vitro antitumor efficacy
In consideration of the fact that endocytosis is a critical step in determining the intranuclear accumulation of nanoparticles, we evaluated the cellular uptake mechanism of D-KCD/A. As shown in Fig. 4A, compared with the control group, the cellular uptake of D-KCD/A nanoparticles was significantly reduced at 4 °C, or by pretreatment with sodium azide (an adenosine triphosphate consumptive agent). In addition, a low uptake of D-KCD/A was also observed in cells treated with genistein or methyl-β-cyclodextrin (M-β-CD), indicating that caveolin and lipid raft associated pathways are involved in the cellular internalization process. Taken together, these results suggest that D-KCD/A is actively transported in cells through multiple endocytic pathways, leading to enhanced cellular uptake. Next, we also investigated the nuclear transportation mechanism of D-KCD/A. Cells were preincubated with ivermectin, a specific inhibitor of importin (ImP) mediated nuclear import, followed by incubating D-KCD/A. Minimal inhibition of the nuclear localization of D-KCD/A was observed (Fig. 4B), indicating that the intranuclear entry of D-KCD/A was ImP-independent.To identify the therapeutic potential of D-KCD/A, the generation of singlet oxygen (1O2) under light irradiation (660 nm, 100 mW cm−2, 2 min) was measured using singlet oxygen sensor green (SOSG). It was clear that D-KCD/A had no effects on the singlet oxygen production efficiency of Ce6 in NaCl solution (Fig. 4C). In addition, the fluorescence of Ce6 can be recovered in a high ionic strength environment (Fig. S1†), which further indicated the decomposition of nanoparticles and the release of Ce6, thus achieving effective PDT activation. Moreover, cell survival assay clearly revealed that the anticancer efficacy of both D-KC and D-KC/A was much higher than that of free Ce6 (Fig. 4D). Notably, D-KCD/A exhibited the highest cytotoxicity, confirming the synergistic effects of nucleus-targeted chemo-photodynamic therapy. Meanwhile, no apparent cytotoxicity was observed in any of the treatments without light stimulation (Fig. 4E), suggesting the good biocompatibility of D-K. These data indicate the great potential of self-assembled nanoformulations for cancer treatment.
Biodistribution
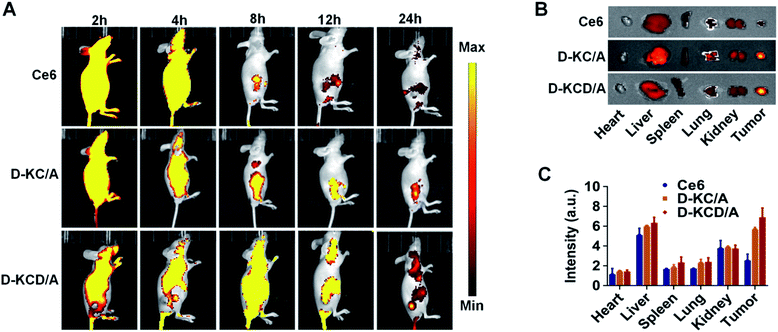
We next investigated the in vivo distribution of D-KCD/A in a 4T1 tumor xenograft mouse model. Free Ce6, D-KC/A, and D-KCD/A were intravenously injected into the mice and the fluorescence was imaged in vivo and ex vivo. Fig. 5A shows that both D-KC/A and D-KCD/A rapidly accumulated in tumor tissues, and generated strong fluorescent signals 8 h after injection. The strong fluorescence could even be observed for up to 24 h, indicating their tumor specificity. In contrast, mice treated with free Ce6 showed weak fluorescence at tumor sites, and the fluorescence greatly attenuated 24 h after injection, due to low tumor uptake and fast clearance of free Ce6. Ex vivo imaging of major organs and tumors harvested 24 h post injection further confirmed the enhanced tumor accumulation and retention of self-assembled Ce6 nanoparticles (Fig. 5B), which was also validated by the quantitative results obtained from the region-of-interest analyses (Fig. 5C).In vivo antitumour activity
Based on the above results, we systematically evaluated the in vivo antitumor efficacy of self-assembled Ce6 nanoparticles in 4T1 tumor-bearing mice. As shown in Fig. 6A, tumor growth was effectively inhibited by Ce6 loaded D-KC/A nanoparticles under laser irradiation (660 nm, 200 mW cm−2, 10 min), while minimal tumor inhibition was observed in mice treated with free Ce6. Importantly, Ce6 and DOX co-loaded D-KCD/A nanoparticles showed the best anticancer effects, suggesting enhanced anticancer effects of synergistic chemo-photodynamic therapy. Furthermore, tumor weights were measured at the end of the experiment, and demonstrated the optimal therapeutic effect of D-KCD/A nanoparticles (Fig. 6B), confirming the importance of combination therapy. Moreover, hematoxylin and eosin (H&E) staining revealed that self-assembled Ce6 nanoparticles including D-KC/A and D-KCD/A caused significant necrosis in the tumor tissue under light irradiation (Fig. 6D). In contrast, negligible damage was observed in healthy organs, such as the heart, liver, spleen, lungs, and kidneys (Fig. S2†). Meanwhile, minimal changes in body weights were observed during treatment (Fig. 6C), indicating their good biocompatibility. All these results suggest that the D-KCD/A multi-component nanoplatform is a promising strategy for cancer treatment.Conclusions
In summary, we present a facile and robust multi-component amphiphilic amino acid based nanoplatform for enhancing simultaneous intranuclear delivery of PS and anticancer drugs. In this strategy, Ce6 and DOX were directly used as building blocks to construct well-defined nanoparticles through cyclooctyne functionalized lysine (D-K) regulated self-assembly, which could avoid the utilization of any organic solvents or other tedious post-processing steps. The self-assembled nanodrugs showed enhanced cellular uptake and nuclear accumulation of Ce6 and DOX, and could not only significantly improve their cytotoxicity in cancer cells, but also minimize side effects. Importantly, D-KCD/A could selectively accumulate and retain in the tumor site, and exhibited optimal tumor growth inhibition via a multi-modal tumor suppressive effect induced by PDT and chemotherapy. Notably, the traditional amino acid protecting group Fmoc was changed to the DIBO functional group, and thus through changing the azide containing crosslinkers, a series of novel nucleus-targeting nanomaterials can be prepared simply and rapidly based on a copper-free click reaction with D-K, which can effectively expand the types of self-assembly material and enhance the self-assembly behavior under mild conditions. Therefore, our work proves the great potential of clickable amino acid derivatives as attractive and powerful tools for developing a self-assembled multi-component nucleus-targeted delivery nanoplatform for cancer treatment.Ethical statement
All animal experiments were carried out in compliance with the requirements of the National Act on the Use of Experimental Animals (People's Republic of China) and were approved by the Experimental Animal Ethical Committee of Chongqing University Cancer Hospital.Author contributions
X. F. and X. N. conceived the project and designed the experiments. L. Y. and X. H. synthesized the chemicals. L. Y. and H. M. performed the cell experiments and analyzed the data. L. Y., Z. Z., J. T. and D. Q. performed the animal experiments. X. F., X. N., and H. C. supervised the project. All of the authors were involved in the analysis and interpretation of data. X. F., X. N., L. Y., X. H., and H. C. wrote the paper, with the help of the co-authors.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21773268, 21877010), Fundamental Research Funds for the Central Universities (2019CDYGYB006), Chongqing Graduate Student Research Innovation Project (CYB18056, CYB19067), and the Startup Funding of Chongqing University (0236011104419).Notes and references
- L. N. Abdullah and E. K.-H. Chow, Clin. Transl. Med., 2013, 2, 3 CrossRef PubMed.
- C. Holohan, S. V. Schaeybroeck, D. B. Longley and P. G. Johnston, Nat. Rev. Cancer, 2013, 13, 714–726 CrossRef CAS PubMed.
- C. He, K. Lu, D. Liu and W. Lin, J. Am. Chem. Soc., 2014, 136, 5181–5184 CrossRef CAS PubMed.
- T.-M. Sun, J.-Z. Du, Y.-D. Yao, C.-Q. Mao, S. Dou, S.-Y. Huang, P.-Z. Zhang, K. W. Leong, E.-W. Song and J. Wang, ACS Nano, 2011, 5, 1483–1494 CrossRef CAS PubMed.
- J. Liu, L. Song, S. Liu, S. Zhao, Q. Jiang and B. Ding, Angew. Chem., Int. Ed., 2018, 130, 15486–15490 CrossRef PubMed.
- D.-W. Zheng, J.-L. Chen, J.-Y. Zhu, L. Rong, B. Li, Q. Lei, J.-X. Fan, M. Z. Zou, C. Li and S.-X. Cheng, et al. , Nano Lett., 2016, 16, 4341–4347 CrossRef CAS PubMed.
- L. Wang, Q. Sun, X. Wang, T. Wen, J.-J. Yin, P. Wang, R. Bai, X.-Q. Zhang, L.-H. Zhang, A.-H. Lu and C. Chen, J. Am. Chem. Soc., 2015, 137, 1947–1955 CrossRef CAS PubMed.
- H. Lee, J. Han, H. Shin, H. Han, K. Na and H. Kim, J. Control Release, 2018, 283, 190–199 CrossRef CAS PubMed.
- T. Wang, D. Wang, J. Liu, B. Feng, F. Zhou, H. Zhang, L. Zhou, Q. Yin, Z. Zhang, Z. Cao, H. Yu and Y. Li, Nano Lett., 2017, 17, 5429–5436 CrossRef CAS PubMed.
- W. Chen, J. Liu, Y. Wang, C. Jiang, B. Yu, Z. Sun and L. Lu, Angew. Chem., Int. Ed., 2019, 58, 6290–6294 CrossRef CAS PubMed.
- Y.-X. Zhu, H.-R. Jia, G.-Y. Pan, N. W. Ulrich, Z. Chen and F.-G. Wu, J. Am. Chem. Soc., 2018, 140, 4062–4070 CrossRef CAS PubMed.
- L. Wu, X. Cai, H. Zhu, J. Li, D. Shi, D. Su, D. Yue and Z. Gu, Adv. Funct. Mater., 2018, 28, 1804324 CrossRef.
- J. F. Lovell, T. W. B. Liu, J. Chen and G. Zheng, Chem. Rev., 2010, 110, 2839–2857 CrossRef CAS PubMed.
- D. Liu, B. Chen, Y. Mo, Z. Wang, T. Qi, Q. Zhang and Y. Wang, Nano Lett., 2019, 19, 6964–6976 CrossRef CAS PubMed.
- S. Yan, X. Zeng, Y. Tang, B.-F. Liu, Y. Wang and X. Liu, Adv. Mater., 2019, 31, 1905825 CrossRef CAS PubMed.
- W. Sun, L. Luo, Y. Feng, Y. Cai, Y. Zhuang, R. Xie, X. Chen and H. Chen, Angew. Chem., Int. Ed., 2020, 59, 9914–9921 CrossRef CAS PubMed.
- C. Xu, J. Nam, H. Hong, Y. Xu and J. J. Moon, ACS Nano, 2019, 13, 12148–12161 CrossRef CAS PubMed.
- G. Pasparakis, T. Manouras, M. Vamvakaki and P. Argitis, Nat. Commun., 2014, 5, 3623 CrossRef PubMed.
- Y. Li, T.-Y. Lin, Y. Luo, Q. Liu, W. Xiao, W. Guo, D. Lac, H. Zhang, C. Feng and S. Wachsmann-Hogiu, et al. , Nat. Commun., 2014, 5, 4712 CrossRef CAS PubMed.
- S. H. Lim, C. Thivierge, P. Nowak-Sliwinska, J. Han, H. Van Den Bergh, G. Wagnières, K. Burgess and H. B. Lee, J. Med. Chem., 2010, 53, 2865–2874 CrossRef CAS PubMed.
- T. Aida, E. W. Meijer and S. I. Stupp, Science, 2012, 335, 813–817 CrossRef CAS PubMed.
- B. H. Northrop, Y.-R. Zheng, K.-W. Chi and P. J. Stang, Acc. Chem. Res., 2009, 42, 1554–1563 CrossRef CAS PubMed.
- C. Rest, R. Kandanelli and G. Fernandez, Chem. Soc. Rev., 2015, 44, 2543–2572 RSC.
- B. O. Okesola and A. Mata, Chem. Soc. Rev., 2018, 47, 3721–3736 RSC.
- D. M. Raymond and B. L. Nilsson, Chem. Soc. Rev., 2018, 47, 3659–3720 RSC.
- X. Zhang, Z.-K. Chen and K. P. Loh, J. Am. Chem. Soc., 2009, 131, 7210–7211 CrossRef CAS PubMed.
- R. Zou, Q. Wang, J. Wu, J. Wu, C. Schmuck and H. Tian, Chem. Soc. Rev., 2015, 44, 5200–5219 RSC.
- J. Wang, K. Liu, R. Xing and X. Yan, Chem. Soc. Rev., 2016, 45, 5589–5604 RSC.
- K. Tao, A. Levin, L. Adler-Abramovich and E. Gazit, Chem. Soc. Rev., 2016, 45, 3935–3953 RSC.
- Q. Zou and X. Yan, Chemistry, 2018, 24, 755–761 CrossRef CAS PubMed.
- M. Kumar, N. L. Ing, V. Narang, N. K. Wijerathne, A. I. Hochbaum and R. V. Ulijn, Nat. Chem., 2018, 10, 696–703 CrossRef CAS PubMed.
- Y. Li, Q. Zou, C. Yuan, S. Li, R. Xing and X. Yan, Angew. Chem., Int. Ed., 2018, 57, 17084–17088 CrossRef CAS PubMed.
- S. Bera, B. Xue, P. Rehak, G. Jacoby, W. Ji, L. J. W. Shimon, R. Beck, P. Král, Y. Cao and E. Gazit, ACS Nano, 2020, 14, 1694–1706 CrossRef CAS PubMed.
- P. Xing, S. Z. F. Phua, X. Wei and Y. Zhao, Adv. Mater., 2018, 30, 1805175 CrossRef PubMed.
- H. Cao, Q. Yuan, X. Zhu, Y.-P. Zhao and M. Liu, Langmuir, 2012, 28, 15410–15417 CrossRef CAS PubMed.
- K. Liu, R. Xing, Q. Zou, G. Ma, H. Möhwald and X. Yan, Angew. Chem., Int. Ed., 2016, 55, 3036–3039 CrossRef CAS PubMed.
- H. Zhang, K. Liu, S. Li, X. Xin, S. Yuan, G. Ma and X. Yan, ACS Nano, 2018, 12, 8266–8276 CrossRef CAS PubMed.
- S. Li, Q. Zou, Y. Li, C. Yuan, R. Xing and X. Yan, J. Am. Chem. Soc., 2018, 140, 10794–10802 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details and supplementary figures. See DOI: 10.1039/d1sc01073e |
| This journal is © The Royal Society of Chemistry 2021 |