Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Simultaneous detection of small molecule thiols with a simple 19F NMR platform†
Zhaofei
Chai‡
a,
Qiong
Wu‡
ab,
Kai
Cheng
a,
Xiaoli
Liu
a,
Ling
Jiang
ab,
Maili
Liu
*ab and
Conggang
Li
 *ab
*ab
aKey Laboratory of Magnetic Resonance in Biological Systems, State Key Laboratory of Magnetic Resonance and Atomic and Molecular Physics, National Center for Magnetic Resonance in Wuhan, Wuhan National Laboratory for Optoelectronics, Wuhan Institute of Physics and Mathematics, Innovation Academy for Precision Measurement Science and Technology, Chinese Academy of Sciences, Wuhan 430071, China. E-mail: conggangli@apm.ac.cn
bGraduate University of Chinese Academy of Sciences, Beijing 100049, China
First published on 13th November 2020
Abstract
Thiols play critical roles in regulating biological functions and have wide applications in pharmaceutical and biomedical industries. However, we still lack a general approach for the simultaneous detection of various thiols, especially in complex systems. Herein, we establish a 19F NMR platform where thiols are selectively fused into a novelly designed fluorinated receptor that has two sets of environmentally different 19F atoms with fast kinetics (k2 = 0.73 mM−1 min−1), allowing us to generate unique two-dimensional codes for about 20 thiols. We demonstrate the feasibility of the approach by reliably quantifying thiol drug content in tablets, discriminating thiols in living cells, and for the first time monitoring the thiol related metabolism pathway at the atomic level. Moreover, the method can be easily extended to detect the activity of thiol related enzymes such as γ-glutamyl transpeptidase. We envision that the versatile platform will be a useful tool for detecting thiols and elucidating thiol-related processes in complex systems.
Introduction
Small molecule thiols are of great interest to the scientific community owing to their vital roles in many physiological processes and pharmaceutical applications. For example, biothiols of cysteine (Cys), homocysteine (Hcy), glutathione (GSH) and hydrogen sulfide (H2S) are found in abundance and involved in many cellular regulations, and their abnormal levels are directly correlated with many different diseases.1–3 Meanwhile, thiol drugs (e.g. penicillamine, tiopronin, and thiamazole) are present with endogenous thiols to display different curative effects in the clinical field.4–6 Thus, real-time identification and quantification of these thiols would be of considerable significance to provide insights into related pathophysiological changes and pharmacokinetic information. Consequently, various analytical methods have been reported to sense and image these species in living cells and organisms, especially simultaneous detection of two or more species, which could provide more information on their close relationship under specific metabolism and joint action in disease diagnosis.7–9 So far, the noninvasive and selective detection of thiols mainly focuses on the fluorescence technology, involving efficient probes with multiple reaction sites or cleavage paths to different functional groups of the analytes. But the intrinsic property of this strategy makes it difficult for probes to discriminate those containing only one reactive sulfhydryl unit or ones with the same reaction features. To reflect the structural character and concentration of the analytes, the 19F NMR technique, an alternative approach, has recently emerged to identify structurally similar molecules in complex mixtures, benefitting from the similar gyromagnetic ratio of 19F to 1H, 100% natural abundance of 19F, scarcity of naturally occurring background signals, and a broad chemical shift range.10,11 Though the transformation of similar structural information of analytes into resolvable 19F NMR shifts is intriguing, the probe design is still far from being fully explored. Only a few fluorinated compounds have been successfully used to discriminate biological organic compounds in simulated or simple biological samples.12–17 So the systematic design of probes for multiple thiol discrimination is valuable for biological research and 19F NMR probe engineering.In this study, based on the nucleophilic substitution of –SH towards fluorinated sulfoxides, we introduced a simple 19F NMR platform that enabled the recognition of around 20 thiols with high sensitivity and rapid reaction kinetics under physiological conditions. The new method successfully achieved endogenous thiol quantification and the real-time monitoring of their conversion in living HeLa cells. Importantly, we observed a critical intermediate during GSH (or GSH–probe adduct) metabolism in living cells, which would help improve the accuracy of further Cys and GSH measurements. We also extend the proposed strategy to detect thiol-related enzymes, such as γ-glutamyl transpeptidase (γ-GT).
Results and discussion
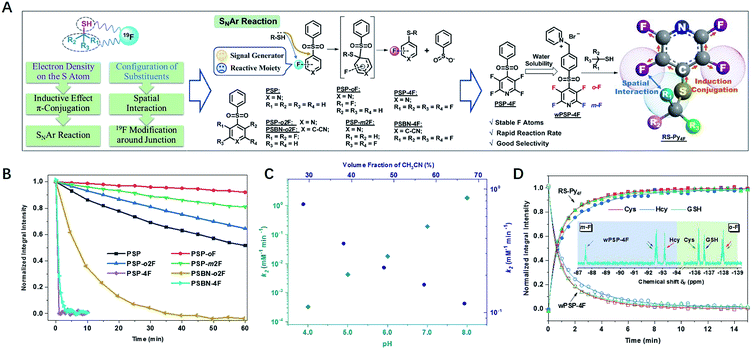
Design of the 19F probe with rapid response, high sensitivity, and reliable quantification
Our 19F NMR approach needed 19F probes with high sensitivity to the local electronic microenvironment induced by thiols (Fig. 1A).18 In order to transmit the electron density on the sulfur atom through both inductive effect and conjugation more efficiently, we chose nucleophilic aromatic substitution (SNAr) that fluorinated aromatic sulfones and thiols formed stable thioethers.19–21,43–47Another characteristic of substitution arrangement around –SH was that it could be perceived by adjacent 19F atoms via spatial proximity. To develop a practical probe with fast response and good selectivity, we synthesized an array of F-substituted sulfones through a one-step process (ESI†), and evaluated their practicability with three typical biothiols of Cys, Hcy, and GSH. The results indicated that the modification of electron-withdrawing units (F atoms, pyridyl, and benzonitrile) could significantly increase the reactivity of sulfones (Fig. 1B, S1–S8†). Both PSP-4F and PSBN-4F outstood among these candidates because of their short reaction times (<3 min). Owing to the strongest electron-deficient ability, PSBN-4F displayed a very complicated 19F NMR spectrum upon the addition of Cys due to the activated F–C bonds and the special S → N rearrangement,22–25 while PSP-4F formed a stable product and exhibited two explicit peaks under the same conditions, which was much more straightforward for further analysis. Besides suitable reactivity, PSP-4F possessing two types of environmentally different 19F atoms also showed superior recognition ability towards thiols (Fig. S9†). Two factors contributed to this recognition ability: the meta-fluorine (m-F) could sense the change of electron density (Cys vs. Hcy) and meanwhile kept the thiol signals away from the probe itself (Δδ > 4 ppm), and the ortho-fluorine (o-F) was able to precisely reflect the structural variation around –SH owing to different interactions with substituents (Cys vs. GSH). Further investigation of PSP-4F and Cys showed that high pH values and water fraction were also favorable to this reaction, possibly due to the enhanced nucleophilicity of –SH and the stabilization of the intermediate, respectively (Fig. 1C, S10 and S11†).25In order to be more compatible with biological samples, PSP-4F was converted into a water-soluble pyridinium salt (wPSP-4F), which likewise achieved satisfactory discrimination with fast reaction kinetics under physiological conditions (Fig. 1D, S12 and S13†). wPSP-4F also manifested good selectivity towards –SH (Fig. S14†), and common inorganic ions and amino acids except for Cys did not induce any observable changes. Only H2S elicited a set of sharply distinguishable peaks compared with three biothiols (ΔδmF > 7 and ΔδoF > 3 ppm). Moreover, two distinct peaks (Δδ > 50 ppm) were beneficial to further multi-thiol determination through selective peaks (m-F or o-F) with better resolutions. In addition, the integral intensities of wPSP-4F and corresponding thioether (RS–Py4F) were linearly proportional to the thiol concentration (R2 > 99.7%) in the 0.1–10 mM range (GSH for instance), in favor of quantitative detection (Fig. S15†). Compared with reported 19F NMR probes for multi-component detection, our probe reacted with thiols in a stoichiometric proportion of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and formed stable thioester without any exchange with analogs, which improved the detection limit (<5 μM) under the same conditions (Fig. S16†) and widened its potential application in complex environments.13–16
1 and formed stable thioester without any exchange with analogs, which improved the detection limit (<5 μM) under the same conditions (Fig. S16†) and widened its potential application in complex environments.13–16
As an illustration of reliable quantitative detection, the content of thiols in commercial N-acetylcysteine (NAC) effervescent tablets was determined (Table 1, Fig. S17 and S18†).26–28 By simply mixing the probe and samples, the NMR approach gave similar accuracy to the standard HPLC method,29 and NMR could determine the NAC at a lower concentration (0.5 mg mL−1vs. 0.01 mg mL−1) of NAC, suggesting wPSP-4F to be a promising and practical tool for thiol quantification in vitro.
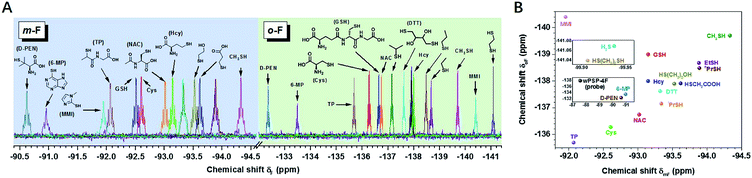
Distinct discriminatory ability of the 19F probe towards small molecule thiols
After the initial test of the probe with typical biothiols, we assessed its discriminatory ability with a variety of other biothiols, thiol drugs, and pollutants that may coexist or have closely related structures. As shown in Fig. 2A, most of the thiols showed well-resolved m- or o-F peaks, except a few similar structures that had overlapped ones. However, the ones with overlapped m-F peaks had separated o-F peaks, and vice versa. For example, nPrSH and EtSH displayed almost the same δmF because the electronic densities of –SH in them were similar, but they had different δoF because nPrSH had slightly larger steric hindrance than EtSH, resulting in a downfield chemical shift of δoF (ΔδoF > 0.2 ppm). We found that the increased spatial interactions from the primary thiols to the secondary and tertiary ones led to a similar trend of chemical shifts in each region (e.g. δD-PEN > δTP > δCys; δi-PrSH > δn-PrSH), but had a more profound influence on the o-F. In contrast, 3-mercaptopropionic acid and 2-mercaptoethanol exhibited a similar interaction with o-F after the reaction, but the different inductive effects of hydroxyl and carboxyl groups resulted in the change of electronic densities on the S atom and induced distinguishable δmFs. The aromatic thiols also had characteristic fingerprints, of which 6-mercaptopurine (6-MP) with larger steric hindrance showed downfield chemical shifts in each region compared with those of methimazole (MMI). We also noticed that thiols with larger steric hindrance (D-penicillamine, D-PEN) and decreased electron density (6-MP and MMI) required more time or higher temperature to equilibrium because of their slower reaction rates (Fig. S19 and S20†). The good capacity of wPSP-4F for multi-thiol discrimination could be presented by a two-dimensional plot (2D code) with the chemical shifts of m-F and o-F as the axes, which clearly showed no overlapped signals of the tested thiols (Fig. 2B). Certainly, wPSP-4F could discriminate more small lipophilic thiols in conjunction with organic solvents to improve the solubility of thiols and corresponding thioesters (more suitable for the PSP-4F system) (Fig. S21 and S22†).Real-time monitoring of thiols in HeLa cells
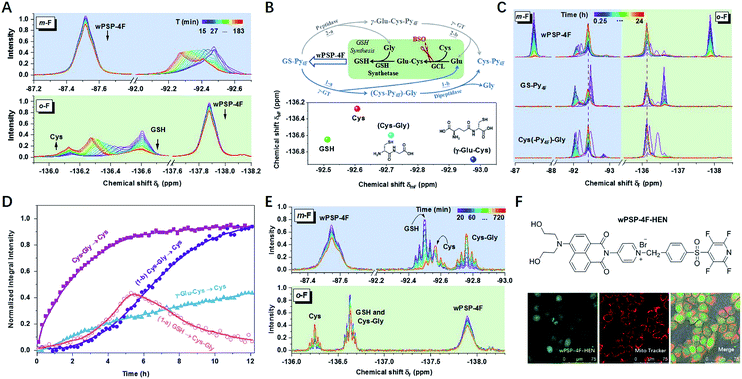
To further test the suitability of wPSP-4F for differentiating small molecule thiols in biological systems, we tried to measure biothiols in HeLa cells. Before the measurement, the influence of free cysteine residues in proteins was assessed with cell lysates (6 mg mL−1) after dialysis with a 3k cut-off membrane to remove small molecule thiols (Fig. S23†). The results indicated 15.3% consumption of the probe (0.5 mM) without any 19F resonances. This observation was perhaps due to both the low abundance of –SH containing proteins and the line broadening effect of macromolecules, while the 19F spectrum of remaining dialysate with only small molecules exhibited the 19F chemical shift of GSH (29.4%) and Cys (3.0%), consistent with the reported ratio range.30 Other thiols accounted for 0.07% of the probe without any characteristic peaks because of low content. Therefore, the 19F NMR approach could potentially detect intracellular Cys and GSH, which had not been realized using NMR techniques before.31In living HeLa cells, the integral intensity of the probe leveled off after 2 h at high concentrations (5.0 mM) (Fig. 3A and S24†). As expected, Cys (−136.15 ppm) and GSH (−136.60 ppm) were obviously observed in the initial stage. However, GSH underwent a rapid decline, accompanied by the increase of an unknown thiol (UT) and Cys, which was rarely addressed in any thiol-sensing system in living cells. Moreover, the long-term tracking of this process showed almost full conversion of thiols into Cys under a lower concentration (2.0 mM) with high cell viability (Fig. 3C and S25†). After ruling out the degradation of thioesters (Fig. S26†), UT was supposed to be the intermediate product during GSH (or GS–Py4F) metabolism. In addition, only one set of invariable peaks of Cys was detected after the cells were incubated with buthionine sulfoximine (BSO) to inhibit the activity of γ-glutamylcysteine ligase (GCL) and then GSH regeneration,11,32 which further confirmed our hypothesis (Fig. S27†).
Theoretically, GS–Py4F had two possible degradation pathways, emerging from cleavage of the γ-glutamyl bond (γ-GT) and peptide bond (peptidase) in different orders (Fig. 3B).33–36 Fortunately, wPSP-4F could well distinguish the possible intermediates of γ-Glu–Cys and Cys–Gly (Fig. 3B and S28†). To reduce the line-broadening and the influence of the complex intracellular environment on NMR spectra in cells, we used diluted cell lysates to monitor the degradation of exogenous adducts (Fig. 3D and S29†). With the catalysis of endogenous enzymes, the conversion of (Cys–Py4F)–Gly was much more efficient than that of γ-Glu–Cys–Py4F, and only signals of (Cys–Py4F)–Gly were detected during the degradation of GS–Py4, in accordance with the endogenous GSH degradation in cell lysates after being treated with wPSP-4F (Fig. 3E). This process also could be stuck in the first stage of (Cys–Py4F)–Gly if γ-GT was added into GS–Py4 in dilute solution (Fig. S30†).37 In the further experiment, the variation trend was recovered in living cells with these adducts, though the time courses were different owing to different catalytic rates and cell permeability (Fig. 3C). The above results implied that the irreversible GSH adduct quickly decomposed to (Cys–Py4F)–Gly (UT) (t1/2 ≈ 76 min under our conditions) under the catalysis of γ-GT and then Cys–Py4F with dipeptidase. The variable upfield shifts of (Cys–Py4F)–Gly perhaps stemmed from environmental changes induced by the cell membrane enzyme of γ-GT.13 Although the probe with the lipophilic cation of pyridinium was generally assumed to accumulate in mitochondria, the time-varying 19F NMR spectra were found to reflect the whole-cell thiols using the fluorescent wPSP-4F–HEN, a derivative of wPSP-4F, presumably owing to the high concentration in our case (Fig. 3F and S31†).38
Thiol related enzyme activity detection
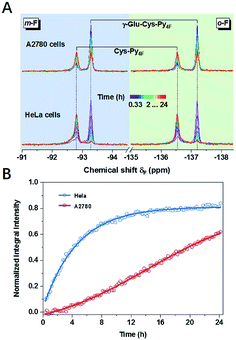
The concept of thiol discrimination could also be easily extended to detect enzyme activities based on suitable thiol or thiol–Py4F substrates. For example, γ-Glu–Cys–Py4F could be used to detect the activity of the aforementioned γ-GT, which was extensively studied as a diagnostic index for drug-induced liver injury or some types of cancer.39 Instead of continuous degradation of GS–Py4 with ambiguous signals (1-a and 1-b, Fig. 3E), γ-Glu–Cys–Py4F exhibited a single reliable response towards different concentrations of γ-GT (Fig. S32†). As illustrated, evidently different activities of γ-GT were observed in two cell lines of HeLa and A2780 under the same conditions, which suggested that the probe has the potential to diagnose related diseases (Fig. 4). We believed that this method could easily detect other kinds of enzyme activities by rational fusion of Cys (Cys–Py4F) into recognition sequences.40–42Conclusions
In summary, we have rationally developed a practical platform (PSP-4F and derivatives) that enabled selective and rapid trapping of thiols and outputting characteristic 19F NMR signals for easy quantification. Based on the perturbations of 19F atoms induced by different electron densities and spatial structures around the sulfhydryl group, the probe discriminated multiple small molecule thiols through unique “2D codes”. The method showed powerful discriminatory ability that can detect up to 20 small molecule thiols simultaneously. The 19F NMR platform presented here potentially had wide applications, illustrated by reliable determination of drug content, long-term tracking of biothiols in living cells, and detection of γ-GT activities. We expect that our concept could further realize the monitoring of thiol-related physiological activities for disease diagnosis.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the National Natural Science Foundation of China (21804136, 21925406, 21991080 and 21921004), Ministry of Science and Technology of China (2017YFA0505400) and K. C. Wong Education Foundation.References
- X. Chen, Y. Zhou, X. Peng and J. Yoon, Chem. Soc. Rev., 2010, 39, 2120–2135 RSC.
- T. V. Mishanina, M. Libiad and R. Banerjee, Nat. Chem. Biol., 2015, 11, 457–464 CrossRef CAS.
- C. Yin, F. Huo, J. Zhang, R. Martinez-Manez, Y. Yang, H. Lv and S. Li, Chem. Soc. Rev., 2013, 42, 6032–6059 RSC.
- J. Aaseth, O. P. Ajsuvakova, A. V. Skalny, M. G. Skalnaya and A. A. Tinkov, Coord. Chem. Rev., 2018, 358, 1–12 CrossRef CAS.
- M. D. Hall, T. S. Marshall, A. D. Kwit, L. M. Miller Jenkins, A. E. Dulcey, J. P. Madigan, K. M. Pluchino, A. S. Goldsborough, K. R. Brimacombe, G. L. Griffiths and M. M. Gottesman, J. Biol. Chem., 2014, 289, 21473–21489 CrossRef.
- M. Cazzola, L. Calzetta, C. Page, P. Rogliani and M. G. Matera, Trends Pharmacol. Sci., 2019, 40, 452–463 CrossRef CAS.
- C. X. Yin, K. M. Xiong, F. J. Huo, J. C. Salamanca and R. M. Strongin, Angew. Chem., Int. Ed., 2017, 56, 13188–13198 CrossRef CAS.
- Z. Xu, T. Qin, X. Zhou, L. Wang and B. Liu, Trends Anal. Chem., 2019, 121, 115672 CrossRef CAS.
- X. Jiao, Y. Li, J. Niu, X. Xie, X. Wang and B. Tang, Anal. Chem., 2018, 90, 533–555 CrossRef CAS.
- Z. Xu, C. Liu, S. Zhao, S. Chen and Y. Zhao, Chem. Rev., 2019, 119, 195–230 CrossRef CAS.
- Y. Takaoka, T. Sakamoto, S. Tsukiji, M. Narazaki, T. Matsuda, H. Tochio, M. Shirakawa and I. Hamachi, Nat. Chem., 2009, 1, 557–561 CrossRef CAS.
- Y. Zhao, L. Chen and T. M. Swager, Angew. Chem., Int. Ed., 2016, 55, 917–921 CrossRef CAS.
- J. F. Teichert, D. Mazunin and J. W. Bode, J. Am. Chem. Soc., 2013, 135, 11314–11321 CrossRef CAS.
- J. Axthelm, S. H. C. Askes, M. Elstner, G. Upendar Reddy, H. Gorls, P. Bellstedt and A. Schiller, J. Am. Chem. Soc., 2017, 139, 11413–11420 CrossRef CAS.
- J. Axthelm, H. Görls, U. S. Schubert and A. Schiller, J. Am. Chem. Soc., 2015, 137, 15402–15405 CrossRef CAS.
- Y. Zhao, G. Markopoulos and T. M. Swager, J. Am. Chem. Soc., 2014, 136, 10683–10690 CrossRef CAS.
- S. Yang, Q. Zeng, Q. Guo, S. Chen, H. Liu, M. Liu, M. T. McMahon and X. Zhou, Talanta, 2018, 184, 513–519 CrossRef CAS.
- J. X. Yu, R. R. Hallac, S. Chiguru and R. P. Mason, Prog. Nucl. Magn. Reson. Spectrosc., 2013, 70, 25–49 CrossRef CAS.
- N. Toda, S. Asano and C. F. Barbas, Angew. Chem., Int. Ed., 2013, 52, 12592–12596 CrossRef CAS.
- P. Zhou, J. Yao, G. Hu and J. Fang, ACS Chem. Biol., 2016, 11, 1098–1105 CrossRef CAS.
- M. R. Bauer, A. C. Joerger and A. R. Fersht, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E5271–E5280 CrossRef CAS.
- C. Zhang, E. V. Vinogradova, A. M. Spokoyny, S. L. Buchwald and B. L. Pentelute, Angew. Chem., Int. Ed., 2019, 58, 4810–4839 CrossRef CAS.
- G. X. Yin, T. T. Niu, Y. B. Gan, T. Yu, P. Yin, H. M. Chen, Y. Y. Zhang, H. T. Li and S. Z. Yao, Angew. Chem., Int. Ed., 2018, 57, 4991–4994 CrossRef CAS.
- G. Yin, T. Niu, T. Yu, Y. Gan, X. Sun, P. Yin, H. Chen, Y. Zhang, H. Li and S. Yao, Angew. Chem., Int. Ed., 2019, 58, 4557–4561 CrossRef CAS.
- S. Kalhor-Monfared, M. R. Jafari, J. T. Patterson, P. I. Kitov, J. J. Dwyer, J. M. Nuss and R. Derda, Chem. Sci., 2016, 7, 3785–3790 RSC.
- Y. Yu, X. Mao, J. Cheng, Z. Ji, J. Zhuang, J. Liu, Z. Sun and J. You, Microchem. J., 2018, 143, 21–30 CrossRef CAS.
- N. Rastkari, M. Khoobi, A. Shafiee, M. R. Khoshayand and R. Ahmadkhaniha, J. Chromatogr. B, 2013, 932, 144–151 CrossRef CAS.
- Y. Yuan, X. Zhao, S. Liu, Y. Li, Y. Shi, J. Yan and X. Hu, Sens. Actuators, B, 2016, 236, 565–573 CrossRef CAS.
- The United States Pharmacopoeia (USP35), The National Formulary (NF30), United States Pharmacopoeia Convention. Rockville, MD, 2012, p. 2069 Search PubMed.
- X. Jiang, J. Chen, A. Bajic, C. Zhang, X. Song, S. L. Carroll, Z. L. Cai, M. Tang, M. Xue, N. Cheng, C. P. Schaaf, F. Li, K. R. MacKenzie, A. C. M. Ferreon, F. Xia, M. C. Wang, M. Maletic-Savatic and J. Wang, Nat. Commun., 2017, 8, 16087 CrossRef CAS.
- X. Tan, K. Ji, X. Wang, R. Yao, G. Han, F. A. Villamena, J. L. Zweier, Y. Song, A. Rockenbauer and Y. Liu, Angew. Chem., Int. Ed., 2020, 59, 928–934 CrossRef CAS.
- K. Umezawa, M. Yoshida, M. Kamiya, T. Yamasoba and Y. Urano, Nat. Chem., 2017, 9, 279–286 CrossRef CAS.
- A. Meister and M. E. Anderson, Annu. Rev. Biochem., 1983, 52, 711–760 CrossRef CAS.
- M. P. Grillo, F. Hua, K. L. March, L. Z. Benet, C. G. Knutson and J. A. Ware, Chem. Res. Toxicol., 2008, 21, 1933–1938 Search PubMed.
- J. Liu, M. Liu, H. Zhang, X. Wei, J. Wang, M. Xian and W. Guo, Chem. Sci., 2019, 10, 10065–10071 RSC.
- R. E. Hansen, D. Roth and J. R. Winther, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 422–427 CrossRef CAS.
- J. Liu, S. Zhang, B. Zhao, C. Shen, X. Zhang and G. Yang, Biosens. Bioelectron., 2019, 142, 111497 CrossRef CAS.
- J. Zielonka, J. Joseph, A. Sikora, M. Hardy, O. Ouari, J. Vasquez-Vivar, G. Cheng, M. Lopez and B. Kalyanaraman, Chem. Rev., 2017, 117, 10043–10120 CrossRef CAS.
- Y. Urano, M. Sakabe, N. Kosaka, M. Ogawa, M. Mitsunaga, D. Asanuma, M. Kamiya, M. R. Young, T. Nagano, P. L. Choyke and H. Kobayashi, Sci. Transl. Med., 2011, 3, 110–119 Search PubMed.
- R. Hata, H. Nonaka, Y. Takakusagi, K. Ichikawa and S. Sando, Angew. Chem., Int. Ed., 2016, 55, 1765–1768 CrossRef CAS.
- T. Nishihara, H. A. Yoshihara, H. Nonaka, Y. Takakusagi, F. Hyodo, K. Ichikawa, E. Can, J. A. Bastiaansen, Y. Takado, A. Comment and S. Sando, Angew. Chem., Int. Ed., 2016, 55, 10626–10629 CrossRef CAS.
- H. Li, G. Parigi, C. Luchinat and T. J. Meade, J. Am. Chem. Soc., 2019, 141, 6224–6233 CrossRef CAS.
- Y. Yang, J. T. Wang, Y. Y. Pei and X. C. Su, Chem. Commun., 2015, 51, 2824–2827 RSC.
- A. Martorana, Y. Yang, Y. Zhao, Q. F. Li, X. C. Sun and D. Goldfarb, Dalton Trans., 2015, 44, 20812–20816 RSC.
- X. C. Su and J. L. Chen, Acc. Chem. Res., 2019, 52, 1675–1686 CrossRef CAS.
- Y. Yang, F. Yang, Y. J. Gong, J. L. Chen, D. Goldfarb and X. C. Su, Angew. Chem., Int. Ed., 2017, 56, 2914–2918 CrossRef CAS.
- T. Müntener, J. Kottelat, A. Huber and D. Häussinger, Bioconjugate Chem., 2018, 29, 3344–3351 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc04664g |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |