Open Access Article
Open Access ArticleUsing dual exonucleases to finely distinguish structural adjustment of aptamers for small-molecule detection†
Lancheng Wanga,
Huimin Zhoua,
Kun Yanb,
Peng Xuc,
Bin Di b,
Chi Hu
b,
Chi Hu *a and
Mengxiang Su
*a and
Mengxiang Su *b
*b
aDepartment of Pharmaceutical Engineering, School of Engineering, China Pharmaceutical University, Nanjing 211198, P. R. China. E-mail: chihu@cpu.edu.cn
bDepartment of Pharmaceutical Analysis, School of Pharmacy, China Pharmaceutical University, Nanjing 211198, P. R. China. E-mail: sumengxiang@cpu.edu.cn
cKey Laboratory of Drug Monitoring and Control, Drug Intelligence and Forensic Center, Ministry of Public Security, No. 18 Dongbeiwang West Road, Beijing 100193, P. R. China
First published on 6th October 2021
Abstract
The binding of small molecules to their DNA aptamers can modulate their susceptibility to digestion by exonucleases, however, absolute differentiation between digestion and inhibition has never been reported. Here, we show that the digestion of aptamers by T7 exonuclease can be completely inhibited upon binding of small-molecule targets and exploit this finding for the first time to achieve sensitive, label-free small-molecule detection. We use a quinine-binding aptamer to show that target binding entirely halts T7 exonuclease digestion, leaving behind an intact double-stranded product that retains strong target affinity. On the contrary, digestion of nontarget-bound aptamer produces a single-stranded product incapable of target binding. Exonuclease I efficiently eliminates these single-stranded products but is unable to digest the target-bound double-stranded product. The remaining products can be fluorescently quantified with SYBR Gold to determine target concentrations, giving a limit of detection of 100 nM with the linear range from 0 to 8 μM. We demonstrate the first example of a dual-exonuclease-mediated approach capable of producing a concentration-dependent response in terms of aptamer digestion modules, therefore improving performance of the current aptamer-based assay for small-molecule detection.
Introduction
Aptamers offer a number of advantages as molecular sensing reagents relative to antibodies, including greater thermostability, lower cost, excellent reproducibility of manufacturing, and the ease with which they can be engineered or chemically modified.1 Aptamer-based assays have been developed for the detection of a multitude of targets, particularly small molecules, based on various sensing strategies.2–4 Structure-switching aptamers undergo conformational changes to report the presence of the target.5–8 Strand displacement strategies employ a complementary DNA (cDNA) strand to partially block the target-binding domain of an aptamer via hybridization, and target-induced displacement of the cDNA generates sensing signals for small-molecule detection.7Alternatively, nuclease-assisted detection has been utilized to transduce one binding event into multiple signals to achieve ultra-sensitive target detection. For example, nicking endonuclease Nt.BbvCI has been used to develop an enzyme-based signal amplification strategy for cocaine detection, on account of its recognition towards specific DNA sequences.9 T7 exonuclease (T7 Exo) signal amplification in combination with polystyrene nanoparticles has been used to design a fluorescence polarization aptamer sensor.10 Moreover, exonuclease III (Exo III)-aided autocatalytic target recycling strategy was proposed and integrated with the homogeneous electrochemical method for detection of nucleic acid and protein.11
In particular, inhibition of exonuclease activity upon the binding of various types of substances, including drugs and enzymes, to DNA has long been described in literatures.12–14 It is until more recently, however, that this sophisticated modulation of aptamer to exonuclease digestion is applied to the development of label-free assays for small molecule detection.15 For instance, exonuclease I (Exo I)-based fluorescent assay has been reported to detections, proteins and small analytes, relying on the conformational change of structure-switching aptamers in the presence of target molecules.16 Gao et al. described that the digestion of lysozyme-specific aptamers by Exo I was inhibited upon lysozyme binding, and developed an Exo I-assisted electrochemical biosensor.17 Likewise, Exo III can exhibit compromised digestion towards aptamer-target complexes compared to target-free aptamers.18
In this context, dual-exonuclease-assisted systems have been reported to further enhance the detection sensitivity. For example, Xiao and coworkers utilized both Exo III and Exo I to perform digestion of a dehydroisoandrosterone-3-sulfate (DIS)-binding 37 nt aptamer.19 Partial digestion formed a 34 nt product in the presence of DIS, whereas complete digestion of nontarget-bound aptamer afforded mononucleotides. DIS quantification was achieved on account of the differentiation between the 34 nt product and the mononucleotides using SYBR Gold, a DNA-binding dye, as a signal reporter. However, since both unbound and target-bound forms of aptamer are liable to digestion under the reported enzyme systems, sensor performance of the exonuclease-assisted methods is largely undermined.
To overcome the limitations described above, we developed a label-free assay that utilized two exonucleases, T7 Exo and Exo I, to detect small-molecule targets without aptamer engineering requirements. As shown in Fig. 1, T7 Exo catalyzes the stepwise removal of dinucleotide and mononucleotides from the blunt-ended 5′ terminus of duplexed DNA, but not single-stranded DNA.20 When quinine was introduced as a proof-of-concept target, the aptamer–target complex underwent local conformational changes, which completely halted T7 Exo digestion of the 38 nt aptamer (denoted as Apt-38).21 In the absence of quinine, T7 Exo catalyzed digestion of the double-stranded stem of the fully folded aptamer, removing twenty nucleotides to form a 18 nt single-stranded product. Exo I cleaves single-stranded DNA from 3′ to 5′-end, releasing deoxyribonucleoside 5′-monophosphates in a stepwise manner, while leaves double-stranded DNA intact. Therefore, Exo I continued to degrade the 18 nt nontarget-bound digestion product into mononucleotides but was incapable of digesting the aptamer–target complex. Importantly, the local structural adjustment triggered upon target complexation renders the aptamer in a tightly folded state that is completely resistant to T7 Exo and Exo I digestion. As detection signal is generated from intact aptamer versus mononucleotides using SYBR Gold as the reporter dye, assay sensitivity is drastically enhanced. To the best of our knowledge, this is the first example of nuclease-assisted assay that fully discriminates between target-bound and unbound aptamers, affording intact DNA sequence of the former and mononucleotide digestion products of the latter, respectively.
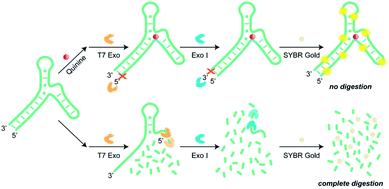 | ||
| Fig. 1 Schematic illustration of the dual-exonuclease-mediated detecting assay using SYBR Gold as a signal reporter. | ||
Experimental
Chemicals and materials
All the oligonucleotide sequences used in this study were synthesized and purified without any modifications by Sangon Biotech Inc. (China). T7 Exo was purchased from HaiGene Biotech Inc. (China). The fluorescent dye SYBR Gold (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00× concentrated) was purchased from Invitrogen (USA). Quinine hydrochloride dihydrate and mefloquine hydrochloride were purchased from Aladdin Reagent (China). Amodiaquine dihydrochloride, primaquine diphosphate, and chloroquine diphosphate were purchased from Shanghai Yuanye Biotechnology Co. Ltd. (China). Cocaine dissolved in acetonitrile was from Cerilliant (USA), and cocaine was separated from acetonitrile by initial dilution in deionized water followed by dehydration under vacuum during centrifugation. Urine samples used in this work were collected from healthy and consenting adult donors, all experiments were performed in accordance with the Declaration of Helsinki guidelines, and approved by the ethics committee at China Pharmaceutical University. Informed consents were obtained from human participants of this study. Exo I, bovine serum albumin (BSA), 2× tris-borate-EDTA (TBE)–urea sample buffer, and all other materials were purchased from Sangon Biotech Inc. (China). The aptamer sequences are reported below (mutated nucleotides are underlined):
00× concentrated) was purchased from Invitrogen (USA). Quinine hydrochloride dihydrate and mefloquine hydrochloride were purchased from Aladdin Reagent (China). Amodiaquine dihydrochloride, primaquine diphosphate, and chloroquine diphosphate were purchased from Shanghai Yuanye Biotechnology Co. Ltd. (China). Cocaine dissolved in acetonitrile was from Cerilliant (USA), and cocaine was separated from acetonitrile by initial dilution in deionized water followed by dehydration under vacuum during centrifugation. Urine samples used in this work were collected from healthy and consenting adult donors, all experiments were performed in accordance with the Declaration of Helsinki guidelines, and approved by the ethics committee at China Pharmaceutical University. Informed consents were obtained from human participants of this study. Exo I, bovine serum albumin (BSA), 2× tris-borate-EDTA (TBE)–urea sample buffer, and all other materials were purchased from Sangon Biotech Inc. (China). The aptamer sequences are reported below (mutated nucleotides are underlined):
Apt-38: 5′-GGG TGA CAA GGA AAA TCC TTC AAT GAA GTG GGT CAC CC-3′
Apt-M: 5′-GGG TGA CAA GGA AAA TCC TTC GAT GAA GTG GGT CAC CC-3′
ATP-33: 5′-CGC ACC TGG GGG AGT ATT GCG GAG GAA GGT GCG-3′
Aptamer digestion
Aptamer digestion experiments were performed using the following procedure unless otherwise specified. Before use, aptamer was incubated at 95 °C for 5 min and then cooled down to room temperature over 30 min. A 2 μL amount of 10 μM aptamer or mutant aptamer was added into 4 μL of reaction buffer (0.9× T7 buffer, 0.5× Exo I buffer, 0.1 mg mL−1 BSA) containing various concentration of target in a 200 μL PCR tube. Reaction mixtures were incubated at room temperature for 15 min. Then, 4 μL of the enzyme(s) was added to the sample tubes and incubated at 29 °C for 90 min. Final enzyme concentrations in the reaction were as follows: quinine (4 U μL−1 Exo I, 2 U μL−1 T7 Exo), cocaine (1.5 U μL−1 Exo I, 0.75 U μL−1 T7 Exo), ATP (1 U μL−1 Exo I, 0.75 U μL−1 T7 Exo). The total reaction volume was 10 μL.PAGE analysis
Digestion products were analyzed by denaturing polyacrylamide gel electrophoresis (PAGE). Specifically, 10 μL of the sample was collected and mixed with 10 μL of 2× TBE–urea sample buffer and incubated at 95 °C for 2 minutes, then immediately cooled on ice and loaded into the wells of a 15% denaturing PAGE gel. Separation was carried out at 20 V cm−1 for 2 h in 1× TBE running buffer. The gel was stained with 1× SYBR Gold for 25 min and imaged using a Tanon-3500 gel imaging system.SYBR Gold fluorescence experiments
The dual-exonuclease-mediated assay was performed using various concentrations of quinine (final concentrations: 0, 0.1, 0.175, 0.25, 0.5, 1, 2, 4, 6, 7, 8, 12.5, 20, 35, 50, and 80 μM). After digestion, 10 μL of digestion products were mixed with 140 μL Tris buffer (20 mM Tris–HCl, 5 mM MgCl2, pH 7.4) and 50 μL 2× SYBR Gold. A 65 μL amount of the mixture was loaded into the wells of a black 384-well microplate. Fluorescence emission spectra were recorded from 535 to 700 nm using a SpectraMax i3x (Molecular Devices) microplate reader with 495 nm excitation. The fluorescence intensity recorded at 545 nm was used to calculate signal gain using the equation (F − F0)/F0, where F and F0 represent fluorescence intensity in the presence and absence of the target, respectively. Calibration curves were constructed by plotting signal gains against target concentrations. Each experiment was performed in triplicate, and error bars represent the standard deviations of three measurements.Target detection in urine
Urine samples were collected from healthy donors and directly used in quinine detections. Specifically, 1 μL of 20 μM aptamer was added into 10 μL of urine sample containing various concentrations of quinine (final concentrations: 0, 0.5, 1, 3, 5, 10, 15, 20, 40, 80 μM) with 0.1 mg mL−1 BSA in a 200 μL PCR tube. Reaction mixtures were incubated at room temperature for 15 min. Then, 4 U μL−1 of Exo I and 2 U μL−1 T7 were added to the sample tubes and incubated at 29 °C for 90 min. The total reaction volume was 20 μL, where the concentration of urine was 50%. After digestion, 20 μL of digestion products were mixed with 40 μL PCR-grade water, 50 μL 2× SYBR Gold, and diluted with 90 μL fresh urine to keep a consistent 50% urine concentration. Afterwards, fluorescent intensities were recorded and data were analyzed with the same procedure as in the previous section.Results and discussion
Label-free fluorescence detection
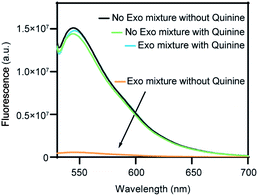
As a proof-of-concept study to demonstrate the T7 Exo/Exo I dual-exonuclease approach for small-molecule detection, we chose quinine as the target on account of its known aptamer structures, and SYBR Gold as a signal reporter, given its selective staining of oligonucleotides but not mononucleotides. Since most systematic evolution of ligands by exponential enrichment (SELEX) methods use unstructured libraries to yield aptamers with unpredictable binding domains, the process of introducing extra structure-switching function normally reduces target-binding affinity of the obtained aptamers.22 Therefore, the quinine-specific aptamer chosen in this work is a 38 nt prefolded oligonucleotide, which features a preformed secondary structure instead of undergoing substantial structure switching upon target binding.23 As shown in Fig. 1, the secondary structure of the aptamer contains three stems arranged around a three-way-junctioned binding domain with a tandem AG mismatch, where the 7-base-pair blunt-ended stem is a preferred substrate for T7 Exo.21,22,24 Quinine was incubated with the aptamer for 15 min at room temperature to form target-aptamer complexes through multiple intermolecular interactions including hydrogen bonding and π–π stacking.25,26 It is generally considered the secondary structure of the Apt-38 is fully folded in the free form, and the blunt end remains after binding to the target.23,24 Upon binding the target, Apt-38 undergoes a minor conformational change, instead of a large-scale change in secondary structure.27 Specifically, the aptamer stiffens and rearranges to a more rigid structure of binding to the target.24,28 Besides, the flexibility of the aptamer was decreased after complexation, with the base pair dynamics at the binding site contributing to the specificity of the recognition.28–30Propensity of the unbound or quinine-bound aptamers towards exonuclease digestion was investigated using a mixture of T7 Exo and Exo I, and fluorescence signal of the remaining oligonucleotide sequences was reported by SYBR Gold staining. As shown in Fig. 2, incubation of the target-bound aptamer with T7 Exo/Exo I mixture yielded a 25-fold signal gain for a sample containing 80 μM quinine, compared to the target-free aptamer. Importantly, the nearly identical fluorescence response of the preformed ligand–aptamer complex before and after T7 Exo/Exo I treatment suggested complete protection of the DNA sequence against digestion in its target-bound format. In contrast, an almost negligible fluorescence signal was observed after exonuclease digestion in the absence of quinine, owing to SYBR Gold with very high sensitivity and the remaining traces amounts of the undigested aptamer, indicating nearly complete degradation of the oligonucleotide into mononucleotides. As a result, a high signal-to-noise ratio with a minimal background fluorescence.
Digestion mechanism
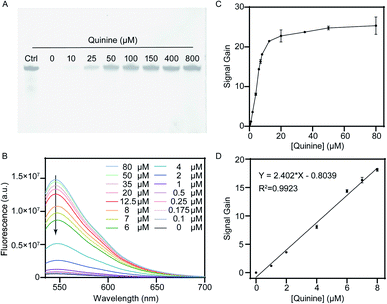
PAGE analysis was carried out to investigate mechanism of the exonuclease-catalyzed digestion of Apt-38 and its mutated variant Apt-M. As shown in Fig. 3A and B, T7 Exo alone was unable to digest Apt-38 into mononucleotides, and Exo I alone was unable to degrade the aptamer, regardless of the presence or absence of quinine. On the contrary, an exonuclease mixture containing both T7 Exo and Exo I (Exo M) fully digested Apt-38 into mononucleotides, as indicated by the complete disappearance of PAGE bands in the absence of quinine. These results indicated that the dual-exonuclease digestion occurred through a two-phase process, where T7 Exo performed its catalytic function prior to Exo I.Importantly, Exo M can well distinguish quinine-complexed Apt-38 from its unbounded form according to the PAGE results, given that no discrete bands except the 38 nt one were observed in the presence of the target. To confirm that enzymatic inhibition was specifically associated with formation of the target–aptamer complex, Apt-M with a single mutant nucleotide adjacent to the binding site was employed and its susceptibility towards exonuclease digestion was investigated. As shown in Fig. 3C and D, Apt-M was resistant to Exo I digestion but prone to T7 Exo degradation.
To validate the generality to other small molecules of the proposed method, we demonstrated the digestion products of Apt-38 in the absence and presence of cocaine by T7 Exo, Exo I, or a mixture of both. As shown in Fig. S2A and B,† PAGE analysis results show that the phenomenon is the same as when the aptamer binding with quinine. We further demonstrated a hairpin-structured ATP-binding aptamer (denoted as ATP-33), ATP-33 is prefolded and forms a hairpin structure with a 7-bp blunt ended stem.19 Despite its structural difference relative to Apt-38, ATP-33 also exhibited a target binding-dependent T7 Exo digestion profile, and similar to Apt-38, nontarget-bound ATP-33 was almost completely digested by the T7 Exo and Exo I, but both enzymes were strongly inhibited by ATP-binding aptamer.
Taken together, the results suggest that target binding plays an imperative role in exonuclease inhibition, presumably on account of the aptamer stiffens results the steric hindrance or distortion of the aptamer structure upon ligand binding, which reduces the range of contact between the aptamer and the enzyme.31
Analytical performance for detection of quinine
Subsequently, we applied the proposed dual-exonuclease-inhibition method to a label-free fluorescence assay that utilized synergistic digestion by T7 Exo and Exo I to detect quinine quantitatively. Experimental parameters, including the concentration of aptamer and SYBR Gold, reaction time and enzyme concentration, were evaluated to achieve optimal analytical performance. The relative fluorescence response was also closely correlated with the concentration of aptamer. Under a certain concentration of enzymes, a low concentration of aptamer leads to a low response, and high concentrations of aptamer lead to partially retained after digestion and cause an increase in background fluorescence. In Fig. S1A,† the relative fluorescence response (F/F0) of the proposed biosensor varied with the changeable aptamer from 0.5 to 6 μM and the relative fluorescence response reached to the maximum at 2 μM. Thus, 2 μM was the optimal concentration. The optimal concentration of SYBR Gold was further investigated, various volumes of 2× SYBR Gold were added to the reaction solution. As shown in Fig. S1B,† the maximum F/F0 ratio was achieved with the addition of 50 μl of 2× SYBR Gold. The enzymatic reaction time was investigated by monitoring the fluorescence intensity of the hydrolysis products after SYBR Gold staining at a time interval of 20 min in 2 hours. As shown in Fig. S1C,† fluorescence intensity of the mixture decreased from 0 to 80 min, and reached a plateau thereafter. To ensure sufficient digestion time, 90 min was selected in the subsequent experiments. Concentration of T7 Exo and Exo I enzymes was also optimized. As displayed in Fig. S1D and E,† when the amount of T7 Exo and Exo I was 4 U μL−1 and 2 U μL−1, respectively, fluorescence intensity of the digestion products reached the lowest value, indicating complete reaction with the given concentration of aptamer. Therefore, 4 U μL−1 of T7 Exo and 2 U μL−1 Exo I were chosen as the optimal amount of enzymes.Next, analytical performance of the dual-exonuclease assay was evaluated under the optimized conditions. As shown in Fig. 4A, after exonuclease digestion with a mixture of T7 Exo and Exo I, we observed the intact 38 nt aptamer in the presence of quinine, which was otherwise completely digested in the absence of quinine. Notably, the 38 nt electrophoresis band became gradually darker with quinine ranging from 0 to 800 μM, reflecting the abundance of remaining Apt-38. Fluorescence studies were carried out to determine whether the response was linear correlated to the target concentration. As shown in Fig. 4B, it could be seen that with the presence of increasing concentration of quinine, intensity of the fluorescence signal was enhanced gradually. We observed a 25-fold signal gain with 80 μM quinine relative to the target-free sample, which exhibited minimal background in the absence of quinine. Fluorescence intensity at 545 nm was calculated versus the target concentration, and a good linear relationship in the range of 0 to 8 μM was obtained with a coefficient of correlation of 0.9923 (Fig. 4C and D). Limit of detection for this “signal on” aptasensor was calculated to be 100 nM according to the 3σ/S method.
We then investigated the selectivity of the dual-exonuclease-mediated approach for quinine analysis. The quinine analogs and other disruptants were measured to replace quinine under the same conditions using 20 μM of each small molecule. As shown in Fig. S3A and B,† our results demonstrated no measurable signal from L-tryptophan, dopamine, glucose, or ATP, some cross-reactivity to mefloquine (14.0%) and amodiaquine (22.1%) and high cross-reactivity to primaquine (73.4%) and chloroquine (68.3%). These results indicate that our method has high specificity and a certain ability to distinguish structural analogs.
Detection in urine
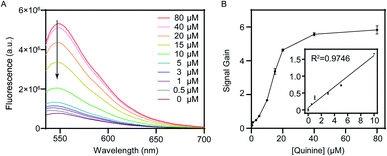
To evaluate the feasibility of our dual-exonuclease-inhibition method in practical applications, the assay was employed to detect quinine in urine samples. Urine is a biological matrix of choice because it can be obtained noninvasively, and an inexpensive and easy assay to detect antimalarial compounds such as quinine from patient samples could improve the collection of drug use and quality data for close monitoring the resistance of the parasite to antimalarial drugs.8 We collected human urine from healthy donors, and spiked the 50% diluted biological samples with different concentrations of quinine. Fluorescence response and signal gain were examined as demonstrated in Fig. 5A and B, respectively, which showed similar pattern as in buffer solution. Although the sensor performance was noticeably lower in urine compared to that in buffer solution, which is often observed in literatures,16,32 we still obtained a 5.8-fold signal gain with 80 μM quinine, confirming the applicability of the proposed assay method in biological matrices. We achieved a linear range from 0 to 10 μM with a limit of detection of 500 nM by the 3σ/S method. In short, these experiments demonstrated that our dual-exonuclease-inhibition method could potentially be used to detect small-molecule in complex systems.Conclusions
In summary, we developed a label-free dual-exonuclease-inhibition method that utilized the synergistic digestion by commercially available T7 Exo and Exo I enzymes for the detection of small-molecule targets. Compared with other assays, our method is the first instance of producing a concentration-dependent response in terms of aptamer digestion modules, and this method can be generally implemented with an unmodified aptamer, so that the sensing performance will not be damaged. As a proof-of-concept study, we demonstrated the detection of quinine, where T7 Exo digests the double-stranded stem of the fully folded aptamer in the absence of quinine, then the remaining single-stranded sequences are completely digested by Exo I into mononucleotides. When quinine was added to the system, however, target complexation triggers a sophisticated local structure adjustment near the binding site, resulting in complete resistance to T7 Exo and Exo I digestion. The remaining intact aptamer was readily quantified by SYBR Gold staining to reflect the concentration of quinine. Sensing performance in real-world samples was demonstrated by detecting the quinine in urine. Our results confirmed that the dual-exonuclease-inhibition approach was capable of enhancing the sensing response to the small-molecule targets, and was still feasible in biological matrices.Author contributions
Lancheng Wang: methodology, investigation, data curation, visualization, writing – original draft. Huimin Zhou: validation, data curation, writing review & editing. Kun Yan: data curation, analysis, writing review & editing. Peng Xu: supervision, project administration, writing review & editing. Bin Di: funding acquisition, project administration, writing review & editing. Chi Hu: conceptualization, supervision, funding acquisition, writing review & editing. Mengxiang Su: conceptualization, funding acquisition, writing review & editing. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by National Natural Science Foundation of China (NSFC 81803485, 81872833) and Key Research and Development Plan of Jiangsu Province (BE2020696).References
- H. Yu, O. Alkhamis, J. Canoura, Y. Liu and Y. Xiao, Angew. Chem., Int. Ed., 2021, 60, 2–26 CrossRef.
- T. Gao, Y. Luo, W. Li, Y. Cao and R. Pei, Analyst, 2020, 145, 701–718 RSC.
- Y. Dong, T. Zhang, X. Lin, J. Feng, F. Luo, H. Gao, Y. Wu, R. Deng and Q. He, Microchim. Acta, 2020, 187, 1–18 CrossRef PubMed.
- O. Alkhamis, J. Canoura, H. Yu, Y. Liu and Y. Xiao, TrAC, Trends Anal. Chem., 2019, 121, 115699 CrossRef CAS PubMed.
- M. N. Stojanovic, P. De Prada and D. W. Landry, J. Am. Chem. Soc., 2001, 123, 4928–4931 CrossRef CAS PubMed.
- M. N. Stojanovic, P. de Prada and D. W. Landry, J. Am. Chem. Soc., 2000, 122, 11547–11548 CrossRef CAS PubMed.
- R. Nutiu and Y. Li, J. Am. Chem. Soc., 2003, 125, 4771–4778 CrossRef CAS PubMed.
- E. S. Coonahan, K.-A. Yang, S. Pecic, M. De Vos, T. E. Wellems, M. P. Fay, J. F. Andersen, J. Tarning and C. A. Long, Sci. Transl. Med., 2021, 13, eabe1535 CAS.
- B. Shlyahovsky, D. Li, Y. Weizmann, R. Nowarski, M. Kotler and I. Willner, J. Am. Chem. Soc., 2007, 129, 3814–3815 CrossRef CAS PubMed.
- Y. Huang, X. Liu, M. Shi, S. Zhao, K. Hu, Z. F. Chen and H. Liang, Chem.–Asian J., 2014, 9, 2755–2760 CrossRef CAS PubMed.
- S. Liu, Y. Lin, L. Wang, T. Liu, C. Cheng, W. Wei and B. Tang, Anal. Chem., 2014, 86, 4008–4015 CrossRef CAS PubMed.
- F. G. Albert, T. T. Eckdahl, D. J. Fitzgerald and J. N. Anderson, Biochemistry, 1999, 38, 10135–10146 CrossRef CAS PubMed.
- L. Tian, J. M. Sayer, H. Kroth, G. Kalena and S. Shuman, J. Biol. Chem., 2003, 278, 9905–9911 CrossRef CAS PubMed.
- L. Yakovleva, C. J. Handy, H. Yagi, J. M. Sayer, D. M. Jerina and S. Shuman, Biochemistry, 2006, 45, 7644–7653 CrossRef CAS PubMed.
- J. Canoura, H. Yu, O. Alkhamis, D. Roncancio and Y. Xiao, J. Am. Chem. Soc., 2021, 143, 805–816 CrossRef CAS PubMed.
- D. Zheng, R. Zou and X. Lou, Anal. Chem., 2012, 84, 3554–3560 CrossRef CAS PubMed.
- X. Gao, L. Qi, K. Liu, C. Meng and H. Z. Yu, Anal. Chem., 2020, 92, 6229–6234 CrossRef CAS PubMed.
- Z. Wang, H. Yu, C. Juan, Y. Liu, A. Obtin, F. Fu and Y. Xiao, Nucleic Acids Res., 2018, 46, e81 CrossRef PubMed.
- J. Canoura, Z. Wang, H. Yu, O. Alkhamis, F. Fu and Y. Xiao, J. Am. Chem. Soc., 2018, 140, 9961–9971 CrossRef CAS PubMed.
- C. Kerr and P. D. Sadowski, J. Biol. Chem., 1972, 247, 311–318 CrossRef CAS.
- D. Roncancio, H. Yu, X. Xu, S. Wu and Y. Xiao, Anal. Chem., 2014, 86, 11100–11106 CrossRef CAS PubMed.
- M. Neves, O. Reinstein, M. Saad and P. E. Johnson, Biophys. Chem., 2010, 153, 9–16 CrossRef CAS PubMed.
- M. Neves, A. A. Shoara, O. Reinstein, O. A. Borhani, T. R. Martin and P. E. Johnson, ACS Sens., 2017, 2, 1539–1545 CrossRef CAS PubMed.
- O. R. Miguel, A. D. Neves and P. E. Johnson, Biochemistry, 2010, 49, 8478–8487 CrossRef PubMed.
- T. Hermann and D. J. Patel, Science, 2000, 287, 820–825 CrossRef CAS PubMed.
- A. A. Shoara, S. Slavkovic, L. W. Donaldson and P. . E. Johnson, Can. J. Chem., 2017, 95, 1253–1260 CrossRef CAS.
- A. Sachan, M. Ilgu, A. Kempema, G. A. Kraus and M. Nilsen-Hamilton, Anal. Chem., 2016, 88, 7715–7723 CrossRef CAS PubMed.
- C. M. Grytz, A. Marko, P. Ce Kan, S. T. Sigurdsson and T. F. Prisner, Phys. Chem. Chem. Phys., 2016, 18, 2993–3002 RSC.
- Z. R. Churcher, M. Neves, H. N. Hunter and P. E. Johnson, J. Biomol. NMR, 2017, 68, 33–39 CrossRef CAS PubMed.
- S. Slavkovic, Z. R. Churcher and P. E. Johnson, Bioorg. Med. Chem., 2018, 26, 5427–5434 CrossRef CAS PubMed.
- A. Obtin, W. Yang, F. Rifat, H. Yu and Y. Xiao, Nucleic Acids Res., 2020, 48, e120 CrossRef PubMed.
- Z. Wu, H. Zhou, Q. Han, X. Lin, D. Han and X. Li, Analyst, 2020, 145, 4664–4670 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra05551h |
| This journal is © The Royal Society of Chemistry 2021 |