Open Access Article
Open Access ArticlePenicacids H–J, three new mycophenolic acid derivatives from the marine-derived fungus Rhizopus oryzae†
Amine Dilara Pilevneli‡
 a,
Sherif S. Ebada‡
a,
Sherif S. Ebada‡ *bc,
Banu Kaşkateped and
Belma Konuklugil
*bc,
Banu Kaşkateped and
Belma Konuklugil *e
*e
aDepartment of Pharmacognosy, Faculty of Pharmacy, Ankara University, 06560 Ankara, Turkey
bDepartment of Pharmacognosy, Faculty of Pharmacy, Ain Shams University, 11566 Abbassia, Cairo, Egypt. E-mail: sherif_elsayed@pharma.asu.edu.eg; Fax: +20-2405-1107; Tel: +20-2405-1180
cDepartment of Pharmacognosy, Faculty of Pharmacy, Sinai University, Ismailia, Egypt. E-mail: sherif.elsayed@su.edu.eg
dDepartment of Pharmaceutical Microbiology, Faculty of Pharmacy, Ankara University, 06560 Ankara, Turkey
eDepartment of Pharmacognosy, Faculty of Pharmacy, Lokman Hekim University, Söğütözü, 06510 Çankaya, Ankara, Turkey. E-mail: konuklug@pharmacy.ankara.edu.tr
First published on 1st November 2021
Abstract
Chemical investigation of secondary metabolites in crude methanol extract of a solid rice medium of a marine-derived fungus, Rhizopus oryzae, has enriched the metabolic profile of this genus by affording three mycophenolic acid derivatives recognized as new fungal metabolites trivially named as penicacids H–J (1–3), along with two known naphtho-γ-pyrone dimers, asperpyrone A (4) and dianhydroaurasperone C (5). Structure elucidation of isolated compounds was unambiguously determined based on extensive 1D and 2D NMR spectroscopic analyses together with comparing coupling constant and optical rotation values with those reported for related congeners in literature. All isolated compounds were assessed for their antibacterial activity against four different bacterial microorganisms and they revealed moderate to weak activities with minimum inhibitory concentration (MIC) values ranging from 62.5 to 250 μg mL−1.
Introduction
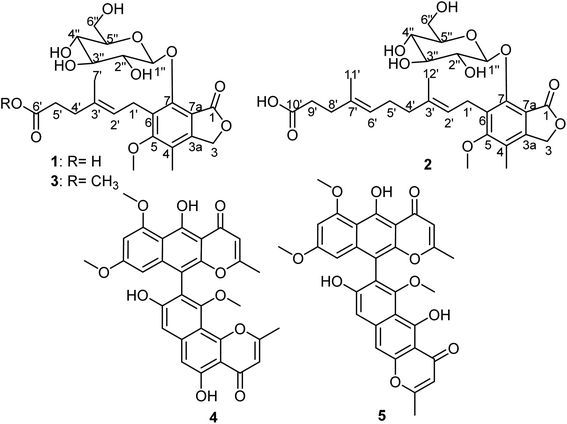
Marine-derived fungi represent a ubiquitous group of microorganisms that continue to be a prolific source of new natural products featuring novel chemical scaffolds with intriguing pharmacological activities including antimicrobial, antiviral, antiproliferative, and anti-inflammatory activities.1–3 In the course of our ongoing research for the discovery of bioactive secondary metabolites, we have explored and reported several new and/or novel bioactive entities from marine-derived fungi of different regions such as the Mediterranean Sea,4–6 the Red Sea7–9 and a hypersaline lake sediment.10 During the past few decades, it was found that the drastic emergence of resistant microbes was related to the immense irresponsible use of antibiotics yielding difficult-to-treat or untreatable infections including life-threatening nosocomial ones caused by methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa.11 Therefore, it is conceivable to say that the enrichment of antimicrobial pipeline with new entities is of an everlasting importance to keep as much reservoir of potential antimicrobial agents as possible to be implemented whenever needed. Moreover, it is known that mycophenolic acid (MPA) is a phenyl-terpenoid fungal secondary metabolite, first reported in 1893 as a fungal product, contaminating corn crops and causing pellagra upon its consumption.12 It was mainly reported as a fungal metabolite in different species of the genus Penicillium.13 Since its first report, MPA and its derivatives have been hot research topics due to their vast array of bioactivities ranging from antibacterial, antifungal, antiviral, antitumor to immunosuppressive aspects.14–17 Mycophenolate mofetil (MMF), a 2-morpholinoethyl ester prodrug of MPA, is a potent inhibitor of inosine-5′-monophosphate dehydrogenase (IMPDH) that is a crucial rate-limiting enzyme in the purine metabolic pathway. IMPDH influences numerous cellular processes such as replication, transcription and signaling.18 Hence, it is regarded as an interesting drug target for antiviral, immunosuppression and cancer chemotherapy.19In this current and ongoing study aimed at identifying bioactive fungal metabolites from marine-associated fungi, we investigated a marine-associated fungus, namely Rhizopus oryzae. The fungal strain was derived from a specimen of the Turkish marine sponge Dendrectilla tremitersis collected from the Kömür Limani coastline in 2014. Chromatographic workup of methanol extract obtained from mycelial cells of solid rice culture medium of the fungus was performed implementing various separation techniques. The obtained results (Fig. 1) led to the isolation and identification of three new MPA derivatives (1–3), together with two known naphtho-γ-pyrone dimers, asperpyrone A (4)20–22 and dianhydroaurasperone C (5).23–26
In this study and for the first time, we report the isolation and structural elucidation of three MPA derivatives marked as new natural products, penicacids H–J (1–3) (Fig. 1), together with the results of antimicrobial activity assessment of all isolated compounds against two Gram-positive (Staphylococcus aureus ATCC 29213 and methicillin-resistant S. aureus ATCC 43300) and two Gram-negative (Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853) bacteria.
Materials and methods
General experimental procedures
Several materials and instruments were used in this study to serve the aim and are divided into materials and instruments for purification such as: silica gel and Sephadex LH-20 for column chromatography (E. Merck, Darmstadt, Germany); precoated TLC plates (aluminium sheets, silica gel & RP-18 F254, Merck, Darmstadt, Germany) for thin layer chromatography (TLC) and the detection process was carried out by UV light at 254 and 365 nm wavelengths as a non-destructive technique or after spraying with anisaldehyde reagent and heating. Final purification of fractions was achieved using preparative HPLC (Agilent, U.S.A) on Zorbax Eclipse XDB-C18 preparative column (9.4 mm × 250 mm, L × ID; 5 μm particle size) at a flow rate of 2 mL min−1 and UV screening detection at 210 nm to 330 nm. A standard gradient elution was applied with the use of acetonitrile in water: 0 min, 10% acetonitrile; 5 min, 10% acetonitrile; 15 min, 40% acetonitrile, 30 min, 70% acetonitrile at a flow rate of 2 mL min−1. As for the analysis, the following materials and instruments were used: a Perkin-Elmer-241 MC polarimeter to determine the optical activity; an Agilent 600 MHz spectrometer (U.S.A) for obtaining 1D (1H and 13C NMR) and 2D NMR spectra (chemical shifts in ppm). A methanol-d4 NMR solvent (Sigma Aldrich, Germany) was used to dissolve the isolated compounds; an Agilent Triple Quadrupole 6410 QQQ LC/MS mass spectrometer for ESI† negative and positive scan modes with a nebulizer pressure of 60 psi and gas flow rate of 12 L min−1 while gas temperature was adjusted at 350 °C and ionization was achieved through direct infusion method using CH3OH\H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) (flow rate of 0.2 mL min−1).
1 v/v) (flow rate of 0.2 mL min−1).
Sponge and fungal strain materials
A fungal strain Rhizopus oryzae was isolated from a marine sponge Dendrectilla tremitersis, which was collected at Kömür Limani coastline, Turkey in 2014. This fungal strain was deposited at Department of Pharmacognosy, Ankara University (B. K.). The fungus was identified as Rhizopus oryzae (GenBank accession number MT273952) according to the DNA amplification sequencing using fungal ITS (Internal Transcribed Spacer) region as reported before.27Fermentation, extraction and isolation
Cultivation of the fungal strain was conducted in ten different 2.0 L Erlenmeyer flasks, each containing a 100 mL solid rice medium prepared by autoclaving rice (100 g) and distilled water (100 mL) enriched with 3.5% artificial sea salt. Fermentation was obtained with a waiting period of 30 days at room temperature in static conditions. Each flask was then terminated using ethyl acetate (EtOAc) (3 × 350 mL) to stop the growth of cells. Then, the flasks were shaken for 12 h and filtered. Lastly, the EtOAC filtrates were pooled and evaporated under a reduced pressure to yield a crude solid extract (2.6 g).The crude extract was then fractionated between n-hexane and 90% aqueous MeOH by liquid–liquid partitioning where both fractions were separated, evaporated under reduced pressure and weighed. The aqueous 90% MeOH phase (1.1 g) was then subjected to vacuum liquid chromatography (VLC) using silica gel 60 as a stationary phase and the mobile phase used was n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc (1
EtOAc (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), followed by 2
4), followed by 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 and 1
3 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and for each fraction 300 mL mobile phase was used. A gradient elution development of DCM
1, and for each fraction 300 mL mobile phase was used. A gradient elution development of DCM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (100
MeOH (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 90
0, 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 80
10, 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 70
20, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, 60
30, 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40, 50
40, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 and 0
50 and 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100) afforded ten fractions (RO-1–RO-10), respectively.
100) afforded ten fractions (RO-1–RO-10), respectively.
All obtained fractions were assessed based on their TLC and analytical HPLC chromatograms. Fractions RO-2, RO-4 and RO-5 were chosen for further preparative TLC and HPLC purification procedures. Fraction RO-2 (237 mg), eluted with n-hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc (2
EtOAc (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), was applied to column chromatography using a stationary phase Sephadex LH-20 column (100 × 2.5 cm) and 100% methanol as a mobile phase to afford 4 (3.4 mg) and 5 (1.8 mg). Fractions RO-4 (153 mg) and RO-5 (92 mg) from silica gel column chromatography were separately rechromatographed by column chromatography using Sephadex LH-20 as a stationary phase and 100% methanol as a mobile phase, followed by preparative HPLC for final purification to yield 2 (2.1 mg) and 3 (1.2 mg) from RO-4 and 1 (1.7 mg) from RO-5.
3), was applied to column chromatography using a stationary phase Sephadex LH-20 column (100 × 2.5 cm) and 100% methanol as a mobile phase to afford 4 (3.4 mg) and 5 (1.8 mg). Fractions RO-4 (153 mg) and RO-5 (92 mg) from silica gel column chromatography were separately rechromatographed by column chromatography using Sephadex LH-20 as a stationary phase and 100% methanol as a mobile phase, followed by preparative HPLC for final purification to yield 2 (2.1 mg) and 3 (1.2 mg) from RO-4 and 1 (1.7 mg) from RO-5.
| Pos. | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δHa (multi, J in Hz) | δCb,e, type | δHa (multi, J in Hz) | δCb,e, type | δHc (multi, J in Hz) | δCd,e, type | |
| a Measured in methanol-d4 at 600 MHz.b Measured in methanol-d4 at 150 MHz.c Measured in methanol-d4 at 400 MHz.d Measured in methanol-d4 at 100 MHz.e Assigned based on gHMQC and gHMBC spectra. | ||||||
| 1 | 172.1, C | 172.2, C | 172.1, C | |||
| 3 | 5.27, br s (2H) | 70.2, CH2 | 5.27, br s (2H) | 70.2, CH2 | 5.27, br s (2H) | 70.2, CH2 |
| 3a | 148.8, C | 148.8, C | 148.8, C | |||
| 4 | 122.7, C | 122.8, C | 122.7, C | |||
| 5 | 164.6, C | 164.6, C | 164.6, C | |||
| 6 | 130.9, C | 130.9, C | 130.9, C | |||
| 7 | 153.8, C | 153.9, C | 153.8, C | |||
| 7a | 113.2, C | 113.2, C | 113.2, C | |||
| Me-4 | 2.21, s (3H) | 11.6, CH3 | 2.22, s (3H) | 11.6, CH3 | 2.21, s (3H) | 11.6, CH3 |
| MeO-5 | 3.79, s (3H) | 61.6, CH3 | 3.80, s (3H) | 61.5, CH3 | 3.79, s (3H) | 61.6, CH3 |
| 1′ | 3.45, m | 25.0, CH2 | 3.45, m | 25.0, CH2 | 3.45, m | 25.0, CH2 |
| 3.65, dd (14.3, 6.5) | 3.69, dd (14.0, 7.3) | 3.65, dd (14.2, 7.2) | ||||
| 2′ | 5.28, t (6.5) | 124.9, CH | 5.26, t (7.3) | 124.3, CH | 5.28, t (7.2) | 124.9, CH |
| 3′ | 134.9, C | 136.6, C | 134.9, C | |||
| 4′ | 2.28, t (7.4, 2H) | 36.0, CH2 | 2.00, t (7.4, 2H) | 40.6, CH2 | 2.28, t (7.5, 2H) | 36.0, CH2 |
| 5′ | 2.34, t (7.4, 2H) | 35.5, CH2 | 2.08, m (2H) | 35.9, CH2 | 2.41, t (7.5, 2H) | 35.5, CH2 |
| 6′ | 178.2, C | 5.09, t (7.4) | 125.5, CH | 176.9, C | ||
| 7′ | 1.82, d (1.2, 3H) | 16.5, CH3 | 134.8, C | 1.82, d (1.2, 3H) | 16.5, CH3 | |
| 6′-OMe | 3.57, s (3H) | 52.0, CH3 | ||||
| 8′ | 2.18, m (2H) | 34.0, CH2 | ||||
| 9′ | 2.16, m (2H) | 35.6, CH2 | ||||
| 10′ | 179.0, C | |||||
| 11′ | 1.57, d (1.3, 3H) | 16.1, CH3 | ||||
| 12′ | 1.80, d (1.3, 3H) | 16.5, CH3 | ||||
| 1′′ | 5.33, d (7.7) | 106.5, CH | 5.29, d (7.7) | 106.7, CH | 5.33, d (7.7) | 106.5, CH |
| 2′′ | 3.50, m | 75.8, CH | 3.53, m | 75.9, CH | 3.50, m | 75.8, CH |
| 3′′ | 3.23, ddd (9.7, 5.5, 2.3) | 78.5, CH | 3.22, ddd (9.8, 5.4, 2.2) | 78.5, CH | 3.23, ddd (9.7, 5.5, 2.3) | 78.4, CH |
| 4′′ | 3.37, dd (9.8, 8.8) | 71.4, CH | 3.37, t (9.0) | 71.4, CH | 3.37, dd (9.8, 8.8) | 71.4, CH |
| 5′′ | 3.46, t (9.0) | 78.4, CH | 3.45, t (9.0) | 78.4, CH | 3.46, t (9.0) | 78.4, CH |
| 6′′ | 3.65, m | 62.6, CH2 | 3.64, m | 62.7, CH2 | 3.65, m | 62.6, CH2 |
| 3.76, m | 3.78, m | 3.76, m | ||||
Antibacterial assay
Antibacterial activities of pure compounds were assessed via broth microdilution method using a 96-well plate according to CLSI documents M27-A3.28 Two Gram-positive bacteria, namely Staphylococcus aureus (ATCC 29213) and methicilin-resistant Staphylococcus aureus (ATCC 43300), were used in an antibacterial assay in addition to two Gram-negative bacteria, namely Eschericia coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 27853), using ciprofloxacin as a positive control.Results and discussion
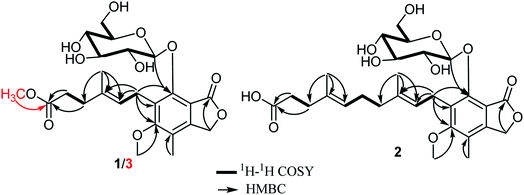
Compound 1 was isolated as a white amorphous solid. When isolated, its HPLC chromatogram revealed maximum absorption peaks (λmax) at 220, 250 and 295 nm. HRESIMS displayed two pseudomolecular ion peaks at m/z 505.25462 [M + Na]+ (calcd for C23H30O11Na, 505.25442) and at m/z 481.28388 [M − H]− (calcd for C23H29O11, 481.28404). Hence, the molecular formula was deduced to be C23H30O11 indicating the existence of nine degrees of unsaturation. The 13C NMR spectrum of 1 (Table 1) exhibited the existence of 23 different carbon resonances distinguished via 2D NMR spectra namely HMBC and HMQC into nine quaternary carbons including two carbonyl carbons (δC 178.2 and δC 172.1), six methines (δC 124.9, δC 106.5, δC 78.5, δC 78.4, δC 75.8 and δC 71.4), five methylenes (δC 70.2, δC 62.6, δC 36.0, δC 35.5 and δC 25.0) and three methyls classified into one methoxy carbon at δC 61.6, one aromatic methyl carbon at δC 11.6 and one olefinic methyl carbon at δC 16.5 ppm. The 1H NMR data (Table 1) and 1H–1H COSY spectrum (Fig. 2) of 1 revealed the presence of three spin systems as follows: one between an olefinic proton at δH 5.28 (t, J = 6.5, 1H) and one methylene group at δH 3.45/δH 3.65, while the second spin system occurred between two methylene groups at δH 2.28 (t, J = 7.4, 2H) and δH 2.34 (t, J = 7.4, 2H). The third spin system is rather characteristic ranging from an anomeric proton at δH 5.33 (d, J = 7.7, 1H) to four successive methine protons at δH 3.50, δH 3.23, δH 3.37 and δH 3.46 and then ending with hydroxymethylene protons at δH 3.65/δH 3.76 ppm. The last spin system was linked through the HMQC spectrum to carbon resonances at δC 106.5, δC 75.8, δC 78.5, δC 71.4, δC 78.4 and δC 62.6 ppm, respectively, which confirmed the existence of a glucopyranosyl moiety.29 By searching the reported literature and comparing with NMR data obtained from 1, it was suggested that it is a related derivative to a class of fungal mycophenolic acid derivatives known as penicacids.13,30–32 The configuration of the C-2′/C-3′ double bond was determined to be E-configuration based on the carbon resonance of the olefinic methyl group (Me-7′) that resonated at δC = 16.5 ppm confirming the E-configuration rather than δC = ∼23.0 ppm (expected for a Z-configuration)33,34 and by comparison with reported literature.13,30,31 The HMBC spectrum of 1 (Fig. 2) showed clear correlations that enabled localizing a methoxy group (δH 3.79/δC 61.6) at an oxygenated aromatic carbon (δC 164.6, C-5) and to position a glucopyranosyl moiety at δC 153.8 (C-7) based on its correlation to anomeric proton at δH 5.33 (d, J = 7.7, 1H). In addition, it revealed more key correlations confirming the existence and length of the side chain ending with a free carboxylic acid group at δC 178.2 (C-6′) that showed clear correlations with two methylene groups at δH 2.28 (t, J = 7.4, 2H) and δH 2.34 (t, J = 7.4, 2H). The coupling constant of anomeric proton (H-1′′) recorded in methanol-d4 (J = 7.7 Hz) suggested the β-linkage of the molecule.35–37 The optical rotation of 1 was found to be [α]20D + 31.6 and by comparing 13C NMR data of the sugar moiety with reported literature,29 it was confirmed to be β-D-glucopyranose. To the best of our knowledge, compound 1 was only reported as a hepatic metabolite in plasma of transplant recipients on MMF treatment38 and as a biotransformation product from MPA and glucose using Streptomyces aureofaciens.39 Based on the aforementioned data, compound 1 is for the first time reported as a natural fungal metabolite and unambiguously identified as 7-O-(β-D-glucopyranosyl)-mycophenolic acid that was trivially named as penicacid H.Compound 2 was purified as an amorphous yellowish white solid. The HPLC chromatogram of 2 exhibited maximum absorption peaks (λmax) at 222, 253 and 298 nm. HRESIMS revealed two pseudomolecular ion peaks at m/z 573.31798 [M + Na]+ (calcd for C28H38O11Na, 573.31648) and at m/z 549.35525 [M − H]− (calcd for C28H37O11, 549.35386). Hence, the molecular formula was determined to be C28H38O11 indicating the existence of ten degrees of unsaturation. Molecular weight difference between 1 and 2 is 66 amu that has been interpreted in their molecular formulas by an additional C5H8 moiety including one double bond. The 1H NMR data (Table 1) and 1H–1H COSY spectrum of 2 (Fig. 2) similarly revealed four spin systems resembling those of penicacid H (1), which also suggested it to be a related derivative of penicacids.13,30–32 Apart from those four spin systems, the 1H–1H COSY spectrum of 2 (Fig. 2) exhibited an additional isoprenyl unit (5C) that extends the free side chain of penicacids. The configurations of double bonds at C-2′/C-3′ and C-6′/C-7′ were both determined to be E-configuration based on the carbon resonances recorded for olefinic methyl groups at C-3′ and C-7′ that appeared at δC 16.5 and δC 16.1 ppm, respectively, rather than δC = ∼23.0 ppm (expected for a Z-configuration)33,34 along with comparison with reported literature.13,30,31 The HMBC spectrum of 2 (Fig. 2) showed key correlations that positioned the methoxy group (δH 3.80/δC 61.5) to an oxygenated aromatic carbon at δC 164.6 (C-5) and also localized a glucopyranosyl moiety at δC 153.9 (C-7) via its correlation to an anomeric proton at δH 5.29 (d, J = 7.7, 1H) along with a key correlation confirming existence and length of the side chain ending with a free carboxylic acid group at δC 179.0 (C-10′) showing a clear correlation with two methylene groups at δH 2.18 (m, 2H) and δH 2.16 (m, 2H) ascribed for C-8′ and C-9′, respectively. As in penicacid H (1), the coupling constant of the anomeric proton (H-1′′) recorded in methanol-d4 (J = 7.7 Hz) suggested the β-linkage of the molecule.35–37 The optical rotation of 2 was found to be [α]20D + 29.3 and by comparing 13C NMR data of the sugar moiety with reported literature,29 it was confirmed to be β-D-glucopyranose. Therefore, compound 2 was chemically confirmed to be a derivative of 7-O-(β-D-glucopyranosyl)-mycophenolic acid (penicacid H, 1), which was insignificantly named as penicacid I.
Compound 3 was purified as a white amorphous solid. HPLC chromatogram of 3 exhibited maximum absorptions (λmax) at 223, 251 and 297 nm. HRESIMS revealed two pseudomolecular ion peaks at m/z 505.25462 [M + Na]+ (calcd for C24H32O11Na, 505.25442) and at m/z 496.28388 [M − H]− (calcd for C24H31O11, 496.28404). Hence, the molecular formula was established to be C24H32O11 pointing out the existence of nine degrees of unsaturation. The 1H and 13C NMR spectral data of 3 compared to 1 (Table 1) revealed a close similarity except a molecular weight difference of 14 amu in favour of 3. This difference has been explained through the presence of an additional methoxy group in 3 showing a proton resonance at δH 3.57 in the 1H NMR spectrum that was directly correlated with a carbon resonance at δC 52.0 ppm via HMQC. The position of this additional methoxy group was determined through the HMBC spectrum (Fig. 2) that revealed a long-range correlation with the terminal carboxyl group at δC 176.9 (C-6′) ppm, which was also correlated with two methylene groups at δH 2.28 (t, 7.5, 2H) and assigned to C-4′ and C-5′ at δH 2.41 (t, 7.5, 2H), respectively. These findings unambiguously confirmed that the additional methoxy group is a methyl ester group at the terminal carboxylic acid group in 1. For compounds 1 and 2, the coupling constant of the anomeric proton in 3, optical rotation value ([α]20D + 30.2) and comparing 13C NMR data of sugar moiety with reported literature,29 confirmed the sugar moiety to be β-D-glucopyranose.13,30,31 By searching literature databases, it was found that compound 3 was only patented as a synthetic derivative of mycophenolic acid,40 while in this study, it is reported for the first time as a natural product. Therefore, compound 3 was identified as methyl 7-O-(β-D-glucopyranosyl)-mycophenolate and was given a trivial name penicacid J.
It is noteworthy to mention that, herein, compound 3 was carefully traced in the original HPLC chromatogram of the total methanol extract from mycelial cells in the solid rice medium of R. oryzae and confirmed via obtaining its UV spectrum and LCMS to assure that it is a genuine natural product. In addition, penicacid H with a free carboxylic acid was kept in methanol for more than a month and checked by LCMS to assess whether the methyl ester derivative (3) is a result of an esterification reaction during the sample extraction and workup using methanol. The obtained results revealed no existence of the methyl ester derivative (3) in the sample containing penicacid H (1) dissolved and stored in methanol.
All isolated compounds were tested for their antimicrobial activity against two Gram-positive (S. aureus ATCC 29213 and methicillin-resistant S. aureus ATCC 43300) and two Gram-negative bacteria (E. coli ATCC 25922 and P. aeruginosa ATCC 27853). The obtained results (Table 2) disclosed that all isolated compounds possess moderate to weak antimicrobial activities against the tested organisms with MIC values ranging from 62.5 to 250 μg mL−1.
| Compound | Staphylococcus aureus ATCC 29213 | Methicillin-resistant S. aureus ATCC 43300 | Pseudomonas aeruginosa ATCC 27853 | Escherichia coli ATCC 25922 |
|---|---|---|---|---|
| a n.t.: not tested. | ||||
| Penicacid H (1) | 125 | 250 | 125 | 125 |
| Penicacid I (2) | 62.5 | 250 | 125 | 125 |
| Penicacid J (3) | 62.5 | 250 | 62.5 | 125 |
| Asperpyrone A (4) | 62.5 | 125 | 125 | 125 |
| Dianhydroaurasperone C (5) | 62.5 | 125 | 125 | 250 |
| Ciprofloxacin | 0.25 | n.t. | 0.50 | 0.009 |
Conclusion
Three new mycophenolic acid derivatives namely, penicacids H–J (1–3) along with two known naphtho-γ-pyrone dimers asperpyrone A (4) and dianhydroaurasperone C (5), were purified from the crude methanol extract derived from a solid rice medium of marine-associated fungus Rhizpous oryzae. Chemical structures of isolated compounds were carefully determined via in-depth spectroscopic analyses including HRESIMS, 1D and 2D NMR spectroscopy besides coupling constants and comparing optical rotation values with known related derivatives from literature. In the antibacterial activity assay, all isolated compounds exhibited moderate to weak activities (MIC values between 62.5 and 250 μg mL−1) against the tested microorganisms.Author contributions
SSE and B Konuklugil: conceptualization, writing, reviewing and editing; ADP and B Konuklugil: methodology, purification and isolation of pure metabolites; ADP, SSE and B Konuklugil: NMR analysis, data curation and structure elucidation; B Kaşkatepe: antimicrobial activity assay; SSE and B Konuklugil; supervision and editing the manuscript.Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
S. S. E. acknowledges The Scientific and Technological Research Council of Turkey (TÜBITAK) for financially supporting him via a visiting scientist fellowship. This research was funded by TUBİTAK-BMBF Project No: 114S916. A. D. P. and B. K. would like to acknowledge Dr Muhiddin Çengel for hosting NMR analyses in this study.References
- J. W. Blunt, A. R. Carroll, B. R. Copp, R. A. Davis, R. A. Keyzers and M. R. Prinsep, Marine Natural Products, Nat. Prod. Rep., 2018, 35, 8–53, 10.1039/c7np00052a.
- B. Yang, W. Sun, J. Wang, S. Lin, X. N. Li, H. Zhu, Z. Luo, Y. Xue, Z. Hu and Y. Zhang, A new breviane spiroditerpenoid from the marine-derived fungus Penicillium sp. TJ403-1, Mar. Drugs, 2018, 16, 110, DOI:10.3390/md16040110.
- J. F. Imhoff, Natural products from marine fungi-still an underrepresented resource, Mar. Drugs, 2016, 14, 19, DOI:10.3390/md14010019.
- I. S. Uras, S. S. Ebada, M. Korinek, A. Albohy, B. S. Abdulrazik, Y. H. Wang, B. H. Chen, J. T. Horng, W. Lin, T. L. Hwang and B. Konuklugil, Anti-inflammatory, antiallergic, and COVID-19 main protease (Mpro) inhibitory activities of butenolides from a marine-derived fungus Aspergillus terreus, Molecules, 2021, 26, 3354, DOI:10.3390/molecules26113354.
- A. M. Elissawy, S. S. Ebada, M. L. Ashour, M. El-Neketi, W. Ebrahim and A. B. Singab, New secondary metabolites from the mangrove-derived fungus Aspergillus sp. AV-2, Phytochem. Lett., 2019, 29, 1–5, DOI:10.1016/j.phytol.2018.10.014.
- A. M. Elissawy, S. S. Ebada, M. L. Ashour, F. C. Özkaya, W. Ebrahim, A. B. Singab and P. Proksch, Spiroarthrinols A and B, two novel meroterpenoids isolated from the sponge derived fungus Arthrinium sp, Phytochem. Lett., 2017, 20, 246–251, DOI:10.1016/j.phytol.2017.05.008.
- R. Orfali, W. Ebrahim, S. Perveen, N. M. Majrashi, K. Alluhayb and S. S. Ebada, Cytotoxic secondary metabolites from mangrove-rhizosphere-associated fungus Emericella sp. Strain SWR1718, J. King Saud Univ., Sci., 2020, 32, 2656–2661, DOI:10.1016/j.jksus.2020.05.008.
- M. S. Elnaggar, S. S. Ebada, M. Ashour, W. Ebrahim, A. N. B. Singab, W. Lin, Z. Liu and P. Proksch, Two new triterpenoids and a new naphthaquinones derivative isolated from a hard coral-derived fungus Scopulariopsis sp, Fitoterapia, 2017, 116, 126–130, DOI:10.1016/j.fitote.2016.12.003.
- M. S. Elnaggar, S. S. Ebada, M. L. Ashour, W. Ebrahim, W. E. G. Müller, A. Mándi, T. Kurtán, A. Singab, W. Lin, Z. Liu and P. Proksch, Xanthones and sesquiterpene derivatives from a marine-derived fungus Scopulariopsis sp, Tetrahedron, 2016, 72, 2411–2419, DOI:10.1016/j.tet.2016.03.073.
- S. S. Ebada and W. Ebrahim, Quinoisobutyride A, an acyclic antibacterial tetrapeptide incorporating an unprecedented heterocyclic amino acid residue from the hypersaline lake-derived fungus Penicillium simplicissimum strain WSH17, Phytochem. Lett., 2020, 36, 95–98, DOI:10.1016/j.phytol.2020.01.022.
- M. Laws, A. Shaaban and K. M. Rahman, Antibiotic resistance breakers: current approaches and future directions, FEMS Microbiol. Rev., 2019, 43, 490–516, DOI:10.1093/femsre/fuz014.
- R. Bentley, Mycophenolic acid: A one hundred year odyssey from antibiotic to immunosuppressant, Chem. Rev., 2000, 100, 3801–3826, DOI:10.1021/cr990097b.
- X. H. Chen, G. L. Zhou, C. X. Sun, X. M. Zhang, G. J. Zhang, T. J. Zhu, J. Li, Q. Che and D. H. Li, Penicacids E-G, three new mycophenolic acid derivatives from the marine-derived fungus Penicillium parvum HDN17-478, Chin. J. Nat. Med., 2020, 18, 850–854, DOI:10.1016/S1875-5364(20)60027-9.
- M. Tremblay-Létourneau, S. Despins, I. Bougie and M. Bisaillon, Virtual high-throughput screening identifies mycophenolic acid as a novel RNA capping inhibitor, PLoS One, 2011, 6, e24806, DOI:10.1371/journal.pone.0024806.
- S. Domhan, S. Muschal, C. Schwager, C. Morath, U. Wirkner, W. Ansorge, C. Maercker, M. Zeier, P. E. Huber and A. Abdollahi, Molecular mechanisms of the antiangiogenic and antitumor effects of mycophenolic acid, Mol. Cancer Ther., 2008, 7, 1656–1668, DOI:10.1158/1535-7163.MCT-08-0193.
- H. Ui, S. Asanuma, H. Chiba, A. Takahashi, Y. Yamaguchi, R. Masuma, S. Omura and H. Tanake, Mycophenolic acid inhibits syncytium formation accompanied by reduction of gp120 expression, J. Antibiot., 2005, 58, 514–518, DOI:10.1038/ja.2005.69.
- C. M. Robertson, L. L. Hermann and K. M. Coombs, Mycophenolic acid inhibits avian reovirus replication, Antiviral Res., 2004, 64, 55–61, DOI:10.1016/j.antiviral.2004.05.004.
- G. Weber, H. Nakamura, Y. Natsumeda, T. Szekeres and M. Nagai, Regulation of GTP biosynthesis, Adv. Enzyme Regul., 1992, 32, 57–69, DOI:10.1016/0065-2571(92)90008-N.
- Q. Shu and V. Nair, Inosine monophosphate dehydrogenase (IMPDH) as a target in drug discovery, Med. Res. Rev., 2004, 28, 219–232, DOI:10.1002/med.20104.
- J. Meng, S. Zhao, P. Dang, Z. Zhou, D. Lai and L. Zhou, Ustilaginoidin M1, a new bis-naphtho-γ-pyrone from the fungus Villosiclava virens, Nat. Prod. Res., 2021, 35, 1555–1560, DOI:10.1080/14786419.2019.1652289.
- X. Cai, Y. Yu, Q. Li, B. K. Chen, Y. Huang, X. W. Zou, J. T. Tang and B. S. Huang, Asperpyrone F, a new dimeric naphtho-γ-pyrone from the edible fungus Pleurotus ostreatus, Nat. Prod. Res., 2019, 33, 1953–1960, DOI:10.1080/14786419.2018.1481844.
- K. Akiyama, S. Teraguchi, Y. Hamasaki, M. Mori, K. Tatsumi, K. Ohnishi and H. Hayashi, New dimeric naphthopyrones from Aspergillus niger, J. Nat. Prod., 2003, 66, 136–139, DOI:10.1021/np020174p.
- A. S. Antonov, E. V. Leshchenko, O. I. Zhuravleva, S. A. Dyshlovoy, G. von Amsberg, R. S. Popov, V. A. Denisenko, N. N. Kirichuk and S. S. Afiyatullov, Naphto-γ-pyrones from the marine-derived fungus Aspergillus foetidus, Nat. Prod. Res., 2021, 35, 131–134, DOI:10.1080/14786419.2019.1610954.
- D. H. Li, T. Han, L. P. Guan, J. Bai, N. Zhao, Z. L. Li, X. Wu and H. M. Hua, New naphthopyrones from marine-derived fungus Aspergillus niger 2HL-M-8 and their in vitro antiproliferative activity, Nat. Prod. Res., 2016, 30, 1116–1122, DOI:10.1080/14786419.2015.1043553.
- J. Xiao, Q. Zhang, Y.-Q. Gao, X.-W. Shi and J.-M. Gao, Antifungal and antibacterial metabolites from an endophytic Aspergillus sp. associated with Melia azedarach, Nat. Prod. Res., 2014, 28, 1388–1392, DOI:10.1080/14786419.2014.904308.
- N. Bouras, F. Mathieu, Y. Coppel and A. Lebrihi, Aurasperone F–a new member of the naphtho-gamma-pyrone class isolated from a cultured microfungus, Aspergillus niger C-433, Nat. Prod. Res., 2005, 19, 653–659, DOI:10.1080/14786410412331286955.
- S. S. Ebada and W. Ebrahim, A new antibacterial quinolone derivative from the endophytic fungus Aspergillus versicolor strain Eich.5.2.2, S. Afr. J. Bot., 2021, 134, 151–155, DOI:10.1016/j.sajb.2019.12.004.
- S. S. Ebada, M. El-Neketi, W. Ebrahim, A. Mándi, T. Kurtán, R. Kalscheuer, W. E. Müller and P. Proksch, Cytotoxic secondary metabolites from the endophytic fungus Aspergillus versicolor KU258497, Phytochem. Lett., 2018, 24, 88–93, DOI:10.1016/j.phytol.2018.01.010.
- L. Y. Foo, Y. Lu, A. L. Molan, D. R. Woodfield and W. C. McNabb, The phenols and prodelphinidins of white clover flowers, Phytochemistry, 2000, 54, 539–548, DOI:10.1016/S0031-9422(00)00124-2.
- X. Song, R. Tu, X. Mei, S. Wu, B. Lan, L. Zhang, X. Luo, J. Liu and M. Luo, A mycophenolic acid derivative from the fungus Penicillium sp. SCSIO sof101, Nat. Prod. Res., 2020, 34, 1206–1212, DOI:10.1080/14786419.2018.1553881.
- Z. Chen, Z. Zheng, H. Huang, Y. Song, X. Zhang, J. Ma, B. Wang, C. Zhang and H. Ju, Penicacids A-C, three new mycophenolic acid derivatives and immunosuppressive activities from the marine-derived fungus Penicillium sp. SOF07, Bioorg. Med. Chem. Lett., 2012, 22, 3332–3335, DOI:10.1016/j.bmcl.2012.02.106.
- J. Rovirosa, A. Diaz-Marrero, J. Darias, K. Painemal and A. San Martin, Secondary metabolites from marine Penicillium brevicompactum, J. Chil. Chem. Soc., 2006, 51, 775–778, DOI:10.4067/S0717-97072006000100004.
- Y. H. Choi, C. Seo, W. Jeong, J. E. Lee, J. Y. Lee, E. K. Ahn, J. S. Kang, J. H. Lee, C. W. Choi, J. S. Oh, D. Lee and S. S. Hong, Glycopentanolones A-D, four new geranylated quinolone alkaloids from Glycosmis pentaphylla, Bioorg. Chem., 2019, 87, 714–719, DOI:10.1016/j.bioorg.2019.03.069.
- G. Belofsky, M. Aronica, E. Foss, J. Diamond, F. Santana, J. Darley, P. F. Dowd, C. M. Coleman and D. Ferreira, Antimicrobial and antiinsectan phenolic metabolites of Dalea searlsiae, J. Nat. Prod., 2014, 77, 1140–1149, DOI:10.1021/np401083g.
- B. Y. Yang, J. T. Guo, Z. Y. Li, C. F. Wang, Z. B. Wang, Q. H. Wang and H. X. Kuang, New thymoquinol glycosides and neuroprotective dibenzocyclooctance lignans from the Rattan stems of Schisandra chinensis, Chem. Biodiversity, 2016, 13, 1118–1125, DOI:10.1002/cbdv.201500311.
- Q. Wen, X. Lin, Y. Liu, X. Xu, T. Liang, N. Zheng, Kintoko and R. Huang, Phenolic and lignin glycosides from the butanol extract of Averrhoa carambola L. Root, Molecules, 2012, 17, 12330–12340, DOI:10.3390/molecules171012330.
- Y. Lu and L. Y. Foo, Flavonoid and phenolic glycosides from Salvia officinalis, Phytochemistry, 2000, 55, 263–267, DOI:10.1016/S0031-9422(00)00309-5.
- M. Shipkova, V. W. Armstrong, E. Wieland, P. D. Niedmann, E. Schutz, G. Brenner-Weiss, M. Voihsel, F. Braun and M. Oellerich, Identification of glucoside and carboxyl-linked glucuronide conjugates of mycophenolic acid in plasma of transplant recipients treated with mycophenolate mofetil, Br. J. Pharmacol., 1999, 126, 1075–1082 CrossRef CAS PubMed.
- B. J. Abbott, D. R. Horton and J. G. Whitney, Glucosylation of mycophenolic acid by Streptomyces aureofaciens, J. Antibiot., 1980, 33, 506–509 CrossRef CAS PubMed.
- R. E. Holmes, Antitumorous glycosylmycophenolic acid derivatives, DE Pat., DE2424119, 1974.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra07196c |
| ‡ These authors equally contributed to the work in this study. |
| This journal is © The Royal Society of Chemistry 2021 |