DOI:
10.1039/D1RA06494K
(Paper)
RSC Adv., 2021,
11, 35893-35897
NIR luminescent detection of quercetin based on an octanuclear Zn(II)–Nd(III) salen nanocluster†
Received
28th August 2021
, Accepted 30th October 2021
First published on 5th November 2021
Abstract
A NIR luminescent octanuclear Zn(II)–Nd(III) nanocluster 1 was constructed by the use of a salen-type Schiff base ligand. 1 exhibits a lanthanide luminescent response to Que with high sensitivity. The quenching constant of Que to the lanthanide emission is 2.6 × 104 M−1, and the detection limit of 1 to Que is 2.5 μM. The response behavior of 1 to Que is not affected by the existence of some potential interferents such as biomolecules.
Introduction
Recently, fluorescence probes have received much attention due to their applications in the detection of various analytes with high sensitivity and selectivity, rapid response, and convenient utilization.1–5 Among all kinds of target objects, quercetin (Que, 3,3′,4′,5,7-penta-hydroxy-flavone) is a kind of flavonoid existing extensively in vegetables, fruits, beverages and herbal medicines, and is a typical active antioxidant with an important role in the treatment of some cardiovascular diseases and prevention of biological cell degradation.6–8 Many traditional analytical techniques, such as chromatography, spectrophotometry, and capillary electrophoresis, have been used to detect Que.9–11 The fluorescence probes for the detection of Que mainly focus on quantum dots (QDs)12 and carbon nanodots (CNDs).13 Lanthanide complexes may display interesting visible and near-infrared (NIR) emissions with sharp emission spectra, large antenna-generated shifts and long lifetimes. Currently, Bin Zhao et al. first reported a visible luminescent sensor of Que based on a Tb(III)-based metal–organic framework (Tb-MOF).14 Heterometallic d-4f complexes with Nd(III), Er(III) and Yb(III) ions can show superior NIR luminescence properties due to the efficient transfer energy from the light-absorbing units containing d-block metal ions such as Zn2+,15 Cd2+,16 Ru2+,17 Pt2+,18 and Cr2+,19 to lanthanide ions. However, so far there are few reports on the use of polynuclear d-4f complexes for the NIR luminescent detection of Que.
Salen-type Schiff bases are typical ligands used in the construction of luminescent d-4f complexes.20,21 Our recent studies have focused on the construction of lanthanide complexes with luminescent response toward small molecules and ions.22–25 In this work, we report the construction of an octanuclear d-4f complex [Zn6Nd2L4(OAc)2(DMF)EtOH] (1) from a Schiff base ligand N,N′-bis(3-hydroxysalicylidene)(4-methoxyphenylene)-1,2-diamine (H4L, Scheme 1), which has a phenyl backbone. Compared with visible fluorescence, lanthanide NIR luminescence has advantages in the detection of biological system, due to the low absorption of biological matrixes in 1000–1700 nm. Interestingly, 1 shows NIR Nd(III) emission and exhibits high luminescent response to Que.
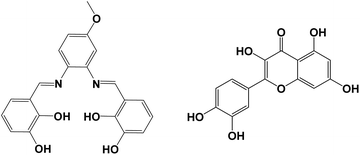 |
| | Scheme 1 The structures of salen-type Schiff base ligand H4L (left) and Que (right). | |
Results and discussion
Synthesis and crystal structure of the Zn(II)–Nd(III) nanocluster
The salen-type Schiff base ligand H4L was synthesized from the reaction of 2,3-dihydroxybenzaldehyde and 4-methoxy-o-phenylenediamine (ESI†).26 1 was synthesized from the reaction of H4L with Nd(OAc)3·4H2O and Zn(OAc)2·2H2O. The molecular dimensions of 1 are about 1.1 × 1.6 × 1.8 nm (Fig. 1). In 1, six Zn(II) and two Nd(III) ions are coordinated with four Schiff base ligands. Four Zn(II) ions are located in the N2O2 cavities of the Schiff base ligands with a square based pyramidal geometry, while other two Zn(II) ions are coordinated with phenolic oxygen atoms of the ligands with a tetrahedral geometry. Each Nd(III) ion is bound to two O2O2 donor sets of the ligands, and the coordination number is eight. Each Nd(III) ion is surrounded by eight O atoms from three Schiff base ligands, one OAc− anion and one solvent molecule EtOH (or DMF). The neighboring Zn(II) and Nd(III) ions are bridged by the phenolic oxygen atoms of four Schiff base ligands and two OAc− anions, with an average separation of 3.76 Å. The bond lengths of Nd–O, Zn–O and Zn–N are 2.28–2.52 Å, 1.91–2.08 Å and 1.99–2.18 Å, respectively. Intramolecular π–π stacking interactions exist between the phenyl rings of Schiff base ligands (distances: 3.371 Å to 3.750 Å),27,28 which helps the formation of 1. Compared with other reported d-4f complexes with similar salen-type conjugated Schiff base ligands,15,20,21,29 the Zn(II) ions in 1 are not only bound to the N2O2 donor sets of the ligands but also to their O2O2 sets. In Fig. 2, one image of scanning electron microscopy (SEM) displays the crystalline nature of 1, and in energy dispersive X-ray (EDX) spectroscopy the molar ratio of Zn/Nd is about 3, consistent with its structure. The measured powder XRD pattern of 1 is similar to the simulated one generated from the crystal structure, indicating the purity of the solid product (Fig. S3†). Thermogravimetric analysis indicates that 1 loses about 3% weight before 100 °C, due to the escaping of uncoordinated solvent molecules such as MeOH, EtOH and H2O in the sample. 1 starts to decompose from about 180 °C (Fig. S4†), which is also confirmed by the melting point measurement (see Experimental section).
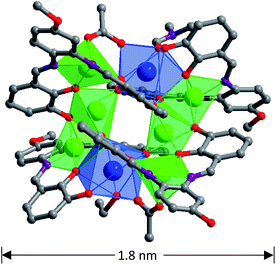 |
| | Fig. 1 The crystal structure of 1 (Zn(II): green; Nd(III): blue). | |
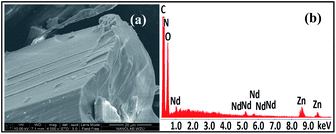 |
| | Fig. 2 The image of scanning electron microscopy (a) and energy dispersive X-ray spectroscopy (b) of 1. | |
Photophysical properties and NIR luminescent response to Que
The free H4L has three bands at 214 nm, 276 nm and 339 nm, which are assigned to n → π* or π → π* transitions associated with imine and phenyl functions of the Schiff base ligand. These bands are red-shifted in 1 due to the disturbance of metal ions in the structure (Fig. S7†). Upon the excitation in ligand-centered absorption band, 1 displays the NIR luminescence of Nd(III) (4F3/2 → 4Ij/2 transitions, j = 9, 11 and 13) at 880, 1064 and 1337 nm (Fig. 3). The emission lifetime (τ) is 5.6 μs (Fig. S8†), so the intrinsic quantum yield (ΦLn) of Nd(III) is calculated to be 2.2%, using equation ΦLn = τ/τ0 (τ0 = 250 μs, the natural lifetime of Nd(III)).30 The whole luminescence quantum yield (Φem) of 1 is 0.14%, thus, the energy transfer efficiency (ηsens = Φem/ΦLn) from the Schiff base ligands to Nd(III) ions can be calculated as 6.4%.31
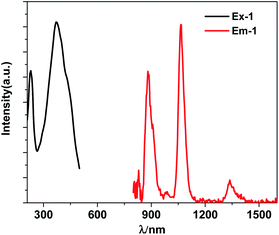 |
| | Fig. 3 The excitation (λem = 1064 nm) and NIR lanthanide emission (λex = 370 nm) spectra of 1 (30 μM) in CH3CN. | |
The NIR luminescent sensing behaviour of 1 to Que was studied in CH3CN. The results show that, as the concentrations of added Que increase, the intensities of lanthanide luminescence of 1 are decreased gradually (Fig. 4). The quenching constant (KSV) of Que to the lanthanide emission is calculated to be 2.6 × 104 M−1, by using Stern–Volmer equation.32 Also, the limit of detection (LOD = 3σ/KSV (ref. 33)) of 1 to Que is obtained to be 2.5 μM, which is among the lowest ones reported for the fluorescent detection of Que so far.12–14,34–37 It is noticeable that, the shapes of NIR emission peaks of 1 are same before and after the addition of Que (Fig. 4). Meanwhile, conductivity study confirms that 1 is neutral in CH3CN, in accordance with the crystal structure. These results suggest that 1 is stable during the sensing experiments.
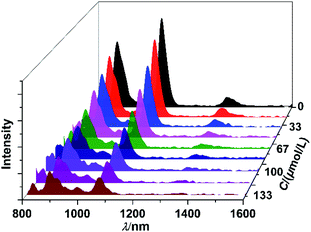 |
| | Fig. 4 The quenching of NIR luminescence of 1 (30 μM) to the addition of Que with different concentrations in CH3CN. | |
To assess the response selectivity of 1 to Que in biological system, we investigated its sensing behaviour to potential biomolecule interferents such as apigenin (Api), flavanone (Fla), naringin (Nar), hesperidin (Hes), phloroglucinol (Phl), daidzein (Dai), hydroquinone (Hyd), resorcinol (Res) and puerarin (Pue) (Scheme S1†). Compared to Que, the addition of these biomolecules causes much smaller changes to the lanthanide emission of 1 (Fig. 5 and S9†). For instance, the lanthanide emission quenching of 1 caused by 60 μM biomolecules is less than 30% (Fig. S9†), indicating the sensing selectivity of 1 to Que. Additionally, the response behaviour of 1 to Que in the presence of the interferents was examined. The results show that the quenching of the lanthanide emission of 1 aroused by Que is not affected by the existence of the interferents (Fig. 6). The luminescence selectivity of 1 to Que was further confirmed by the step-by-step addition of the interferents. For instance, the luminescence decreases slowly when Api is added twice firstly (Fig. 7). While, it decreases fast by the subsequent addition of equal amount of Que. This trend happens again in the following additions of Api and Que, suggesting the high selectivity of 1 to Que even in the existence of the interferents.
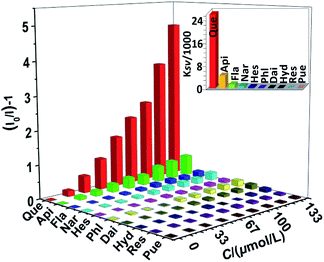 |
| | Fig. 5 The lanthanide luminescence quenching of 1 (30 μM) at 1064 nm to Que and interferents in CH3CN. Inset: the KSV values for the luminescence quenching of 1. (λex = 370 nm). | |
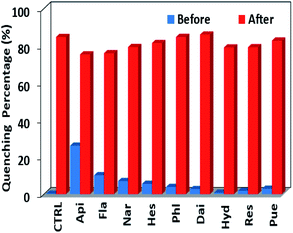 |
| | Fig. 6 The lanthanide luminescence quenching of 1 (30 μM) at 1064 nm before and after the addition of Que (60 μM) with the existence of the interferents (60 μM) in CH3CN. | |
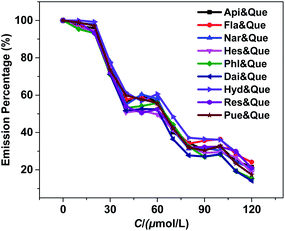 |
| | Fig. 7 The luminescence intensity changes of 1 (30 μM) with the step-by-step addition of interferents and Que (10 μM each time). | |
In 1, the Schiff base ligands can absorb and transfer energy to Nd(III) ions, resulting in the lanthanide luminescence (“antenna effect”, Scheme 2).38 It is noticeable that, Que has a much higher absorption coefficient at λex = 370 nm than 1 (1.18 × 105 M−1 cm−1 vs. 0.21 × 105 M−1 cm−1, Fig. 8 and S7†). Thus, the added Que can compete with 1 for the energy absorption at λex, resulting in the quenching of lanthanide luminescence (“inner filter effect”, Scheme 2).39–41 As shown in Fig. 8, the absorptions of biomolecule interferents at λex are much weaker than Que, and they have little effect on the lanthanide luminescence of 1. The “inner filter effect” of Que to the lanthanide luminescence can be confirmed by comparing the quenching constants of Api and other biomolecule interferents. The absorption coefficient of Api at λex is 0.15 × 105 M−1 cm−1, which is higher than those of other interferents (less than 0.07 × 105 M−1 cm−1) (Fig. 8), thus, Api can absorb more light energy than other interferents. Unsurprisingly, Api also shows larger KSV value (0.44 × 104 M−1) than other interferents (less than 0.13 × 104 M−1), demonstrating the “inner filter effect” to the lanthanide luminescence. Furthermore, the addition of Que results in the quenching of visible ligand-centered emission of 1 at 545 nm (Fig. S10†). This is also due to the decrease of excitation energy of Schiff base ligands caused by the “inner filter effect” of Que.
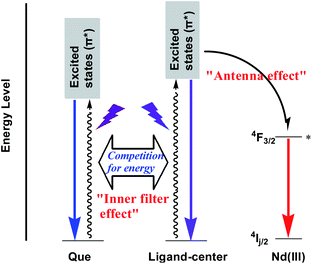 |
| | Scheme 2 The “antenna effect” of the Schiff base ligand and “inner filter effect” of Que to the lanthanide luminescence in 1. | |
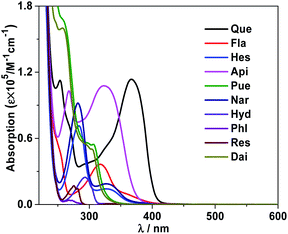 |
| | Fig. 8 Absorption spectra of Que and interferents in CH3CN at room temperature. | |
Experimental
Materials and general methods
All chemical materials were obtained from commercial sources and directly used without further purified. The Schiff base ligand and the lanthanide complex were synthesized using Schlenk system. Elemental analyses were performed on a EURO EA3000. Melting points were obtained on an XT-4 electrothermal micromelting point apparatus. IR spectra of the Schiff base ligand and the complex were measured on a FTIR-650 spectrometer. 1H NMR spectrum of the ligand was obtained using AVANCE III AV500 at 298 K. The mass spectrum (ESI) was measured on a MicroOTOF-QII. The conductivity measurement was carried out with a DDS-11 conductivity bridge. The powder XRD spectrum of the complex was recorded on a D8 Advance, and its scanning electron microscopy image was obtained using a Nova NanoSEM 200 microscope.
Synthesis of [Zn6Nd2L4(OAc)2(DMF)EtOH] (1)
At room temperature, Zn(OAc)2·2H2O (0.20 mmol, 0.0438 g), Nd(OAc)3·4H2O (0.20 mmol, 0.0823 g) and H4L (0.20 mmol, 0.0756 g) were dissolved in 20 mL MeOH, and then triethylamine in EtOH (1.0 mol L−1, 2 mL) was added into the solution. The mixture was heated for 6 h under reflux with stirring, and cooled to room temperature. The clear solution was obtained by filtered, and slow diffusion of diethyl ether into the solution for one week gave the yellow crystalline product of 1. Yield (based on Zn(OAc)2·2H2O): 0.0302 g (35%). Mp >178 °C (decompose). EA: C, 45.06; H, 3.37; N, 5.46% (found). Calc. for C97H86N10Nd2O32Zn6: C, 45.04; H, 3.33; N, 5.42%. IR (KBr, cm−1): 1578.34 (s), 1448.64 (s), 1249.94 (s), 1183.24 (s), 1083.26 (s), 879.42 (m), 788.13 (m), 839.04 (s), 731.12 (m).
Photophysical studies
The UV-vis absorbance spectra of the Schiff base ligand and the complex were determined on an UV-3600 spectrophotometer. The wavelength range is 200 nm–600 nm. The NIR emission and excitation spectra of the complex were carried out on a FLS 980 fluorimeter. Liquid nitrogen cooled Ge PIN diode detector is used to detect the NIR emissions (800 nm–1700 nm). An integrating sphere is used to obtain the quantum yield, and the luminescence lifetime is obtained by using the attached storage digital oscilloscope. Systematic errors are deducted through the standard instrument corrections.
Crystallography
The X-ray data of the complex were collected by a Smart APEX CCD diffractometer (Mo Kα radiation). The structure was solved using SHELX 97 program with direct method. The empirical absorption was corrected by SADABS program. Non-hydrogen atoms were refined anisotropically, and hydrogen atoms were calculated by geometrical methods and refined isotropically. The CCDC number of 1 is 2102148 (see www.ccdc.cam.ac.uk/data_request/cif). The bond lengths and angles are displayed in Table S1.†
For 1: C97H86N10O32Zn6Nd2, monoclinic (P21/n), a = 20.327(15), b = 23.139(16), c = 25.376(19) Å, α = 90°, β = 94.941(13)°, γ = 90°, V = 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 891(15) Å3, Dc = 1.444 g cm−3, μ(Mo Kα) = 2.118 mm−1, T = 190 K, Z = 4, F(000) = 5176, GOF = 1.163, R1 = 0.0913, wR2 = 0.1995.
891(15) Å3, Dc = 1.444 g cm−3, μ(Mo Kα) = 2.118 mm−1, T = 190 K, Z = 4, F(000) = 5176, GOF = 1.163, R1 = 0.0913, wR2 = 0.1995.
Conclusions
In conclusion, an octanuclear Zn(II)–Nd(III) cluster 1 was synthesized from a salen-type Schiff base ligand. The molecular sizes of 1 are 1.1 × 1.6 × 1.8 nm. It shows NIR lanthanide emission, which can be decreased gradually by the addition of Que. The emission quenching of 1 is due to the “inner filter effect” of Que. The limit of the detection of 1 to Que is 2.5 μM, and its luminescent response behaviour is not affected by the existence of some potential biomolecule interferents.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
The work was supported by the National Natural Science Foundation of China (No. 21771141).
Notes and references
- C. Papatriantafyllopoulou, E. E. Moushi, G. Christou and A. J. Tasiopoulos, Chem. Soc. Rev., 2016, 45, 1597–1628 RSC.
- A. Baniodeh, C. E. Anson and A. K. Powell, Chem. Sci., 2013, 4, 4354–4361 RSC.
- Y. Ning, M. Zhu and J.-L. Zhang, Coord. Chem. Rev., 2019, 399, 213028 CrossRef CAS.
- P. Shi, H. Hu, Z. Zhang, G. Xiong and B. Zhao, Chem. Commun., 2015, 51, 3985–3988 RSC.
- Y. Wang, G. Zhang, F. Zhang, T. Chu and Y. Yang, Sens. Actuators, B, 2017, 251, 667–673 CrossRef CAS.
- Y. H. Li and W. S. Huang, Anal. Methods, 2015, 7, 2537–2541 RSC.
- Y. Zou, F. Y. Yan, T. C. Zheng, D. C. Shi, F. Z. Sun, N. Yang and L. Chen, Talanta, 2015, 135, 145–148 CrossRef CAS PubMed.
- C. Y. Huang, T. K. Chen, D. M. Zhu and Q. Q. Huang, Front. Chem., 2020, 8, 225 CrossRef CAS PubMed.
- G. Chen, H. W. Zhang and J. N. Ye, Anal. Chim. Acta, 2000, 423, 69–76 CrossRef CAS.
- F. Buiarelli, F. Bernardini, P. D. Filippo, C. Riccardi, D. Pomata, G. Simonetti and R. Risoluti, Food Anal. Methods, 2018, 11, 3558–3562 CrossRef.
- M. Arvand and M. Anvari, J. Iran. Chem. Soc., 2013, 10, 841–849 CrossRef CAS.
- D. D. Wu and Z. Chen, Luminescence, 2014, 29, 307–313 CrossRef CAS PubMed.
- Y. Zou, F. Yan, L. Dai, Y. Luo, Y. Fu, N. Yang, J. Wun and L. Chen, Carbon, 2014, 77, 1148–1156 CrossRef CAS.
- T.-Q. Song, K. Yuan, W.-Z. Qiao, Y. Shi, J. Dong, H.-L. Gao, X.-P. Yang, J.-Z. Cui and B. Zhao, Anal. Chem., 2019, 91, 2595–2599 CrossRef CAS PubMed.
- W.-K. Wong, H. Liang, W.-Y. Wong, Z. Cai, K.-F. Li and K.-W. Cheah, New J. Chem., 2002, 26, 275–278 RSC.
- Y.-X. Chi, S.-Y. Niu, J. Jin, R. Wang and Y. Li, J. Chem. Soc., Dalton Trans., 2009, 47, 7653–7659 RSC.
- S. G. Baca, H. Adams, C. S. Grange, A. P. Smith, I. Sazanovich and M. D. Ward, Inorg. Chem., 2007, 46, 9779–9789 CrossRef CAS PubMed.
- H.-B. Xu, L.-X. Shi, E. Ma, L.-Y. Zhang, Q.-H. Wei and Z.-N. Chen, Chem. Commun., 2006, 1601–1603 RSC.
- S. Torelli, D. Imbert, M. Cantuel, G. Bernardinelli, S. Delahaye, A. Hauser, J.-C. G. Bünzli and C. Piguet, Chem.–Eur. J., 2005, 11, 3228–3242 CrossRef CAS PubMed.
- W.-K. Wong, X.-J. Zhu and W.-Y. Wong, Coord. Chem. Rev., 2007, 251, 2386–2399 CrossRef CAS.
- J. Zhang, L. Xu and W.-Y. Wong, Coord. Chem. Rev., 2018, 355, 180–198 CrossRef CAS.
- C. Wang, X. Yang, S. Wang, T. Zhu, L. Bo, L. Zhang, H. Chen, D. Jiang, X. Dong and S. Huang, J. Mater. Chem. C, 2018, 6, 865–874 RSC.
- D. Jiang, X. Yang, X. Zheng, L. Bo, T. Zhu, H. Chen, L. Zhang and S. Huang, J. Mater. Chem. C, 2018, 6, 8513–8521 RSC.
- D. Shi, X. Yang, Z. Xiao, X. Liu, H. Chen, Y. Ma, D. Schipper and R. A. Jones, Nanoscale, 2020, 12, 1384–1388 RSC.
- Y. Ma, X. Yang, M. Niu, D. Shi and D. Schipper, J. Lumin., 2022, 241, 118494 CrossRef CAS.
- F. Lam, J.-X. Xu and K.-S. Chan, J. Org. Chem., 1996, 61, 8414–8418 CrossRef CAS.
- V. Chandrasekhar, A. Athimoolam, N. D. Reddy, S. Nagendran, A. Steiner, S. Zacchini and R. Butcher, Inorg. Chem., 2003, 42, 51–59 CrossRef CAS PubMed.
- A. M. Rahman, R. Bishop, D. C. Craig and M. L. Scudder, CrystEngComm, 2003, 5, 422–428 RSC.
- W. K. Wong, X. Yang, R. A. Jones, J. Rivers, V. Lynch, W. K. Lo, D. Xiao, M. M. Oye and A. Holmes, Inorg. Chem., 2006, 45, 4340–4345 CrossRef CAS PubMed.
- S. I. Klink, L. Grave, D. N. Reinhoudt and F. C. J. M. v. Veggel, J. Phys. Chem. A, 2000, 104, 5457–5468 CrossRef CAS.
- J.-C. G. Bünzli and C. Piguet, Chem. Soc. Rev., 2005, 34, 1048–1077 RSC.
- Y. Xiao, Y. Cui, Q. Zheng, S. Xiang, G. Qian and B. Chen, Chem. Commun., 2010, 46, 5503–5505 RSC.
- X. Qi, Y. Jin, N. Li, Z. Wang, K. Wang and Q. Zhang, Chem. Commun., 2017, 53, 10318–10321 RSC.
- H.-W. Yang, P. Xu, B. Ding, X.-G. Wang, Z.-Y. Liu, H.-K. Zhao, X.-J. Zhao and E.-C. Yang, Cryst. Growth Des., 2020, 20, 7615–7625 CrossRef CAS.
- Z.-X. Wang, Y.-F. Gao, X. Jin, X.-H. Yu, X. Tao, F.-Y. Kong, D.-H. Fan and W. Wang, Analyst, 2019, 144, 2256–2263 RSC.
- C. Li, W. Liu, X. Sun, W. Pan, G. Yu and J. Wang, Sens. Actuators, B, 2018, 263, 1–9 CrossRef CAS.
- H. Qiu, C. Luo, M. Sun, F. Lu, L. Fan and X. Li, Food Chem., 2012, 134, 469–473 CrossRef CAS.
- J.-C. G. Bünzli and C. Piguet, Chem. Soc. Rev., 2005, 34, 1048–1077 RSC.
- S. Xu, J. Shi, B. Ding, Z. Liu, X. Wang, X. Zhao and E. Yang, Dalton Trans., 2019, 48, 1823–1834 RSC.
- F. Zhang, H. Yao, T. Chu, G. Zhang, Y. Wang and Y. Yang, Chem.–Eur. J., 2017, 23, 10293–10300 CrossRef CAS PubMed.
- Y. Zhang, B. Li, H. Ma, L. Zhang, H. Jiang, H. Song, L. Zhang and Y. Luo, J. Mater. Chem. C, 2016, 4, 7294–7301 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental and characterization details and additional figures. CCDC 2102148 for 1. See DOI: 10.1039/d1ra06494k |
|
| This journal is © The Royal Society of Chemistry 2021 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *ab,
Yanan Maab,
Chengri Wangab and
Desmond Schipperc
*ab,
Yanan Maab,
Chengri Wangab and
Desmond Schipperc







![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 891(15) Å3, Dc = 1.444 g cm−3, μ(Mo Kα) = 2.118 mm−1, T = 190 K, Z = 4, F(000) = 5176, GOF = 1.163, R1 = 0.0913, wR2 = 0.1995.
891(15) Å3, Dc = 1.444 g cm−3, μ(Mo Kα) = 2.118 mm−1, T = 190 K, Z = 4, F(000) = 5176, GOF = 1.163, R1 = 0.0913, wR2 = 0.1995.


