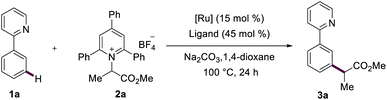Open Access Article
Open Access ArticleDeaminative meta-C–H alkylation by ruthenium(II) catalysis†
Wen
Wei
a,
Hao
Yu
 a,
Agnese
Zangarelli
a and
Lutz
Ackermann
a,
Agnese
Zangarelli
a and
Lutz
Ackermann
 *ab
*ab
aInstitut für Organische und Biomolekulare Chemie, Georg-August-Universität Göttingen, Tammannstraße 2, 37077 Goettingen, Germany. E-mail: Lutz.Ackermann@chemie.uni-goettingen.de
bWoehler Research Institute for Sustainable Chemistry (WISCh), Georg-August-Universität Göttingen, Tammannstrasse 2, 37077 Göttingen, Germany
First published on 9th April 2021
Abstract
Precise structural modifications of amino acids are of importance to tune biological properties or modify therapeutical capabilities relevant to drug discovery. Herein, we report a ruthenium-catalyzed meta-C–H deaminative alkylation with easily accessible amino acid-derived Katritzky pyridinium salts. Likewise, remote C–H benzylations were accomplished with high levels of chemoselectivity and remarkable functional group tolerance. The meta-C–H activation approach combined with our deaminative strategy represents a rare example of selectively converting C(sp3)–N bonds into C(sp3)–C(sp2) bonds.
Introduction
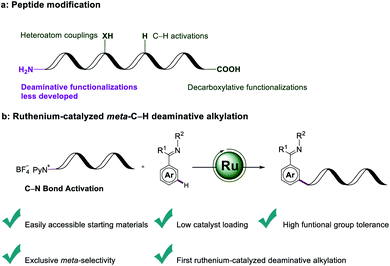
The straightforward formation of modified amino acids from native amino acid precursors1 is conceptually appealing in order to increase the step- and atom-economy associated with the preparation of orthogonally protected molecular scaffolds. Synthetic methods have in the past mainly focused on the modification of limited nucleophilic residues of amino acids, such as found in lysine, tyrosine or cysteine.2 The prevalence of carboxylic acids motifs abundant in peptidic structures of aspartic acid, glutamic acid and α-carboxylic acids, renders them as ideal candidates for targeted peptide modifications. Beyond the traditional amidation and esterification, proteinogenic alkyl carboxylic acids have generally been utilized as handles for transition-metal-catalyzed decarboxylative functionalizations, either by employing activated redox-active esters3 or the direct manipulation of native peptides.4 C–H functionalizations of inert C–H bonds of peptides are of current topical importance for selective late-stage diversifications.5 However, deaminative functionalizations of amino acids as a complementary strategy for the late-stage modifications of amino acids and peptides continue to be underdeveloped (Fig. 1a). By means of activating kinetically stable C(sp3)–N bonds via the in situ formation of α-diazoesters, Wang developed the transition-metal-free deaminative coupling with boronic acids for the synthesis of α-aryl esters.6 Very recently, Rovis reported on a challenging photoredox-catalyzed deaminative alkylation with sterically encumbered α-primary amines.7 Similarly, bench-stable redox-active alkylpyridinium salts—also known as Katritzky pyridinium salts—were utilized by Watson as deaminative reagents for elegant Suzuki–Miyaura cross-coupling reactions.8 Glorius concurrently found a visible-light-mediated Minisci reaction with Katritzky salts.9 Photo-induced deaminative borylations10 and Giese reactions11 were further made possible via the formation of electron-donor–acceptor complex by Aggarwal. The popularity of Katritzky salts as a functional handle was likewise demonstrated by visible-light-mediated transformations, such as Mizoroki–Heck-type reactions,12 allylations,13 alkynylations14 and nickel-catalyzed couplings.15Positional selectivity is paramount to synthetically useful C–H transformations, but challenging because of the close bond dissociation energies.16 Proximity-induced ortho-C–H functionalization by chelation assistance17 proved powerful for late-stage diversification. In stark contrast, remote C–H functionalizations of arenes18 continue to be challenging. Especially, strategies for meta-C–H functionalizations19 continue to be scarce, even though major progress has been achieved by steric control, template assistance, transient mediators and weak hydrogen bonding.20 In this context, we have now merged ruthenium-catalyzed meta-C–H transformations21 with a deaminative bond formation to disclose herein unprecedented ruthenium-catalyzed meta-C–H alkylations with Katritzky salt. Notable features of our findings (Fig. 1b) include (1) easily accessible and bench-stable alkylating agent for C–H functionalizations, (2) low catalyst loading, (3) high functional group tolerance, and (4) selective ruthenium-catalyzed deaminative meta-C–H alkylation and benzylation.
Results and discussion
Optimization of the reaction conditions
We initiated our studies by probing the secondary alkylation of arene 1a with Katritzky salt 2a (Table 1). Among a variety of phosphine ligands, electron-deficient P(4-CF3C6H4)3 proved to be optimal (entries 1–5). Subsequently, we tested different ruthenium catalysts, and [RuCl2(p-cymene)]2 led to the desired meta-alkylated product 3a in high yield (entries 8 and 9). Control experiments verified the key role of the phosphine ligand and the ruthenium catalyst (entries 6 and 7). Notably, decreasing the amount of the catalyst and the phosphine ligand did not significantly alter the reaction performance (entry 9).| Entry | Catalyst | Ligand | Yieldb/% |
|---|---|---|---|
| a Reaction conditions: 1a (0.2 mmol), 2a (0.4 mmol), [Ru(O2CMes)2(p-cymene)2] (15 mol%), P(4-C6H4CF3)3 (45 mol%), Na2CO3 (0.4 mmol) 1,4-dioxane (2.0 mL) at 100 °C for 24 h. b Yield of isolated products. c [RuCl2(p-cymene)2]2 (2.5 mol%), P(4-C6H4CF3)3 (10 mol%). d 2,4,6-Triphenyl pyridine was isolated in 89%. | |||
| 1 | [Ru(O2CMes)2(p-cymene)] | PPh3 | 37 |
| 2 | [Ru(O2CMes)2(p-cymene)] | P(4-F-C6H4)3 | 45 |
| 3 | [Ru(O2CMes)2(p-cymene)] | P(4-MeOC6H4)3 | 24 |
| 4 | [Ru(O2CMes)2(p-cymene)] | P(4-C6H4CF3)3 | 63 |
| 5 | [Ru(O2CMes)2(p-cymene)] | PCy3 | 7 |
| 6 | [Ru(O2CMes)2(p-cymene)] | — | Trace |
| 7 | — | P(4-C6H4CF3)3 | Trace |
| 8 | [RuCl2(p-cymene)]2 | P(4-C6H4CF3)3 | 79 |
| 9 | [RuCl 2 (p-cymene)] 2 | P(4-C 6 H 4 CF 3 ) 3 | 77 , |
Versatility
With the optimized reaction conditions for the ruthenium-catalyzed deaminative C–H alkylation in hand, we next examined its versatility (Scheme 1). The ruthenium catalysis was not limited to pyridine guidance. Indeed, the meta-alkylation also allowed for the use of pyrazoles (3g–3l), pyrimidines (3m), oxazolines (3n and 3o) and benzoquinoline (3p) as orienting motifs, while the functionalization of heteroarenes provided thus far less satisfactory results. A range of synthetically useful functional groups, such as halides (3c, 3k and 3l), ester (3j) and ketone (3f), were well tolerated and furnished the desired meta-alkylated products 3 in good to excellent yields and high levels of meta-selectivity.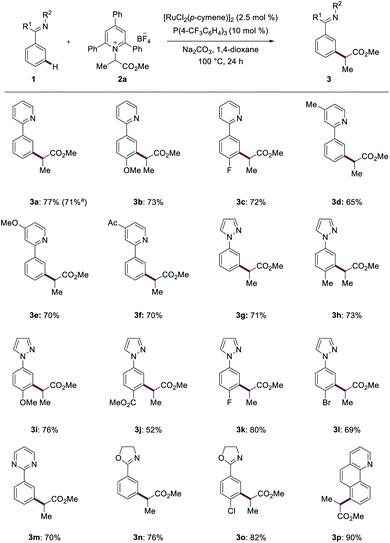 | ||
| Scheme 1 Ruthenium-catalyzed meta-C–H alkylations with heteroarene orienting groups. a Gram scale reaction at 1 mmol. | ||
Next, Katritzky salts 2 derived from functionalized amino acids were tested (Scheme 2). A wide range of aliphatic amino acids, including alanine, valine, leucine, and phenylalanine, could be selectively converted into the desired products 4a–4d. Furthermore, the corresponding products 4e and 4f were obtained in 77% and 69% yield, respectively, when aspartic acid- and glutamic acid-derived Katritzky salts were employed. Notably, functional groups, including the free hydroxyl group in tyrosine (4g) and the free amino group in lysine (4j) as well as free NH-indole in tryptophan (4k), were well tolerated. Derivatives from protected tyrosine and phenylalanine containing easily transformable functional groups furnished the desired products 4h and 4i efficiently. Structurally more complex dipeptide-derived Katritzky salt ([PyN]–Phe–Ala–OMe) selectively underwent the meta-ligation, delivering the desired product 4l. In addition, heteroarenes (4o–4q) were compatible with the ruthenium catalysis. Importantly, the transformative power of our approach was harnessed for late-stage diversifications. Marketed drugs and natural product-like compounds, such as indomethacin (4r), dehydrocholic acid (4s) and elaidic acid (4t), proved to be amenable, indicating the potential for drug discovery programs.
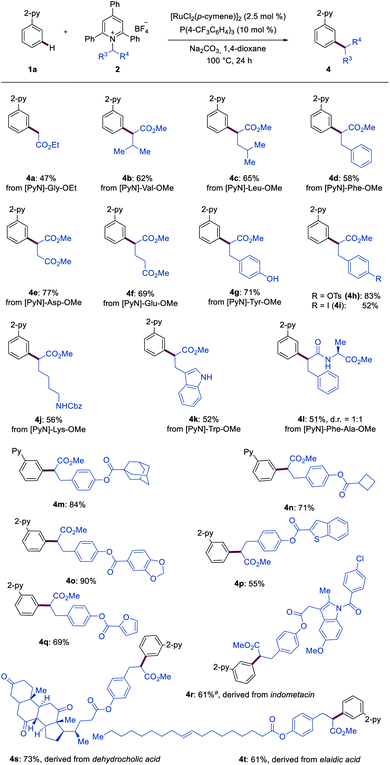 | ||
| Scheme 2 Ruthenium-catalyzed meta-C–H secondary alkylation. a 5 mol% of [RuCl2(p-cymene)2]2 was used. | ||
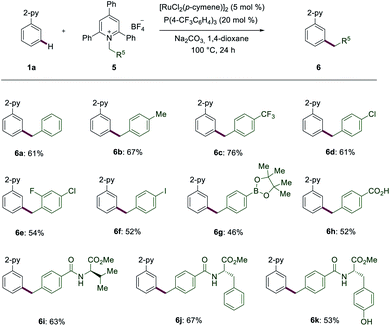
In addition to amino acid derivatives, we also explored Katritzky salts of primary amines to achieve benzylations (Scheme 3). Electron-donating and electron-withdrawing benzyl groups, such as products 6b and 6c were well obtained. Halogen-containing substrates also resulted in good yields of the corresponding meta-benzylated products 6d–6f. Notably, highly labile functional groups, such as Bpin (6g), also proved to be applicable. Katritzky salts bearing a free acid chemo-selectively led to the desired product 6h. Products 6i–6k were obtained in a synthetically useful yield and with high levels of chemo-selectivity from the amino acids-derived Katritzky salts.
Mechanistic studies
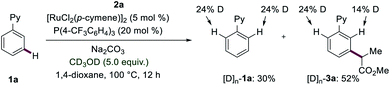
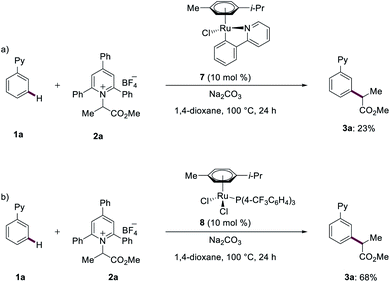
In order to elucidate the reaction mechanism of the ruthenium(II)-catalyzed meta-C–H transformations, we subsequently conducted mechanistic studies. Competition experiments highlighted that electron-rich substrates are inherently less reactive than the electron-deficient counterparts. Katritzky salts with electron-withdrawing groups turned out to be more effective for the meta-transformation (see ESI†). Reactions with isotopically labelled co-solvent CD3OD provided strong support for facile reversible C–H activation at the ortho-position (Fig. 2). Given this exclusive meta-selectivity, we were inspired to identify key intermediates. First, the ruthenium complexes 7 and 8 were synthesized and isolated. When catalytic amounts of complex 7 were utilized, the product 3a was obtained in 23% (Fig. 3a). Instead, ruthenium–phosphine catalyst 8 displayed a significantly improved performance (Fig. 3b).Subsequently, experiments with the typical radical trapping reagent TEMPO led to an inhibition of reactivity (see ESI†). A radical clock experiment with allyl-containing Katritzky salt afforded the corresponding ring closure product observed by ESI-MS spectrometry (see ESI†), being supportive of a radical pathway being operative.
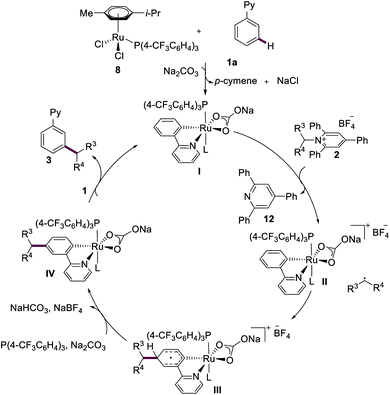
On the basis of our mechanistic findings, a plausible reaction mechanism for the ruthenium-catalyzed meta-C–H alkylation is put forward in Scheme 4, which commences by a chelation-assisted C–H ruthenation and dissociation of p-cymene ligand, forming ruthenacycle I.22 Single-electron transfer (SET) from the ruthenium(II) complex I to the Katritzky salt 2 delivers the ruthenium(III) intermediate II, along with isolated triphenylpyridine (12) (see ESI†). The newly formed secondary alkyl radical attacks the aromatic motif at the position para to the C–Ru bond, generating the triplet ruthenium intermediate III. Rearomatization then leads to the formation of ruthenacycle IV. Finally, proto-demetalation and ligand exchange affords the desired meta-functionalized product 3 and regenerates ruthenium(II) complex I.
Conclusion
In summary, we have reported on ruthenium-catalyzed meta-selective C–H secondary alkylations and benzylations with easily accessible pyridinium salts. The ruthenium catalysis featured excellent chemo- and position-selectivities as well as a broad functional group tolerance. Importantly, the deaminative strategy set the stage for late-stage diversification of bioactive molecules and marketed drugs by deaminative transformations of amino acids and peptides.Author contributions
L. A. and Y. H. conceived the project. W. W. performed the experiments, analyzed and interpreted the experimental data. W. W. and A. Z. drafted the paper. All of the authors discussed the results and contributed to the preparation of the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Generous support by the DFG (Gottfried-Wilhelm-Leibniz award to L. A.) and the CSC (fellowship to W. W.) is gratefully acknowledged.Notes and references
-
(a) C. T. Walsh, S. Garneau-Tsodikova and G. J. Gatto, Jr., Angew. Chem., Int. Ed., 2005, 44, 7342–7372 CrossRef CAS PubMed
; (b) J. M. Chalker, G. J. Bernardes, Y. A. Lin and B. G. Davis, Chem.–Asian J., 2009, 4, 630–640 CrossRef CAS PubMed
; (c) C. D. Spicer and B. G. Davis, Nat. Commun., 2014, 5, 4740 CrossRef CAS PubMed
; (d) J. N. deGruyter, L. R. Malins and P. S. Baran, Biochemistry, 2017, 56, 3863–3873 CrossRef CAS PubMed
; (e) C. Bottecchia and T. Noel, Chem.–Eur. J., 2019, 25, 26–42 CrossRef CAS PubMed
.
-
(a) B. A. Vara, X. Li, S. Berritt, C. R. Walters, E. J. Petersson and G. A. Molander, Chem. Sci., 2018, 9, 336–344 RSC
; (b) A. J. Rojas, C. Zhang, E. V. Vinogradova, N. H. Buchwald, J. Reilly, B. L. Pentelute and S. L. Buchwald, Chem. Sci., 2017, 8, 4257–4263 RSC
; (c) H. G. Lee, G. Lautrette, B. L. Pentelute and S. L. Buchwald, Angew. Chem., Int. Ed., 2017, 56, 3177–3181 CrossRef CAS PubMed
; (d) C. Bottecchia, M. Rubens, S. B. Gunnoo, V. Hessel, A. Madder and T. Noel, Angew. Chem., Int. Ed., 2017, 56, 12702–12707 CrossRef CAS PubMed
; (e) J. Willwacher, R. Raj, S. Mohammed and B. G. Davis, J. Am. Chem. Soc., 2016, 138, 8678–8681 CrossRef CAS PubMed
; (f) J. Ohata, M. B. Minus, M. E. Abernathy and Z. T. Ball, J. Am. Chem. Soc., 2016, 138, 7472–7475 CrossRef CAS PubMed
; (g) E. V. Vinogradova, C. Zhang, A. M. Spokoyny, B. L. Pentelute and S. L. Buchwald, Nature, 2015, 526, 687–691 CrossRef CAS PubMed
; (h) D. T. Cohen, C. Zhang, B. L. Pentelute and S. L. Buchwald, J. Am. Chem. Soc., 2015, 137, 9784–9787 CrossRef CAS PubMed
.
-
(a) J. M. Smith, T. Qin, R. R. Merchant, J. T. Edwards, L. R. Malins, Z. Liu, G. Che, Z. Shen, S. A. Shaw, M. D. Eastgate and P. S. Baran, Angew. Chem., Int. Ed., 2017, 56, 11906–11910 CrossRef CAS PubMed
; (b) T. Qin, L. R. Malins, J. T. Edwards, R. R. Merchant, A. J. Novak, J. Z. Zhong, R. B. Mills, M. Yan, C. Yuan, M. D. Eastgate and P. S. Baran, Angew. Chem., Int. Ed., 2017, 56, 260–265 CrossRef CAS PubMed
; (c) C. Li, J. Wang, L. M. Barton, S. Yu, M. Tian, D. S. Peters, M. Kumar, A. W. Yu, K. A. Johnson, A. K. Chatterjee, M. Yan and P. S. Baran, Science, 2017, 356, eaam7355 CrossRef PubMed
; (d) J. T. Edwards, R. R. Merchant, K. S. McClymont, K. W. Knouse, T. Qin, L. R. Malins, B. Vokits, S. A. Shaw, D. H. Bao, F. L. Wei, T. Zhou, M. D. Eastgate and P. S. Baran, Nature, 2017, 545, 213–218 CrossRef CAS PubMed
; (e) W. M. Cheng, R. Shang and Y. Fu, ACS Catal., 2017, 7, 907–911 CrossRef CAS
; (f) Y. Jin, M. Jiang, H. Wang and H. Fu, Sci. Rep., 2016, 6, 20068 CrossRef PubMed
.
-
(a) M. Garreau, F. Le Vaillant and J. Waser, Angew. Chem., Int. Ed., 2019, 58, 8182–8186 CrossRef CAS PubMed
; (b) S. Bloom, C. Liu, D. K. Kolmel, J. X. Qiao, Y. Zhang, M. A. Poss, W. R. Ewing and D. W. C. MacMillan, Nat. Chem., 2018, 10, 205–211 CrossRef CAS PubMed
; (c) S. J. McCarver, J. X. Qiao, J. Carpenter, R. M. Borzilleri, M. A. Poss, M. D. Eastgate, M. M. Miller and D. W. MacMillan, Angew. Chem., Int. Ed., 2017, 56, 728–732 CrossRef CAS PubMed
.
-
(a) H.-R. Tong, B. Li, G. Li, G. He and G. Chen, CCS Chem., 2020, 1797–1820 Search PubMed
; (b) D. G. Rivera, G. M. Ojeda-Carralero, L. Reguera and E. V. Van der Eycken, Chem. Soc. Rev., 2020, 49, 2039–2059 RSC
; (c) Z. B. Bai and H. Wang, Synlett, 2020, 31, 199–204 CrossRef CAS
; (d) B.-B. Zhan, M.-X. Jiang and B.-F. Shi, Chem. Commun., 2020, 56, 13950–13958 RSC
; (e) W. Wang, M. M. Lorion, J. Shah, A. R. Kapdi and L. Ackermann, Angew. Chem., Int. Ed., 2018, 57, 14700–14717 CrossRef CAS PubMed
; (f) A. Correa and M. Segundo, Synthesis, 2018, 50, 2853–2866 CrossRef
; (g) T. Brandhofer and O. G. Mancheno, Eur. J. Org. Chem., 2018, 2018, 6050–6067 CrossRef CAS
; (h) S. Sengupta and G. Mehta, Tetrahedron Lett., 2017, 58, 1357–1372 CrossRef CAS
; (i) G. He, B. Wang, W. A. Nack and G. Chen, Acc. Chem. Res., 2016, 49, 635–645 CrossRef CAS PubMed
; (j) A. F. Noisier and M. A. Brimble, Chem. Rev., 2014, 114, 8775–8806 CrossRef CAS PubMed
.
- G. Wu, Y. Deng, C. Wu, Y. Zhang and J. Wang, Angew. Chem., Int. Ed., 2014, 53, 10510–10514 CrossRef CAS PubMed
.
- M. A. Ashley and T. Rovis, J. Am. Chem. Soc., 2020, 142, 18310–18316 CrossRef CAS PubMed
.
-
(a) C. H. Basch, J. Liao, J. Xu, J. J. Piane and M. P. Watson, J. Am. Chem. Soc., 2017, 139, 5313–5316 CrossRef CAS PubMed
; (b) M. E. Hoerrner, K. M. Baker, C. H. Basch, E. M. Bampo and M. P. Watson, Org. Lett., 2019, 21, 7356–7360 CrossRef CAS PubMed
.
- F. J. R. Klauck, M. J. James and F. Glorius, Angew. Chem., Int. Ed., 2017, 56, 12336–12339 CrossRef CAS PubMed
.
-
(a) J. Wu, L. He, A. Noble and V. K. Aggarwal, J. Am. Chem. Soc., 2018, 140, 10700–10704 CrossRef CAS PubMed
; (b) J. Hu, G. Wang, S. Li and Z. Shi, Angew. Chem., Int. Ed., 2018, 57, 15227–15231 CrossRef CAS PubMed
.
- J. Wu, P. S. Grant, X. Li, A. Noble and V. K. Aggarwal, Angew. Chem., Int. Ed., 2019, 58, 5697–5701 CrossRef CAS PubMed
.
- X. Jiang, M. M. Zhang, W. Xiong, L. Q. Lu and W. J. Xiao, Angew. Chem., Int. Ed., 2019, 58, 2402–2406 CrossRef CAS PubMed
.
- M. M. Zhang and F. Liu, Org. Chem. Front., 2018, 5, 3443–3446 RSC
.
- M. Ociepa, J. Turkowska and D. Gryko, ACS Catal., 2018, 8, 11362–11367 CrossRef CAS
.
-
(a) C. G. Yu and Y. Matsuo, Org. Lett., 2020, 22, 950–955 CrossRef CAS PubMed
; (b) J. Wang, M. E. Hoerrner, M. P. Watson and D. J. Weix, Angew. Chem., Int. Ed., 2020, 59, 13484–13489 CrossRef CAS PubMed
; (c) F. T. Pulikottil, R. Pilli, R. V. Suku and R. Rasappan, Org. Lett., 2020, 22, 2902–2907 CrossRef CAS PubMed
; (d) H. Yue, C. Zhu, L. Shen, Q. Geng, K. J. Hock, T. Yuan, L. Cavallo and M. Rueping, Chem. Sci., 2019, 10, 4430–4435 RSC
; (e) J. Yi, S. O. Badir, L. M. Kammer, M. Ribagorda and G. A. Molander, Org. Lett., 2019, 21, 3346–3351 CrossRef CAS PubMed
; (f) S. Z. Sun, C. Romano and R. Martin, J. Am. Chem. Soc., 2019, 141, 16197–16201 CrossRef CAS PubMed
; (g) S. Plunkett, C. H. Basch, S. O. Santana and M. P. Watson, J. Am. Chem. Soc., 2019, 141, 2257–2262 CrossRef CAS PubMed
; (h) S. Ni, C. X. Li, Y. Mao, J. Han, Y. Wang, H. Yan and Y. Pan, Sci. Adv., 2019, 5, eaaw9516 CrossRef CAS PubMed
; (i) R. Martin-Montero, V. R. Yatham, H. Yin, J. Davies and R. Martin, Org. Lett., 2019, 21, 2947–2951 CrossRef CAS PubMed
; (j) J. Liao, C. H. Basch, M. E. Hoerrner, M. R. Talley, B. P. Boscoe, J. W. Tucker, M. R. Garnsey and M. P. Watson, Org. Lett., 2019, 21, 2941–2946 CrossRef CAS PubMed
; (k) K. M. Baker, D. Lucas Baca, S. Plunkett, M. E. Daneker and M. P. Watson, Org. Lett., 2019, 21, 9738–9741 CrossRef CAS PubMed
; (l) J. Liao, W. Guan, B. P. Boscoe, J. W. Tucker, J. W. Tomlin, M. R. Garnsey and M. P. Watson, Org. Lett., 2018, 20, 3030–3033 CrossRef CAS PubMed
.
-
(a) R. Gramage-Doria and C. Bruneau, Coord. Chem. Rev., 2021, 428, 213602 CrossRef CAS
; (b) Q. Zhao, G. Meng, S. P. Nolan and M. Szostak, Chem. Rev., 2020, 120, 1981–2048 CrossRef CAS PubMed
; (c) S. Rej, Y. Ano and N. Chatani, Chem. Rev., 2020, 120, 1788–1887 CrossRef CAS PubMed
; (d) F. Kakiuchi and T. Kochi, Chem. Lett., 2020, 49, 1256–1269 CrossRef CAS
; (e) S. Rej and N. Chatani, Angew. Chem., Int. Ed., 2019, 58, 8304–8329 CrossRef CAS PubMed
; (f) S. M. Khake and N. Chatani, Trends Chem., 2019, 1, 524–539 CrossRef CAS
; (g) Y. Park, Y. Kim and S. Chang, Chem. Rev., 2017, 117, 9247–9301 CrossRef CAS PubMed
; (h) B. Li and P. H. Dixneuf, Chem. Soc. Rev., 2013, 42, 5744–5767 RSC
; (i) P. B. Arockiam, C. Bruneau and P. H. Dixneuf, Chem. Rev., 2012, 112, 5879–5918 CrossRef CAS PubMed
.
- L. Ackermann, Acc. Chem. Res., 2014, 47, 281–295 CrossRef CAS PubMed
.
-
(a) A. R. A. Spencer, R. Korde, M. Font and I. Larrosa, Chem. Sci., 2020, 11, 4204–4208 RSC
; (b) S. Porey, X. Zhang, S. Bhowmick, V. Kumar Singh, S. Guin, R. S. Paton and D. Maiti, J. Am. Chem. Soc., 2020, 142, 3762–3774 CrossRef CAS PubMed
; (c) S. Bag, K. Surya, A. Mondal, R. Jayarajan, U. Dutta, S. Porey, R. B. Sunoj and D. Maiti, J. Am. Chem. Soc., 2020, 142, 12453–12466 CrossRef CAS PubMed
; (d) M. Font, A. R. A. Spencer and I. Larrosa, Chem. Sci., 2018, 9, 7133–7137 RSC
; (e) P. Wang, P. Verma, G. Xia, J. Shi, J. X. Qiao, S. Tao, P. T. W. Cheng, M. A. Poss, M. E. Farmer, K. S. Yeung and J. Q. Yu, Nature, 2017, 551, 489–493 CrossRef CAS PubMed
; (f) M. Simonetti and I. Larrosa, Nat. Chem., 2016, 8, 1086–1088 CrossRef CAS PubMed
; (g) X. C. Wang, W. Gong, L. Z. Fang, R. Y. Zhu, S. Li, K. M. Engle and J. Q. Yu, Nature, 2015, 519, 334–338 CrossRef CAS PubMed
; (h) Y. Kuninobu, H. Ida, M. Nishi and M. Kanai, Nat. Chem., 2015, 7, 712–717 CrossRef CAS PubMed
; (i) J. Luo, S. Preciado and I. Larrosa, J. Am. Chem. Soc., 2014, 136, 4109–4112 CrossRef CAS PubMed
; (j) D. Leow, G. Li, T. S. Mei and J. Q. Yu, Nature, 2012, 486, 518–522 CrossRef CAS PubMed
; (k) R. J. Phipps and M. J. Gaunt, Science, 2009, 323, 1593–1597 CrossRef CAS PubMed
; (l) J. Y. Cho, M. K. Tse, D. Holmes, R. E. Maleczka, Jr. and M. R. Smith, 3rd, Science, 2002, 295, 305–308 CrossRef CAS PubMed
.
-
(a) G. Meng, N. Y. S. Lam, E. L. Lucas, T. G. Saint-Denis, P. Verma, N. Chekshin and J. Q. Yu, J. Am. Chem. Soc., 2020, 142, 10571–10591 CrossRef CAS PubMed
; (b) A. Dey, S. K. Sinha, T. K. Achar and D. Maiti, Angew. Chem., Int. Ed., 2019, 58, 10820–10843 CrossRef CAS PubMed
; (c) M. T. Mihai, G. R. Genov and R. J. Phipps, Chem. Soc. Rev., 2018, 47, 149–171 RSC
; (d) J. A. Leitch and C. G. Frost, Chem. Soc. Rev., 2017, 46, 7145–7153 RSC
; (e) N. Della Ca, M. Fontana, E. Motti and M. Catellani, Acc. Chem. Res., 2016, 49, 1389–1400 CrossRef CAS
; (f) C. G. Frost and A. J. Paterson, ACS Cent. Sci., 2015, 1, 418–419 CrossRef CAS PubMed
; (g) L. Ackermann and J. Li, Nat. Chem., 2015, 7, 686–687 CrossRef CAS
; (h) F. Julia-Hernandez, M. Simonetti and I. Larrosa, Angew. Chem., Int. Ed., 2013, 52, 11458–11460 CrossRef CAS PubMed
.
-
(a) G. Meng, N. Y. S. Lam, E. L. Lucas, T. G. Saint-Denis, P. Verma, N. Chekshin and J.-Q. Yu, J. Am. Chem. Soc., 2020, 142, 10571–10591 CrossRef CAS PubMed
; (b) Y. Kuninobu and T. Torigoe, Org. Biomol. Chem., 2020, 18, 4126–4134 RSC
; (c) A. Dey, S. K. Sinha, T. K. Achar and D. Maiti, Angew. Chem., Int. Ed., 2019, 58, 10820–10843 CrossRef CAS PubMed
; (d) M. T. Mihai, G. R. Genov and R. J. Phipps, Chem. Soc. Rev., 2018, 47, 149–171 RSC
; (e) L. Ackermann and R. Vicente, in C–H Activation, ed. J.-Q. Yu and Z. Shi, Springer Berlin Heidelberg, Berlin, Heidelberg, 2010, pp. 211–229 Search PubMed
.
-
(a)
K. Korvorapun, R. C. Samanta, T. Rogge and L. Ackermann, Ruthenium-Catalyzed Remote C–H Functionalizations, in Remote C–H bond Functionalizations, ed. D. Maiti and S. Guin, Wiley-VCH, Weinheim, 2021, pp. 137–165 Search PubMed
; (b) G. Li, J. An, C. Jia, B. Yan, L. Zhong, J. Wang and S. Yang, Org. Lett., 2020, 22, 9450–9455 CrossRef CAS PubMed
; (c) K. Korvorapun, M. Moselage, J. Struwe, T. Rogge, A. M. Messinis and L. Ackermann, Angew. Chem., Int. Ed., 2020, 59, 18795–18803 CrossRef CAS PubMed
; (d) K. Korvorapun, R. Kuniyil and L. Ackermann, ACS Catal., 2020, 10, 435–440 CrossRef CAS
; (e) X. G. Wang, Y. Li, H. C. Liu, B. S. Zhang, X. Y. Gou, Q. Wang, J. W. Ma and Y. M. Liang, J. Am. Chem. Soc., 2019, 141, 13914–13922 CrossRef CAS PubMed
; (f) A. Sagadevan and M. F. Greaney, Angew. Chem., Int. Ed., 2019, 58, 9826–9830 CrossRef CAS PubMed
; (g) P. Gandeepan, J. Koeller, K. Korvorapun, J. Mohr and L. Ackermann, Angew. Chem., Int. Ed., 2019, 58, 9820–9825 CrossRef CAS
; (h) K. Korvorapun, N. Kaplaneris, T. Rogge, S. Warratz, A. C. Stückl and L. Ackermann, ACS Catal., 2018, 8, 886–892 CrossRef CAS
; (i) J. A. Leitch and C. G. Frost, Chem. Soc. Rev., 2017, 46, 7145–7153 RSC
; (j) S. Warratz, D. J. Burns, C. Zhu, K. Korvorapun, T. Rogge, J. Scholz, C. Jooss, D. Gelman and L. Ackermann, Angew. Chem., Int. Ed., 2017, 56, 1557–1560 CrossRef CAS PubMed
; (k) J. Li, K. Korvorapun, S. De Sarkar, T. Rogge, D. J. Burns, S. Warratz and L. Ackermann, Nat. Commun., 2017, 8, 15430 CrossRef
; (l) G. B. Li, D. Z. Li, J. Y. Zhang, D. Q. Shi and Y. S. Zhao, ACS Catal., 2017, 7, 4138–4143 CrossRef CAS
; (m) B. Li, S. L. Fang, D. Y. Huang and B. F. Shi, Org. Lett., 2017, 19, 3950–3953 CrossRef CAS PubMed
; (n) C. J. Teskey, A. Y. Lui and M. F. Greaney, Angew. Chem., Int. Ed., 2015, 54, 11677–11680 CrossRef CAS PubMed
; (o) A. J. Paterson, S. St John-Campbell, M. F. Mahon, N. J. Press and C. G. Frost, Chem. Commun., 2015, 51, 12807–12810 RSC
; (p) J. Li, S. Warratz, D. Zell, S. De Sarkar, E. E. Ishikawa and L. Ackermann, J. Am. Chem. Soc., 2015, 137, 13894–13901 CrossRef CAS PubMed
; (q) N. Hofmann and L. Ackermann, J. Am. Chem. Soc., 2013, 135, 5877–5884 CrossRef CAS PubMed
; (r) L. Ackermann, N. Hofmann and R. Vicente, Org. Lett., 2011, 13, 1875–1877 CrossRef CAS PubMed
; (s) O. Saidi, J. Marafie, A. E. Ledger, P. M. Liu, M. F. Mahon, G. Kociok-Kohn, M. K. Whittlesey and C. G. Frost, J. Am. Chem. Soc., 2011, 133, 19298–19301 CrossRef CAS PubMed
.
-
(a) L. Ackermann, Chem. Rev., 2011, 111, 1315–1345 CrossRef CAS PubMed
; (b) L. Ackermann, R. Vicente and A. Althammer, Org. Lett., 2008, 10, 2299–2302 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc00986a |
| This journal is © The Royal Society of Chemistry 2021 |