Open Access Article
Open Access ArticleDeep-red fluorogenic cyanine dyes carrying an amino group-terminated side chain for improved RNA detection and nucleolar RNA imaging†
Yusuke Sato *,
Yugo Igarashi,
Michiyuki Suzuki,
Kei Higuchi and
Seiichi Nishizawa
*,
Yugo Igarashi,
Michiyuki Suzuki,
Kei Higuchi and
Seiichi Nishizawa *
*
Department of Chemistry, Graduate School of Science, Tohoku University, Aoba-ku, Sendai 980-8578, Japan. E-mail: yusuke.sato.a7@tohoku.ac.jp; seiichi.nishizawa.c8@tohoku.ac.jp; Fax: +81-22-795-6552; Tel: +81-22-795-6549
First published on 17th November 2021
Abstract
The introduction of an amino-group-terminated side chain into deep-red emissive benzo[c,d]indole–quinoline monomethine cyanine dye has led to the improved detection of RNAs as well as the imaging of nucleolar RNAs in cells.
Small fluorescent probes capable of binding to RNAs have been powerful and versatile tools for the study of RNA functions.1 Compared to DNA-targeting probes, there are limited reports on the probes suitable for the practical use toward RNA analysis since small probes usually have a better selectivity toward DNAs than RNAs.1,2 SYTO RNA select is one of the commercially available probes with RNA selectivity. SYTO RNA select enables sensitive RNA detection due to the remarkable increase in its emission intensity upon binding to RNA. Nevertheless, the usefulness of SYTO RNA select has been not proved widely as its green emission (λem = ∼530 nm) suffers from background autofluorescence in the cellular imaging experiments. Small fluorescent probes having longer emission wavelength are highly desirable for the analysis of cellular or in vivo RNAs.3
We recently reported on benzo[c,d]indole–quinoline monomethine cyanine dye (BIQ, Fig. 1) as a new class of RNA-selective fluorescent dye in the deep-red spectral region (λem = 657 nm).4 BIQ features a large light-up signaling ability when binding to RNA (more than 100-fold), which stands in sharp contrast to coumarin–pyronin (CP: λem = 658 nm, ϕbound = 0.024)3a and Nile blue derivative (NBE: λem = 705 nm, ϕfree = 0.04)3b that were previously developed as deep-red emissive small probes for live cell imaging (less than 5-fold). We demonstrated that BIQ served as a useful fluorescent probe for detection of in vitro RNAs as well as for imaging of nucleolar RNAs in both living and fixed cells. Significantly, these 3 kinds of deep-red emissive probes (CP, NBE and BIQ) are rare example of small molecules with a clear selectivity for RNAs over DNAs in both solutions and living cells, but fluorogenic “off–on” signaling ability of cyanine-based BIQ (ϕfree < 0.0001, ϕbound = 0.0085) is outstanding compared to CP and NBE with rigid chemical structures. With the virtue of its light-up signaling ability, the nucleolar RNAs can be visualized even without washing steps after incubation with BIQ. In addition, BIQ is superior in terms of high photostability and negligible cytotoxicity. We believe that BIQ can be the most useful candidate as a deep-red small probe suitable for the imaging of nucleolar RNAs and their biological processing in living cells. However, further efforts are desirable to improve its properties such as the binding affinity and the brightness of the complex with RNA (ϕbound = 0.0085). The improvement of such BIQ functions is expected to make it more valuable for RNA analysis.
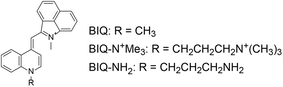 | ||
| Fig. 1 Chemical structures of benzo[c,d]indole–quinoline monomethine cyanine dye (BIQ) and its derivatives carrying an amino group-terminated side chain. | ||
In this work, we explored the introduction of amino group-terminated side chain into BIQ for the enhanced binding affinity with a view toward improved RNA sensing ability. Such a chemical modification was demonstrated to be effective for various nucleic acid-binding ligands because the amino groups show the favorable contribution into the binding with target nucleic acids through electrostatic interactions.5 Quaternary (BIQ–N+Me3) or primary amino group (BIQ–NH2) was thus introduced into the quinoline ring of BIQ scaffold through a propyl spacer (Fig. 1). As expected, such simple modification resulted in the effective increase in the binding affinity for RNAs, accompanied by the more pronounced “off–on” response as well as the improved ϕbound value. In addition, we demonstrated their applicability toward the fluorescence imaging of nucleolar RNAs in the fixed and living cells.
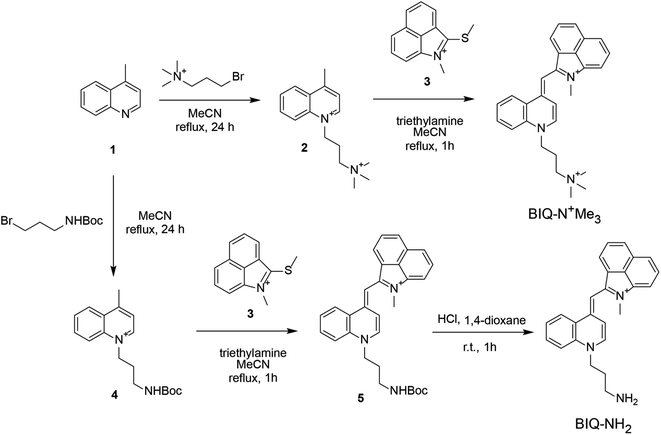
Synthesis scheme of BIQ–N+Me3 and BIQ–NH2 was shown in Fig. 2. Briefly, BIQ–N+Me3 was successfully synthesized in two key steps, where the quinoline derivative carrying the trimethylammonium group-terminated side chain (2) was reacted with benzo[c,d]indole unit (3). On the other hand, the attempt to obtain BIQ–NH2 by the similar synthetic scheme failed. Thus, primary amino group in the side chain of quinolone derivative was first protected with Boc group (4). After conjugation with benzo[c,d]indole unit, the obtain compound was deprotected to afford BIQ–NH2. Absorption and fluorescence spectra of BIQ–N+Me3, BIQ–NH2 and BIQ were shown in Fig. S1.†
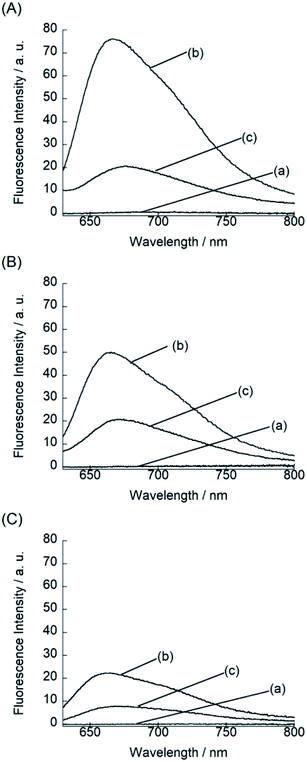
We examined the fluorescence response of BIQ derivatives for RNAs or DNAs at 25 °C in solutions buffered to pH 7.0 (Fig. 3). Here, we utilized E. coli total RNA and calf DNA as target nucleic acids according to the literatures.3a,b,4 This allows the direct comparison of sensing functions with the previously reported deep-red emissive small probes (CP and NBE). BIQ–N+Me3 and BIQ–NH2 exhibited negligible emission in the absence of nucleic acids (fluorescence quantum yield (ϕfree) < 0.001) due to the non-radiative energy loss due to the free rotation between the benzo[c,d]indole and quinoline rings of the BIQ scaffold.4 The addition of E. coli total RNA caused the remarkable light-up response in the deep-red spectral region (λem: BIQ–N+Me3, 666 nm; BIQ–NH2, 665 nm), where the emission intensity increased more than 100-fold. The observed response is attributable to the restricted rotation of BIQ scaffold by intercalation into the base pairs in the RNA, as similar to the parent BIQ where we observed hyperchromicity and a red shift of the absorption band of BIQ when binding to RNA.4 This is further supported by the larger response for double-stranded RNAs over single-stranded RNAs (Fig. S2†). Significantly, the light-up response of BIQ–N+Me3 and BIQ–NH2 was 3.4-fold and 2.2-fold larger than that of the parent BIQ under the identical condition. This is well reflected in the larger ϕfl values of these probes over BIQ in the bound state with RNAs (ϕbound: BIQ–N+Me3, 0.017; BIQ–NH2, 0.020; BIQ,4 0.0085). It should be noted that the ϕbound value of BIQ–NH2 is comparable to CP (ϕbound = 0.024),3a and its light-up signaling ability is much superior to those of CP and NBE (less than 5-fold).3a,3b It strongly suggests the attachment of amino group-terminated side chain into BIQ scaffold results in the improved detection ability for RNAs. The binding affinity of these probes for E. coli total RNA was then assessed by the fluorescence titration experiments (Fig. S3†). The apparent dissociation constants (Kd) of BIQ–N+Me3 and BIQ–NH2 were approximately determined as 360 μM and 210 μM, respectively (N = 2). These values were smaller by at least 2.2-fold compared to BIQ (>800 μM). The enhanced affinity toward RNA is thus responsible for the enhancement of the detection ability for RNAs. Importantly, the introduction of the positively charged side chain did not cause any loss of the selectivity for RNAs over DNAs (Fig. 3), indicating that the binding event is not governed simply by electrostatic interactions. The observed RNA-selectivity is much better than the commercial RNA-selective dye, SYTO RNA select, as examined in the literature,3a this commercial dye has no selectivity between RNAs and DNAs at low nucleic equivalents (<50), and the response intensity ratio of RNA/DNA is only ca. 1.6 (at 100 equivalent). In addition, BIQ derivatives feature high photostability compared to SYTO RNA select. We examined the photostability of BIQ–N+Me3 and BIQ–NH2 in the bound state with RNAs under continuous irradiation of excitation light for 120 scans (Fig. S4†). The emission intensity of BIQ–NH2 showed almost no change, where its photostability is comparable to BIQ.4 Meanwhile, the emission of BIQ–N+Me3 slightly decreased (10%) under the identical conditions. These BIQ derivatives had much superior photostability compared to SYTO RNA select whose emission decreased as much as 52%.4 Taken together, both BIQ–N+Me3 and BIQ–NH2 are promising probes for the robust and improved sensitivity for RNA sensing in solutions.
We then applied these BIQ derivatives for the fluorescence imaging of living MCF7 cells obtained from Cell Resource Center for Biomedical Research at Tohoku University. After incubation with BIQ–NH2 (5.0 μM) for 20 min, we observed the emission in the deep-red region in the cytoplasm and nucleolus (Fig. 4A). The emission in the cytoplasm would be explained by the accumulation within the negatively charged mitochondrial matrix due to the cationic property of BIQ–NH2, as observed for mitochondria-targeting triphenylphosphonium (TPP+).6 Similar behavior was also found in the parent BIQ4 and other monomethine cyanines.7 Meanwhile, it is highly likely that the fluorescence signal in the nucleolus arises from the binding of BIQ–NH2 to RNAs that were abundant in the nucleolus, such as ribosomal RNAs (rRNAs).3,4 This was supported by the examination of nuclease digestion in the fixed-permeabilized cells (Fig. 5). In the case of deoxyribonuclease (DNase), the emission in the nucleolus was almost unaffected. In contrast, the treatment with ribonuclease (RNase) resulted in the disappearance of the strong emission in the nucleolus. These results strongly indicate that BIQ–NH2 is able to stain the nucleolus based on the binding to nucleolar RNAs. In addition, BIQ–NH2 has a good counterstaining compatibility with Hoechst 33342 dye (Fig. S5†). Note that the nucleolus can be well stained when BIQ–NH2 was incubated for 24 h (Fig. S6†). This indicate BIQ–NH2 displayed little cytotoxicity toward living MCF7 cells like BIQ.4 Meanwhile, it was found that BIQ–N+Me3 failed to stain nucleolus in the living cells (Fig. S7†). Considering its light-up signaling for RNA (cf. Fig. 3A) and staining for the nucleolus in fixed-permeabilized cells (Fig. S8†), this would be attributed to the poor membrane permeability of BIQ–N+Me3 for the living cells due to the increased hydrophobicity compared to BIQ and BIQ–NH2.8,9
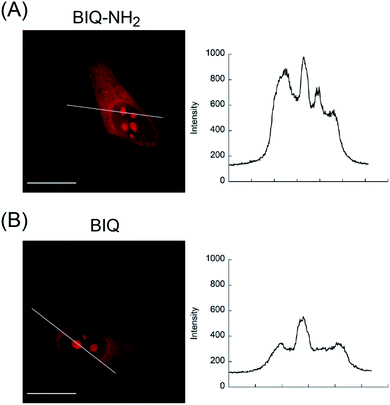 | ||
| Fig. 4 Fluorescence images of living MCF7 cells stained by the probes (5.0 μM: (A) BIQ–NH2 and (B) BIQ). Scale bar: 15 μm. Fluorescence intensity profiles along the white line are also shown. | ||
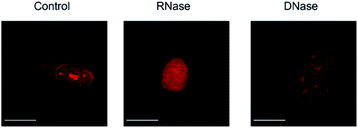 | ||
| Fig. 5 Fluorescence images of fixed-permeabilized MCF7 cells stained by BIQ–NH2 (5.0 μM) before (control) and after treatment of RNase or DNase. Scale bar: 15 μm. | ||
The comparison with BIQ reveals the usefulness of BIQ–NH2 for nucleolus staining in the living cells. The emission intensity of BIQ–NH2 in the nucleolus was found to be more than 2 times higher than that of BIQ (Fig. 4). It is highly likely that the enhanced binding affinity of BIQ–NH2 for RNAs relative to BIQ leads to the strong emission signal in the nucleolus. We found that even 500 nM BIQ–NH2 enables the nucleolus imaging in the living cells whereas almost no staining was observed for BIQ (Fig. S9†). Accordingly, BIQ–NH2 facilitates more sensitive imaging of nucleolar RNAs in the living cells compared to BIQ.
In summary, we described a simple, but effective modification of deep-red emissive benzo[c,d]indole–quinoline monomethine cyanine dye (BIQ) for the improved RNA sensing and imaging in the cells. The introduction of amino groups into the BIQ scaffold through the propyl spacer led to the improved fluorescence response upon binding to RNAs due to the enhanced binding affinity. The fluorogenic “off–on” signaling ability of the modified BIQ (BIQ–N+Me3 and BIQ–NH2) was further polished while the RNA-selectivity over DNA was maintained, which is much better than the commercial RNA-selective dye, SYTO RNA select. As for the cellular imaging, we found that BIQ–NH2 allowed more sensitive detection of nucleolar RNA in the living cells compared to the parent BIQ. The failure of BIQ–N+Me3 for live-cell imaging suggests that the slight difference in the chemical structure significantly affects the membrane permeability. Considering the lack of commercial dyes for RNA imaging in living cells and/or the moderate light-up properties of reported deep-red probes for RNA imaging (CP and NBE), we envision that BIQ–NH2 is a useful tool for the imaging of the dynamics of nucleolar RNAs. Also, considering the lack of rational design strategy for RNA-selective small molecules,1,10 the BIQ scaffold would be valuable and its further modification would offer novel deep-red probes for a wide range of RNA-related biomedical researches.3,11 We are now undertaking further studies in this direction.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Grant-in-Aid for Scientific Research (B) (No. 20H02761) from Japan Society for the Promotion of Science (JSPS).References
- Y. Xia, R. Zhang, Z. Wang, J. Tian and X. Chen, Chem. Soc. Rev., 2017, 46, 2824–2843 RSC.
- (a) W. D. Wilson, L. Ratmeyer, M. Zhao, L. Strekowski and D. Boykin, Biochemistry, 1993, 32, 4098–4104 CrossRef CAS PubMed; (b) Y. Sato, T. Ichihashi, S. Nishizawa and N. Teramae, Angew. Chem., Int. Ed., 2012, 51, 6369–6372 CrossRef CAS PubMed; (c) Y. Sato, Y. Toriyabe, S. Nishizawa and N. Teramae, Chem. Commun., 2013, 49, 9983–9985 RSC.
- (a) B. Zhou, W. Liu, H. Zhang, J. Wu, S. Liu, H. Xu and P. Wang, Biosens. Bioelectron., 2015, 68, 189–196 CrossRef CAS PubMed; (b) Q. Yao, H. Li, L. Xian, F. Xu, J. Xia, J. Fan, J. Du, J. Wang and X. Peng, Biomaterials, 2018, 177, 78–87 CrossRef CAS PubMed; (c) W. Liu, B. Zhou, G. Niu, K. Ge, J. Wu, H. Zhang, H. Xu and P. Wang, ACS Appl. Mater. Interfaces, 2015, 7, 7421–7427 CrossRef CAS PubMed; (d) Z. Li, S. Sun, Z. Yang, S. Zhang, H. Zhang, M. Hu, J. Cao, J. Wang, F. Liu, F. Song, J. Fan and X. Peng, Biomaterials, 2013, 34, 6473–6481 CrossRef CAS PubMed; (e) A. Sin-Yee, L. C.-C. Lee, K. K.-W. Lo and V. W.-W. Yam, J. Am. Chem. Soc., 2021, 143, 5396–5405 CrossRef PubMed.
- Y. Yoshino, Y. Sato and S. Nishizawa, Anal. Chem., 2019, 91, 14254–14260 CrossRef CAS PubMed.
- (a) V. Thiagarajan, A. Rajendran, H. Satake, S. Nishizawa and N. Teramae, ChemBioChem, 2010, 11, 94–100 CrossRef CAS PubMed; (b) R. E. McKnight, E. Reisenauer, M. V. Pintado, S. R. Polasani and D. W. Dixon, Bioorg. Med. Chem. Lett., 2011, 21, 4288–4291 CrossRef CAS PubMed; (c) Y. Abe, O. Nakagawa, R. Yamaguchi and S. Sasaki, Bioorg. Med. Chem., 2012, 20, 3470–3479 CrossRef CAS PubMed.
- (a) J. Zielonka, J. Joseph, A. Sikora, M. Hardy, O. Ouari, J. Vasquez-Vivar, G. Cheng, M. Lopez and B. Kalyanaraman, Chem. Rev., 2017, 117, 10043–10120 CrossRef CAS PubMed; (b) P. C. Saha, T. Chatterjee, R. Pattanayak, R. S. Das, A. Mukherjee, M. Bhattacharyya and S. Guha, ACS Omega, 2019, 4, 14579–14588 CrossRef CAS PubMed.
- (a) J. R. Carreon, K. M. Stewart, K. P. Mahon Jr, S. Shin and S. O. Kelly, Bioorg. Med. Chem. Lett., 2007, 17, 5182–5185 CrossRef CAS PubMed; (b) A. Kurutos, I. Orehovec, D. Saftic, L. Horvat, I. Crnolatac, I. Piantanida and T. Deligeorgiev, Dyes Pigm., 2018, 158, 517–525 CrossRef CAS.
- C. Cao, P. Wei, R. Li, Y. Zhong, X. Li, F. Xue, Y. Shi and T. Yi, ACS Sens., 2019, 4, 1409–1416 CrossRef CAS PubMed.
- B. L. Timney, B. Raveh, R. Mironska, J. M. Trivedi, S. J. Kim, S. R. Wente, A. Sali and M. P. Rout, J. Cell Biol., 2016, 215, 57–76 CrossRef CAS PubMed.
- A. U. Juru and A. E. Hargrove, J. Biol. Chem., 2021, 296, 100191 CrossRef PubMed.
- (a) Y. Sato, S. Yajima, A. Taguchi, K. Baba, M. Nakagomi, Y. Aiba and S. Nishizawa, Chem. Commun., 2019, 55, 3183–3186 RSC; (b) Y. Sato, Y. Aiba, S. Yajima, T. Tanabe, K. Higuchi and S. Nishizawa, ChemBioChem, 2019, 20, 2752–2756 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra05872j |
| This journal is © The Royal Society of Chemistry 2021 |