DOI:
10.1039/D1RA05240C
(Paper)
RSC Adv., 2021,
11, 30020-30029
Aminomalononitrile inspired prebiotic chemistry as a novel multicomponent tool for the synthesis of imidazole and purine derivatives with anti-influenza A virus activity†
Received
7th July 2021
, Accepted 30th August 2021
First published on 8th September 2021
Abstract
Amino imidazole carbonitrile derivatives decorated with α-amino acid side-chains have been synthesized by a multicomponent microwave assisted reaction inspired by the prebiotic chemistry of aminomalononitrile as a tool for generating high chemical diversity. These compounds were used as annulation synthons for the preparation of 8,9-disubstituted-6,9-dihydro-1H-purin-6-ones by reaction with formic acid as a simple C-1 donor reagent. The novel heterocycles were characterized by significant activity against influenza A virus, amino imidazole carbonitrile derivatives showing the highest activity. Thus, the chemical complexity generated by prebiotic chemistry furnished a general tool for the identification of novel antiviral agents.
Introduction
It is well recognized that Multi-Component Chemistry (MCC) played a key role in molecular evolution generating high chemical diversity from a limited number of reagents.1 The MCC of the Origins of Life generated an incredible variety of organic structures in the pristine Earth, setting the molecular evolution conditions for the emergence of the Last Universal Common Ancestor (LUCA).2 The first documented prebiotic MCC encompasses the Strecker reaction for the synthesis of amino acids,3 the Butlerow condensation of formaldehyde to sugars,4 and the Orò transformation of cyanuric acid (HCN) to purine nucleobases.5 More recently, prebiotic MCC involving formamide (NH2CHO), a product of hydrolysis of HCN,6 yielded a robust chemical framework for the synthesis of sugars, carboxylic acids, amino acids and heterocyclic compounds, including purine and pyrimidine nucleobases and nucleosides.7–13 A number of unnatural derivatives were also synthesized by the combination of the initial products, including peptide nucleic acids14 and modified acyclic nucleosides,15 which belong to well-known antiviral drug families.16 In particular, peptide nucleic acid derivatives are characterized by high antiviral activity against influenza A, HIV and HBV viruses.17,18 Today, the modernization of the MCC by innovative techniques (e.g. microwave-assisted procedures) furnishes an impressive combination of tools to force and speed-up the production of complex heterocyclic derivatives encompassing the so called “Molecular Evolution-Inspired Synthesis”.19 In this context, aminomalononitrile (AMN)20 has been identified as a common intermediate in the prebiotic chemistry of HCN and NH2CHO, besides to diaminomaleonitrile.21,22 AMN is involved in the formation of imidazole intermediates, such as amino imidazole carbonitrile and amino imidazole carboxyamide, which are precursors of purine and pyrimidine nucleobases and analogues.23 Imidazoles are important pharmacophores in natural products and drugs, including antiviral agents24 with anti-influenza virus activity.25–27 In addition, imidazoles are synthons in the preparation of bioactive purines by annulation with compounds as simple as urea, guanidine, formic acid and NH2CHO.28 Here we report that AMN inspired multicomponent prebiotic chemistry yields amino imidazole carbonitrile derivatives decorated with different α-amino acid side-chains. The reaction involves the microwave-assisted condensation between aminomalononitrile para-toluenesulfonate salt (AMNS), α-amino acids and trimethyl orthoacetate (TOA). The chimera between nucleobases and amino acids has been largely explored in the case of peptide-nucleic acids as a possible alternative to RNA in the origin of life scenarios.29 Imidazole derivatives were successively annulated into the corresponding purines by a solvent free microwave assisted pyrimidine ring-closing procedure involving formic acid. Formic acid is a ubiquitous component in the pristine Earth, often involved in the prebiotic chemistry of AMN.29 The novel derivatives showed appreciable activity against influenza A virus as a consequence of the block of the viral budding from the cell, impairing the viral release to other cells.
Results and discussion
Synthesis of amino imidazole carbonitrile derivatives 4a–f and 5a–f
Amino imidazole carbonitrile derivatives were prepared by a multicomponent procedure involving the microwave assisted condensation of AMNS 1, TOA 2 and a panel of protected α-amino acids (methyl ester derivatives) 3a–f, including glycine 3a, alanine 3b, valine 3c, serine 3d, phenylalanine 3e, and tyrosine 3f (Scheme 1). TOA was preferred to trimethyl orthoformate in order to avoid undesired side-processes.30 The reaction was performed by two sequential steps without intermediate workup and purification as follows: (a) AMNS 1 (5.9 mmol) dissolved in THF (30 mL) and triethylamine TEA (7.1 mmol, 1.0 mL) was stirred at room temperature for 30 min, followed by the addition of 2 (8.3 mmol) and microwave irradiation (250 W, 250 psi) for 2.0 min at 200 °C; (b) the solution was cooled down at 25 °C and the crude treated with TEA (7.1 mmol, 1.0 mL) and α-amino acids 3a–f (7.1 mmol) under previously reported microwave conditions. The reaction proceeded with a quantitative conversion of 1 to afford amino imidazole carbonitrile methyl ester derivatives 4a–f in a yield ranging from 25% to 60% (Table 1, entries 1–6) depending on the nature of selected α-amino acid (Scheme 1; Pathway A). The closure of the imidazole ring proceeded via imino-ether intermediate followed by nucleophilic addition of the amino acid residue.31 In this latter case, compounds 4a–f were selectively decorated by the α-amino acid side-chain at the N-1 position of the imidazole ring. The possible formation of bis-imidazole derivatives by reaction of compounds 4a–f with trimethyl orthoformate was not observed under our experimental conditions.28 On the other hand, the low value of the mass balance was probably due to the competitive self-oligomerization of AMN to HCN polymers.32 As a general trend, the yield of the reaction products decreased by increasing the structural complexity of the α-amino acid side-chain (Table 1, entry 1 vs. entries 2–3), with the only exception of aromatic derivatives phenyl alanine and tyrosine (Table 1, entries 5–6). The beneficial effect of the aromatic moiety in the annulation of AMN with simple amine has been previously reported.28 Compounds 4a–f were successively de-protected (NaOH 1 M for 18 h at 25 °C)33 to afford the corresponding amino imidazole carbonitriles (AIC) 5a–f (Scheme 1, Pathway B) (Table 1, entries 7–12).
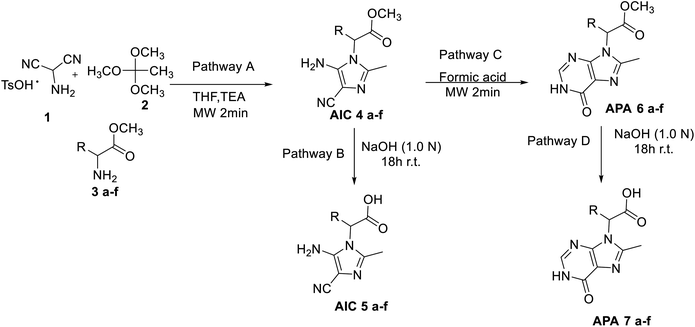 |
| | Scheme 1 Synthesis of amino imidazole carbonitrile derivatives 4a–f and 5a–f, and 8,9-disubstituted-6,9-dihydro-1H-purin-6-ones 6a–f and 7a–f. | |
Table 1 Amino imidazole carbonitrile methyl esters 4a–f and amino imidazole carbonitriles 5a–f from three component condensation of AMN, trimethyl orthoacetate and α-amino acids
| Entry |
α-Amino acid |
Condition |
Products |
R |
Yieldc (%) |
| Reaction conditions: (a) compound 1 (5.9 mmol) in THF (30 mL) and triethylamine (7.1 mmol) for 30 min at 25 °C; (b) compound 2 (8.3 mmol) and amino-acids 3a–f (7.1 mmol) under MW irradiation. NaOH (1 N, 1.0 mL) stirring for 18 h at 25 °C. The yield was calculated on the basis of the amount of the recovered product. Reactions were performed in triplicate. |
| 1 |
Glycine |
THF, 2.0 min, 250 W, 250 psi at 200 °Ca |
4a |
H |
60 |
| 2 |
Alanine |
4b |
CH3 |
34 |
| 3 |
Valine |
4c |
CH(CH3)2 |
32 |
| 4 |
Serine |
4d |
CH2OH |
25 |
| 5 |
Phenylalanine |
4e |
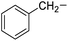 |
51 |
| 6 |
Tyrosine |
4f |
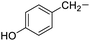 |
48 |
| 7 |
Glycine |
NaOH, 18 h, 25 °Cb |
5a |
H |
92 |
| 8 |
Alanine |
5b |
CH3 |
95 |
| 9 |
Valine |
5c |
CH(CH3)2 |
98 |
| 10 |
Serine |
5d |
CH2OH |
93 |
| 11 |
Phenylalanine |
5e |
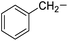 |
90 |
| 12 |
Tyrosine |
5f |
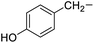 |
94 |
Synthesis of 8,9-disubstituted-6,9-dihydro-1H-purin-6-one derivatives 6a–f and 7a–f
The role of amino imidazole carbonitriles as synthons for the synthesis of a large diversity of heterocyclic derivatives has been reported, and the mechanism of the condensation discussed in detail.34–36 For example, C-1 containing reagents as simple as urea, guanidine, formic acid and NH2CHO, have been applied to yield di-amino purines, hypoxanthine, adenine, 1H-purin-2-(3H)-ones and other analogues, respectively. On the basis of these data, compounds 4a–f were annulated with formic acid to yield the corresponding 8,9-disubstituted-6,9-dihydro-1H-purin-6-one methyl esters 6a–f. Briefly, compounds 4a–f (3.8 mmol) were dissolved in formic acid (3.0 mL) under microwave irradiation (250 W, 250 psi) for 2.0 min at 200 °C (Scheme 1, Pathway C) to afford 6a–f in quantitative conversion of substrate and yield (Table 2, entries 1–6). In addition, 6a–f (0.1 mmol) were successively de-protected (NaOH 1 M for 18 h at 25 °C) to corresponding 8,9-disubstituted-6,9-dihydro-1H-purin-6-ones 7a–f (Scheme 1, Pathway D) (Table 2, entries 7–12).
Table 2 Synthesis of amino purine analogues 6a–f and 7a–f
| Entry |
α-Amino acid |
Condition |
Products |
R |
Yieldc (%) |
| Reaction conditions: compounds 4a–f (3.8 mmol) were dissolved in formic acid (3.0 mL) under MW irradiation. NaOH (1 N, 1.0 mL) stirring for 18 h at 25 °C. The yield was calculated on the basis of the amount of the recovered product. Reactions were performed in triplicate. |
| 1 |
Glycine |
Formic acid, 2.0 min, 250 W, 250 psi at 200 °Ca |
6a |
H |
88 |
| 2 |
Alanine |
6b |
CH3 |
85 |
| 3 |
Valine |
6c |
CH(CH3)2 |
87 |
| 4 |
Serine |
6d |
CH2OH |
92 |
| 5 |
Phenylalanine |
6e |
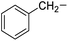 |
90 |
| 6 |
Tyrosine |
6f |
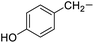 |
95 |
| 7 |
Glycine |
NaOH, 18 h, 25 °Cb |
7a |
H |
98 |
| 8 |
Alanine |
7b |
CH3 |
94 |
| 9 |
Valine |
7c |
CH(CH3)2 |
96 |
| 10 |
Serine |
7d |
CH2OH |
92 |
| 11 |
Phenylalanine |
7e |
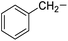 |
94 |
| 12 |
Tyrosine |
7f |
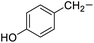 |
88 |
Biological assay
Influenza A virus is one of the most common infectious human respiratory pathogens associated with significant morbidity and mortality, especially in hospitalized patients. Among the influenza viruses, type A strains are responsible for annual epidemics and pandemic events. They belong to the Orthomyxoviridae family and are characterized by segmented, single-stranded negative RNA genome. RNA segments associate with the nucleoprotein (NP) and the viral RNA-dependent RNA polymerase (RdRp) complex in the replication and transcription cycles.37 Three families of antiviral compounds are active against influenza A virus: (a) the first one blocks the ion-channel activity of the viral matrix (M) 2 protein involved in the virus uncoating; (b) the second family includes inhibitors of the viral neuraminidase (NA), blocking the release of viral particles from infected cells; and (c) inhibitors of the polymerase complex. Among them, baloxavir, marboxil and favirapir are licensed in Japan and United States.38 However, the huge circulation of different influenza viruses and the emergence of drug-resistant strains lead to the search of new antiviral agents. For this reason, the novel imidazole and purine derivatives were tested against influenza A/Puerto Rico/8/34 H1N1 (PR8) virus. In the first set of experiments, A549 cells were infected with 0.001 MOI of PR8 and treated with different concentrations (range 5–80 μg mL−1) of compounds 4a–f, 5a–f, 6a–f and 7a–f (Table 3) for 24 h. Then, infected treated-cells were fixed and stained with antibodies specific for the viral protein hemagglutinin (HA), as described in the experimental part. The cytotoxicity of the compounds was evaluated by standard MTT assay. Table 3 shows the values of IC50, CC50, and relative selective index (SI) obtained on A549 cells. Compounds 4e, 5a, 4b, 4d, 5e, 7c, 6c, 6f, and 7b showed SI values more than 1.5. These compounds were further investigated to evaluate the ability in impairing viral release from infected cells. Briefly, A549 cells were infected with PR8 and treated with concentrations (100, 150 and 200 μg mL−1) of selected compounds. After 24 h, infected cell monolayers were directly stained with anti-HA antibodies as described above, while supernatants were recovered and used to newly infect fresh monolayers. We found that within the 9 most active derivatives, compounds 5a and 7c decreased the expression of HA on infected monolayers (Fig. 1A) compared to untreated infected cells (Fig. 1; Panel A, part I). The percentage of Relative Fluorescent Units (RFU) of HA was dose-dependently reduced. As a consequence, the supernatants of treated cells contained less viral particles able to infect other monolayers (Fig. 1; Panel B). Overall, the results suggest that both compounds 5a and 7c may block specific steps of viral replication, probably involving the viral budding from the cell, thus impairing viral release from infected cells and the spreading of virions to the other ones.
Table 3 Biological activity of compounds 4a–f, 5a–f, 6a–f and 7a–f against influenza A virus
| Entry |
Class |
Amino acid |
R |
Products |
IC50a |
CC50b |
SIc |
| IC50 is the drug concentration (μmol) causing 50% inhibition of the desired activity. Each experiment was conducted in triplicate. CC50 is the drug concentration (μmol) causing 50% of death of viable cell. SI is the selectivity index defined as the ratio of the CC50 to the IC50. |
| 1 |
Amino-imidazole-carbonitrile derivatives (AIC) |
Glycine |
H |
4a |
nd |
1.02 |
nd |
| 2 |
5a |
0.76 |
2.20 |
2.9 |
| 3 |
Alanine |
CH3 |
4b |
0.76 |
2.75 |
3.6 |
| 4 |
5b |
3.00 |
2.91 |
1 |
| 5 |
Valine |
CH(CH3)2 |
4c |
2.41 |
1.13 |
0.5 |
| 6 |
5c |
0.83 |
0.62 |
0.8 |
| 7 |
Serine |
CH2OH |
4d |
0.68 |
1.99 |
2.9 |
| 8 |
5d |
nd |
nd |
nd |
| 9 |
Phenylalanine |
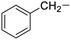 |
4e |
0.39 |
0.84 |
2.1 |
| 10 |
5e |
0.46 |
3.36 |
7.2 |
| 11 |
Tyrosine |
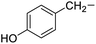 |
4f |
3.20 |
1.45 |
0.5 |
| 12 |
5f |
1.99 |
0.90 |
0.45 |
| 13 |
Amino-purine-analogues (APA) |
Glycine |
H |
6a |
6.96 |
2.11 |
0.3 |
| 14 |
7a |
0.89 |
0.75 |
0.84 |
| 15 |
Alanine |
CH3 |
6b |
nd |
0.47 |
nd |
| 16 |
7b |
1.85 |
2.86 |
1.5 |
| 17 |
Valine |
CH(CH3)2 |
6c |
0.18 |
0.31 |
1.7 |
| 18 |
7c |
0.59 |
1.09 |
1.8 |
| 19 |
Serine |
CH2OH |
6d |
nd |
0.93 |
nd |
| 20 |
7d |
— |
6.11 |
— |
| 21 |
Phenylalanine |
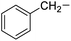 |
6e |
1.87 |
0.51 |
0.3 |
| 22 |
7e |
nd |
6.35 |
nd |
| 23 |
Tyrosine |
 |
6f |
0.48 |
0.80 |
1.7 |
| 24 |
7f |
nd |
nd |
nd |
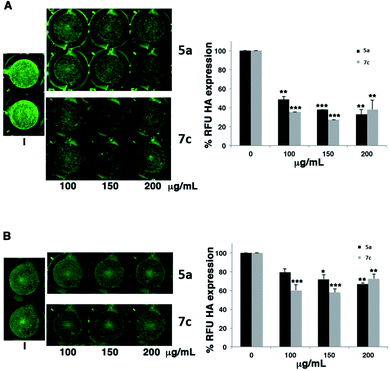 |
| | Fig. 1 The expression of hemagglutinin (HA) by means of In Cell Western (ICW) assay in the presence of most active compounds 5a and 7c. A549 cells were infected with PR8 (0.001 MOI) and treated or not (I) with different concentrations (100, 150 and 200 μg mL−1) of compounds. After 24 h treatment, cells were fixed and stained for HA protein, as described in the Materials and methods section (Panel A); supernatants were recovered and used to infect a fresh monolayer of A549 cells (Panel B). The expression of viral HA was analyzed by ICW assay, using LI-COR Image Studio Software. The percentage of relative fluorescence units (RFU) was calculated in comparison to untreated infected cells (considered 100%). Values are the mean ± S.D. of two replicates of one experiment of two performed (n = 2). Statistical significance of the data vs. untreated infected cells was defined as *p < 0.05 and **p < 0.01 and ***p < 0.001 (unpaired t-test). | |
Experimental section
Materials
Solvents and reagents (analytical grade) were purchased from Aldrich Chemical Co. Silica gel 60 F254 plates and silica gel 60 were furnished from Merck. Chemical reactions were monitored using thin-layer chromatography on precoated aluminum silica gel Merck 60 F254 plates using a UV lamp for visualization. Merck silica gel 60 (230–400 mesh) was used for chromatography. Microwave reactions were performed by a microwave synthesizer CEM Discover (CEM Corporation Italy). NMR spectra were recorded on a Bruker Advance DRX400 (400 MHz/100 MHz) spectrometer. Chemical shifts are reported in parts per million (ppb) using deuterated solvents DMSO-d6, CD3OD, D2O, CDCl3. Coupling constants (J) are reported in Hz. Multiplicities are reported in the conventional form: s = singlet, d = doublet, t = triplet, dd = doublets of doublets, m = multiplet. A 549 (human lung epithelial carcinoma) cell lines (ATCC code no. CCL-185) were purchased from LGC Standards Srl, Sesto San Giovanni, Milan Italy. Influenza A/PR/8/34 (H1N1) virus (ATCC code no. VR-95) purchased from LGC Standards Srl, Sesto San Giovanni, Milan Italy.
General procedure for the synthesis of 5-amino-1,2-disubstituted-1H-imidazole-4-carbonitriles (4a–f)
To a solution of aminomalononitrile p-toluenesulfonate 1 (5.9 mmol) in THF (30 mL) was added triethylamine (7.1 mmol). The mixture was stirred at room temperature for 30 min. To this solution was added trimethyl orthoacetate 2 (8.3 mmol), and the solution was stirred under microwave condition using the following program:
| No. of cycles |
Temperature |
Ramp time |
Hold time |
Pressure (psi) |
Power (W) |
| 1 |
200 °C |
1 min |
2 min |
250 |
250 |
Thereafter, the solution was cooled at room temperature followed by the addition of triethylamine (7.1 mmol) and amino-acids (methyl ester derivatives) 3a–f (7.1 mmol). The solution was stirred under previously reported microwave conditions. The solvent was removed under reduced pressure and the residue dissolved in dichloromethane (30 mL) was washed with Na2CO3 (3 × 20 mL) and NaCl (1 × 20 mL) saturated solutions. The organic layer was dried over Na2SO4, and the solvent was evaporated under reduced pressure. The crude was purified by flash-chromatography eluting with ethyl acetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n-hexane (2
n-hexane (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 4a–f from 15 to 60% yield.
1) to afford 4a–f from 15 to 60% yield.
General procedure for the synthesis of amino-purine-analogues (6a–f)
Compound 4a–f (3.8 mmol) was dissolved in formic acid (3.0 mL) and stirred under microwave condition described for the synthesis of 4a–f. After returning to room temperature, the mixture was poured into water (30 mL). The resulting precipitate was filtered and washed with water. The crude was purified by flash-chromatography eluting with dichloromethane (CH2Cl2)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (CH3OH) (5
methanol (CH3OH) (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 6a–f in quantitative yield.
1) to afford 6a–f in quantitative yield.
General procedure for the synthesis of amino-purine-analogues (5a–f and 7a–f)
The appropriate compound (0.1 mmol) was hydrolyzed in NaOH solution (1 N, 1.0 mL) stirring for 18 h at room temperature. Thereafter, the solution was neutralized with HCl 1 N, freeze-dried and the residue washed with CH3OH. The organic layer was evaporated under reduced pressure to afford 5a–f and 7a–f in quantitative yield. 1H-NMR and 13C-NMR of 5a–f and 7a–f are in ESI#1.†
Spectroscopic data
Compound 4a. Methyl 2-(5-amino-4-cyano-2-methyl-1H-imidazol-1-yl)acetate. 1H-NMR (400 MHz, DMSO-d6, ppm): δ 6.135 (s, 2H, NH2), 4.733 (s, 2H, CH2), 3.706 (s, 3H, O–CH3), 2.078 (s, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 168.515 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 148.883 (C), 140.104 (C), 118.080 (C), 88.533 (C), 52.912 (O–CH3), 43.907 (CH2), 13.297 (CH3). MS (ESI): m/z (M + H) +195.19. Elemental analysis for C8H10N4O2 calculated: C, 49.48; H, 5.19; N, 28.85; O, 16.48. Found: C, 49.45; H, 5.16; N, 28.81; O, 16.43.
O), 148.883 (C), 140.104 (C), 118.080 (C), 88.533 (C), 52.912 (O–CH3), 43.907 (CH2), 13.297 (CH3). MS (ESI): m/z (M + H) +195.19. Elemental analysis for C8H10N4O2 calculated: C, 49.48; H, 5.19; N, 28.85; O, 16.48. Found: C, 49.45; H, 5.16; N, 28.81; O, 16.43.
Compound 4b. Methyl 2-(5-amino-4-cyano-2-methyl-1H-imidazol-1-yl)propanoate. The crude residue was purified by flash-chromatography eluting with ethyl acetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) petroleum ether (2
petroleum ether (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H-NMR (400 MHz, CDCl3, ppm): δ 4.897–4.878 (m, 1H, CH), 4.150 (s, 2H, NH2), 3.847 (s, 3H, O–CH3), 2.336 (s, 3H, CH3), 1.761 (d, J = 7.6 Hz, 3H, CH3). 13C-NMR (100 MHz, CDCl3, ppm): δ 170.573 (C
1). 1H-NMR (400 MHz, CDCl3, ppm): δ 4.897–4.878 (m, 1H, CH), 4.150 (s, 2H, NH2), 3.847 (s, 3H, O–CH3), 2.336 (s, 3H, CH3), 1.761 (d, J = 7.6 Hz, 3H, CH3). 13C-NMR (100 MHz, CDCl3, ppm): δ 170.573 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 145.708 (C), 140.664 (C), 115.587 (C), 96.244 (C), 53.373 (CH), 53.297 (O–CH3), 15.965 (CH3), 14.096 (CH3). MS (ESI): m/z (M + H) +209.22. Elemental analysis for C9H12N4O2 calculated: C, 51.92; H, 5.81; N, 26.91; O, 15.37. Found: C, 51.89; H, 5.77; N, 26.87; O, 15.32.
O), 145.708 (C), 140.664 (C), 115.587 (C), 96.244 (C), 53.373 (CH), 53.297 (O–CH3), 15.965 (CH3), 14.096 (CH3). MS (ESI): m/z (M + H) +209.22. Elemental analysis for C9H12N4O2 calculated: C, 51.92; H, 5.81; N, 26.91; O, 15.37. Found: C, 51.89; H, 5.77; N, 26.87; O, 15.32.
Compound 4c. Methyl 2-(5-amino-4-cyano-2-methyl-1H-imidazol-1-yl)-3-methylbutanoate. The crude residue was purified by flash-chromatography eluting with ethyl acetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n-hexane (2
n-hexane (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H-NMR (400 MHz, DMSO-d6, ppm): δ 6.042 (s, 2H, NH2), 4.577 (d, J = 10.8 Hz, 1H, CH), 3.705 (s, 3H, O–CH3), 2.597–2.553 (m, 1H, CH), 2.123 (s, 3H, CH3), 1.113 (d, J = 6.4 Hz, 3H, CH3), 0.639 (d, J = 6.4 Hz, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 169.805 (C
1). 1H-NMR (400 MHz, DMSO-d6, ppm): δ 6.042 (s, 2H, NH2), 4.577 (d, J = 10.8 Hz, 1H, CH), 3.705 (s, 3H, O–CH3), 2.597–2.553 (m, 1H, CH), 2.123 (s, 3H, CH3), 1.113 (d, J = 6.4 Hz, 3H, CH3), 0.639 (d, J = 6.4 Hz, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 169.805 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 148.935 (C), 139.722 (C), 117.724 (C), 89.925 (C), 61.883 (CH), 53.198 (O–CH3), 28.372 (CH), 20.673 (CH3), 18.860 (CH3), 14.716 (CH3). MS (ESI): m/z (M + H) +238.28. Elemental analysis for C11H16N4O2 calculated: C, 55.92; H, 6.83; N, 23.71; O, 13.54. Found: C, 55.86; H, 6.78; N, 23.65; O, 13.49.
O), 148.935 (C), 139.722 (C), 117.724 (C), 89.925 (C), 61.883 (CH), 53.198 (O–CH3), 28.372 (CH), 20.673 (CH3), 18.860 (CH3), 14.716 (CH3). MS (ESI): m/z (M + H) +238.28. Elemental analysis for C11H16N4O2 calculated: C, 55.92; H, 6.83; N, 23.71; O, 13.54. Found: C, 55.86; H, 6.78; N, 23.65; O, 13.49.
Compound 4d. Methyl 2-(5-amino-4-cyano-2-methyl-1H-imidazol-1-yl)-3-hydroxypropanoate. The crude residue was purified by flash-chromatography eluting with ethyl acetate. 1H-NMR (400 MHz, DMSO-d6, ppm): δ 6.890 (s, 1H, OH), 5.872 (s, 2H, NH2), 5.473–5.092 (m, 2H, CH2), 3.689 (s, 3H, O–CH3), 3.666–3.615 (m, 1H, CH), 2.114 (s, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 168.27 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 148.98 (C), 140.56 (C), 117.99 (C), 90.09 (C), 62.85 (CH2), 59.86 (CH), 53.09 (O–CH3), 14.55 (CH3). MS (ESI): m/z (M + H) +225.22. Elemental analysis for C9H12N4O3 calculated: C, 48.21; H, 5.39; N, 24.99; O, 21.41. Found: C, 48.17; H, 5.35; N, 24.94; O, 21.38.
O), 148.98 (C), 140.56 (C), 117.99 (C), 90.09 (C), 62.85 (CH2), 59.86 (CH), 53.09 (O–CH3), 14.55 (CH3). MS (ESI): m/z (M + H) +225.22. Elemental analysis for C9H12N4O3 calculated: C, 48.21; H, 5.39; N, 24.99; O, 21.41. Found: C, 48.17; H, 5.35; N, 24.94; O, 21.38.
Compound 4e. Methyl 2-(5-amino-4-cyano-2-methyl-1H-imidazol-1-yl)-3-phenylpropanoate. The crude residue was purified by flash-chromatography eluting with ethyl acetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n-hexane (3
n-hexane (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H-NMR (400 MHz, CDCl3, ppm): δ 7.283–7.254 (m, 3H, CH–Ar), 6.995–6.973 (m, 2H, CH–Ar), 4.837–4.799 (dd, J = 4.0, 11.2 Hz, 1H, CH2), 4.323 (s, 2H, NH2), 3.877 (s, 3H, O–CH3), 3.569 (t, J = 12.8 Hz, 1H, CH), 3.385–3.340 (dd, J = 4.4, 13.6 Hz, 1H, CH2), 1.809 (s, 3H, CH3). 13C-NMR (100 MHz, CDCl3, ppm): δ 169.934 (C
1). 1H-NMR (400 MHz, CDCl3, ppm): δ 7.283–7.254 (m, 3H, CH–Ar), 6.995–6.973 (m, 2H, CH–Ar), 4.837–4.799 (dd, J = 4.0, 11.2 Hz, 1H, CH2), 4.323 (s, 2H, NH2), 3.877 (s, 3H, O–CH3), 3.569 (t, J = 12.8 Hz, 1H, CH), 3.385–3.340 (dd, J = 4.4, 13.6 Hz, 1H, CH2), 1.809 (s, 3H, CH3). 13C-NMR (100 MHz, CDCl3, ppm): δ 169.934 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 146.094 (C), 141.435 (C), 135.347 (C), 129.080 (C–Ar ×2), 128.757 (C–Ar ×2), 127.768 (C–Ar), 115.833 (C), 95.757 (C), 60.286 (CH), 53.498 (O–CH3), 35.296 (CH2), 13.456 (CH3). MS (ESI): m/z (M + H) +285.32. Elemental analysis for C15H16N4O2 calculated: C, 63.37; H, 5.67; N, 19.71; O, 11.25. Found: C, 63.31; H, 5.62; N, 19.68; O, 11.20.
O), 146.094 (C), 141.435 (C), 135.347 (C), 129.080 (C–Ar ×2), 128.757 (C–Ar ×2), 127.768 (C–Ar), 115.833 (C), 95.757 (C), 60.286 (CH), 53.498 (O–CH3), 35.296 (CH2), 13.456 (CH3). MS (ESI): m/z (M + H) +285.32. Elemental analysis for C15H16N4O2 calculated: C, 63.37; H, 5.67; N, 19.71; O, 11.25. Found: C, 63.31; H, 5.62; N, 19.68; O, 11.20.
Compound 4f. Methyl 2-(5-amino-4-cyano-2-methyl-1H-imidazol-1-yl)-3-(4-hydroxyphenyl)propanoate. The crude residue was purified by flash-chromatography eluting with ethyl acetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) petroleum ether (4
petroleum ether (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H-NMR (400 MHz, DMSO-d6, ppm): δ 9.249 (s, 1H, OH), 6.846 (d, J = 8.4 Hz, 2H, CH–Ar), 6.593 (d, J = 8.4 Hz, 2H, CH–Ar), 5.700 (s, 2H, NH2), 5.199–5.160 (m, 1H, CH), 3.724 (s, 3H, O–CH3), 3.266–3.219 (m, 2H, CH2), 1.794 (s, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 169.44 (C
1). 1H-NMR (400 MHz, DMSO-d6, ppm): δ 9.249 (s, 1H, OH), 6.846 (d, J = 8.4 Hz, 2H, CH–Ar), 6.593 (d, J = 8.4 Hz, 2H, CH–Ar), 5.700 (s, 2H, NH2), 5.199–5.160 (m, 1H, CH), 3.724 (s, 3H, O–CH3), 3.266–3.219 (m, 2H, CH2), 1.794 (s, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 169.44 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 156.59 (C–OH), 148.22 (C), 140.24 (C), 130.41 (C–Ar ×2), 126.71 (C–Ar), 117.83 (C), 115.60 (C–Ar ×2), 90.92 (C), 60.22 (CH), 53.26 (O–CH3), 34.19 (CH2), 14.56 (CH3). MS (ESI): m/z (M + H) +301.32. Elemental analysis for C15H16N4O3 calculated: C, 59.99; H, 5.37; N, 18.66; O, 15.98. Found: C, 59.95; H, 5.29; N, 18.61; O, 15.94.
O), 156.59 (C–OH), 148.22 (C), 140.24 (C), 130.41 (C–Ar ×2), 126.71 (C–Ar), 117.83 (C), 115.60 (C–Ar ×2), 90.92 (C), 60.22 (CH), 53.26 (O–CH3), 34.19 (CH2), 14.56 (CH3). MS (ESI): m/z (M + H) +301.32. Elemental analysis for C15H16N4O3 calculated: C, 59.99; H, 5.37; N, 18.66; O, 15.98. Found: C, 59.95; H, 5.29; N, 18.61; O, 15.94.
Compound 5a. 2-(5-Amino-4-cyano-2-methyl-1H-imidazol-1-yl)acetic acid. 1H-NMR (400 MHz, D2O, ppm): δ 4.285 (d, J = 8.8 Hz, 2H, CH2), 2.112 (d, J = 11.2 Hz, 3H, CH3). 13C-NMR (100 MHz, D2O, ppm): δ 174.50 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 144.50 (C), 141.60 (C), 109.61 (C), 95.61 (C), 45.88 (CH2), 12.00 (CH3). MS (ESI): m/z (M + H) +181.17. Elemental analysis for C7H8N4O2 calculated: C, 46.67; H, 4.48; N, 31.10; O, 17.76. Found: C, 46.62; H, 4.43; N, 31.05; O, 17.71.
O), 144.50 (C), 141.60 (C), 109.61 (C), 95.61 (C), 45.88 (CH2), 12.00 (CH3). MS (ESI): m/z (M + H) +181.17. Elemental analysis for C7H8N4O2 calculated: C, 46.67; H, 4.48; N, 31.10; O, 17.76. Found: C, 46.62; H, 4.43; N, 31.05; O, 17.71.
Compound 5b. 2-(5-Amino-4-cyano-2-methyl-1H-imidazol-1-yl)propanoic acid. 1H-NMR (400 MHz, MeOD, ppm): δ 4.260 (d, J = 7.6 Hz, 1H, CH), 1.747 (s, 3H, CH3), 1434 (s, 3H, CH3). 13C-NMR (100 MHz, MeOD, ppm): δ 167.11 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 142.78 (C), 139.26 (C), 117.28 (C), 93.45 (C), 58.05 (CH), 22.48 (CH3), 14.60 (CH3). MS (ESI): m/z (M + H) +195.19. Elemental analysis for C8H10N4O2 calculated: C, 49.48; H, 5.19; N, 28.85; O, 16.48. Found: C, 49.43; H, 5.15; N, 28.79; O, 16.41.
O), 142.78 (C), 139.26 (C), 117.28 (C), 93.45 (C), 58.05 (CH), 22.48 (CH3), 14.60 (CH3). MS (ESI): m/z (M + H) +195.19. Elemental analysis for C8H10N4O2 calculated: C, 49.48; H, 5.19; N, 28.85; O, 16.48. Found: C, 49.43; H, 5.15; N, 28.79; O, 16.41.
Compound 5c. 2-(5-Amino-4-cyano-2-methyl-1H-imidazol-1-yl)-3-methylbutanoic acid. 1H-NMR (400 MHz, D2O, ppm): δ 4.384 (d, J = 2.8 Hz, 1H, CH), 2.409–2.340 (m, 1H, CH), 2.229 (s, 3H, CH3), 0.937 (d, J = 6.8 Hz, 3H, CH3), 0.756 (d, J = 6.8 Hz, 3H, CH3). 13C-NMR (100 MHz, D2O, ppm): δ 181.50 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 148.33 (C), 141.22 (C), 117.46 (C), 92.58 (C), 67.34 (CH), 29.76 (CH), 16.76 (CH3), 16.17 (CH3), 13.25 (CH3). MS (ESI): m/z (M + H) +223.25. Elemental analysis for C10H14N4O2 calculated: C, 54.04; H, 6.35; N, 25.21; O, 14.40. Found: C, 53.97; H, 6.31; N, 25.17; O, 14.35.
O), 148.33 (C), 141.22 (C), 117.46 (C), 92.58 (C), 67.34 (CH), 29.76 (CH), 16.76 (CH3), 16.17 (CH3), 13.25 (CH3). MS (ESI): m/z (M + H) +223.25. Elemental analysis for C10H14N4O2 calculated: C, 54.04; H, 6.35; N, 25.21; O, 14.40. Found: C, 53.97; H, 6.31; N, 25.17; O, 14.35.
Compound 5d. 2-(5-Amino-4-cyano-2-methyl-1H-imidazol-1-yl)-3-hydroxypropanoic acid. 1H-NMR (400 MHz, D2O, ppm): δ 4.489–4.415 (m, 1H, CH), 3.119 (s, 2H, CH2), 2.625 (s, 3H, CH3). 13C-NMR (100 MHz, D2O, ppm): δ 168.62 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 145.80 (C), 140.76 (C), 116.44 (C), 94.79 (C), 64.43 (CH2), 61.41 (CH), 12.92 (CH3). MS (ESI): m/z (M + H) +211.19. Elemental analysis for C8H10N4O3 calculated: C, 45.71; H, 4.80; N, 26.66; O, 22.83. Found: C, 45.66; H, 4.75; N, 26.61; O, 22.78.
O), 145.80 (C), 140.76 (C), 116.44 (C), 94.79 (C), 64.43 (CH2), 61.41 (CH), 12.92 (CH3). MS (ESI): m/z (M + H) +211.19. Elemental analysis for C8H10N4O3 calculated: C, 45.71; H, 4.80; N, 26.66; O, 22.83. Found: C, 45.66; H, 4.75; N, 26.61; O, 22.78.
Compound 5e. 2-(5-Amino-4-cyano-2-methyl-1H-imidazol-1-yl)-3-phenylpropanoic acid. 1H-NMR (400 MHz, D2O, ppm): δ 7.160–7.129 (m, 3H, CH–Ar), 6.930–6.925 (m, 2H, CH–Ar), 3.405–3.358 (dd, J = 4.4, 14.4 Hz, 2H, CH2), 3.204 (d, J = 14.4 Hz, 1H, CH), 2.275 (s, 3H, CH3). 13C-NMR (100 MHz, D2O, ppm): δ 166.90 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 154.94 (C), 139.62 (C), 134.76 (C), 129.24 (C–Ar ×2), 128.38 (C–Ar ×2), 127.08 (C–Ar), 108.66 (C), 95.90 (C), 60.23 (CH), 34.70 (CH2), 12.80 (CH3). MS (ESI): m/z (M + H) +271.29. Elemental analysis for C14H14N4O2 calculated: C, 62.21; H, 5.22; N, 20.73; O, 11.84. Found: C, 62.16; H, 5.28; N, 20.69; O, 11.79.
O), 154.94 (C), 139.62 (C), 134.76 (C), 129.24 (C–Ar ×2), 128.38 (C–Ar ×2), 127.08 (C–Ar), 108.66 (C), 95.90 (C), 60.23 (CH), 34.70 (CH2), 12.80 (CH3). MS (ESI): m/z (M + H) +271.29. Elemental analysis for C14H14N4O2 calculated: C, 62.21; H, 5.22; N, 20.73; O, 11.84. Found: C, 62.16; H, 5.28; N, 20.69; O, 11.79.
Compound 5f. 2-(5-Amino-4-cyano-2-methyl-1H-imidazol-1-yl)-3-(4-hydroxyphenyl)propanoic acid. 1H-NMR (400 MHz, MeOD, ppm): δ 6.797 (d, J = 2.0 Hz, 2H, CH–Ar), 6.494 (d, J = 6.4 Hz, 2H, CH–Ar), 4.004–3.961 (m, 1H, CH), 2.885–2.848 (m, 2H, CH2), 1.9876 (s, 3H, CH3). 13C-NMR (100 MHz, MeOD, ppm): δ 167.02 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 156.01 (C–OH), 147.00 (C), 137.88 (C), 129.61 (C–Ar ×2), 120.47 (C–Ar), 118.54 (C–Ar ×2), 108.95 (C), 95.90 (C), 59.81 (CH), 35.99 (CH2), 12.47 (CH3). MS (ESI): m/z (M + H) +287.29. Elemental analysis for C14H14N4O3 calculated: C, 58.74; H, 4.93; N, 19.57; O, 16.77. Found: C, 58.69; H, 4.88; N, 19.52; O, 16.71.
O), 156.01 (C–OH), 147.00 (C), 137.88 (C), 129.61 (C–Ar ×2), 120.47 (C–Ar), 118.54 (C–Ar ×2), 108.95 (C), 95.90 (C), 59.81 (CH), 35.99 (CH2), 12.47 (CH3). MS (ESI): m/z (M + H) +287.29. Elemental analysis for C14H14N4O3 calculated: C, 58.74; H, 4.93; N, 19.57; O, 16.77. Found: C, 58.69; H, 4.88; N, 19.52; O, 16.71.
Compound 6a. Methyl 2-(8-methyl-6-oxo-1,6-dihydro-9H-purin-9-yl)acetate. 1H-NMR (400 MHz, D2O, ppm): δ 8.067 (s, 1H, CH–Ar), 5.051 (s, 2H, CH2), 3.744 (s, 3H, O–CH3), 2.438 (s, 3H, CH3). 13C-NMR (100 MHz, D2O, ppm): δ 169.76 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 155.96 (C), 151.77 (C), 149.80 (C), 145.45 (C), 122.02 (C), 53.42 (O–CH3), 43.91 (CH2), 12.49 (CH3). MS (ESI): m/z (M + H) +221.20. Elemental analysis for C9H10N4O3 calculated: C, 48.65; H, 4.54; N, 25.21; O, 21.60. Found: C, 48.61; H, 4.48; N, 25.16; O, 21.55.
O), 155.96 (C), 151.77 (C), 149.80 (C), 145.45 (C), 122.02 (C), 53.42 (O–CH3), 43.91 (CH2), 12.49 (CH3). MS (ESI): m/z (M + H) +221.20. Elemental analysis for C9H10N4O3 calculated: C, 48.65; H, 4.54; N, 25.21; O, 21.60. Found: C, 48.61; H, 4.48; N, 25.16; O, 21.55.
Compound 6b. Methyl 2-(8-methyl-6-oxo-1,6-dihydro-9H-purin-9-yl)propanoate. 1H-NMR (400 MHz, DMSO-d6, ppm): δ 8.159 (s, 1H, CH–Ar), 4.714–4.697 (m, 1H, CH), 3.652 (s, 3H, O–CH3), 2.265 (s, 3H, CH3), 1.506 (d, J = 7.2 Hz, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 169.21 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 163.66 (C), 156.83 (C), 145.26 (C), 136.91 (C), 111.34 (C), 54.79 (O–CH3), 50.28 (CH), 15.96 (CH3), 13.40 (CH3). MS (ESI): m/z (M + H) +237.23. Elemental analysis for C10H12N4O3 calculated: C, 50.84; H, 5.12; N, 23.72; O, 20.32. Found: C, 50.79; H, 5.06; N, 23.67; O, 20.27.
O), 163.66 (C), 156.83 (C), 145.26 (C), 136.91 (C), 111.34 (C), 54.79 (O–CH3), 50.28 (CH), 15.96 (CH3), 13.40 (CH3). MS (ESI): m/z (M + H) +237.23. Elemental analysis for C10H12N4O3 calculated: C, 50.84; H, 5.12; N, 23.72; O, 20.32. Found: C, 50.79; H, 5.06; N, 23.67; O, 20.27.
Compound 6c. Methyl 3-methyl-2-(8-methyl-6-oxo-1,6-dihydro-9H-purin-9-yl)butanoate. 1H-NMR (400 MHz, DMSO-d6, ppm): δ 7.970 (s, 1H, CH–Ar), 4.970 (d, J = 10.0 Hz, 1H, CH), 3.629 (s, 3H, O–CH3), 2.851–2.793 (m, 1H, CH), 2.268 (s, 3H, CH3), 1.120 (d, J = 6.8 Hz, 3H, CH3), 0.744 (d, J = 6.8 Hz, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 175.20 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 163.69 (C), 145.67 (C), 138.57 (C), 136.98 (C), 111.13 (C), 63.61 (CH), 52.92 (O–CH3), 29.96 (CH), 17.52 (CH3), 16.57 (CH3), 13.88 (CH3). MS (ESI): m/z (M + H) +265.29. Elemental analysis for C12H16N4O3 calculated: C, 54.54; H, 6.10; N, 21.20; O, 18.16. Found: C, 54.49; H, 6.05; N, 21.16; O, 18.11.
O), 163.69 (C), 145.67 (C), 138.57 (C), 136.98 (C), 111.13 (C), 63.61 (CH), 52.92 (O–CH3), 29.96 (CH), 17.52 (CH3), 16.57 (CH3), 13.88 (CH3). MS (ESI): m/z (M + H) +265.29. Elemental analysis for C12H16N4O3 calculated: C, 54.54; H, 6.10; N, 21.20; O, 18.16. Found: C, 54.49; H, 6.05; N, 21.16; O, 18.11.
Compound 6d. Methyl 3-hydroxy-2-(8-methyl-6-oxo-1,6-dihydro-9H-purin-9-yl)propanoate. 1H-NMR (400 MHz, DMSO-d6, ppm): δ 8.090 (s, 1H, CH–Ar), 5.473–5.092 (m, 2H, CH2), 3.689 (s, 3H, O–CH3), 3.666–3.615 (m, 1H, CH), 2.141 (s, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 168.07 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 162.93 (C), 150.65 (C), 146.34 (C), 143.51 (C), 115.97 (C), 63.38 (CH), 61.55 (CH2), 52.59 (O–CH3), 13.11 (CH3). MS (ESI): m/z (M + H) +253.23. Elemental analysis for C10H12N4O4 calculated: C, 47.62; H, 4.80; N, 22.21; O, 25.37. Found: C, 47.57; H, 4.76; N, 22.18; O, 25.32.
O), 162.93 (C), 150.65 (C), 146.34 (C), 143.51 (C), 115.97 (C), 63.38 (CH), 61.55 (CH2), 52.59 (O–CH3), 13.11 (CH3). MS (ESI): m/z (M + H) +253.23. Elemental analysis for C10H12N4O4 calculated: C, 47.62; H, 4.80; N, 22.21; O, 25.37. Found: C, 47.57; H, 4.76; N, 22.18; O, 25.32.
Compound 6e. Methyl 2-(8-methyl-6-oxo-1,6-dihydro-9H-purin-9-yl)-3-phenylpropanoate. 1H-NMR (400 MHz, DMSO-d6, ppm): δ 8.147 (s, 1H, CH–Ar), 7.223–7.197 (m, 3H, CH–Ar), 7.030–7.006 (m, 2H, CH–Ar), 5.068 (t, J = 4.8 Hz, 1H, CH), 3.689 (s, 3H, O–CH3), 3.238 (dd, J = 6.2, 13.6 Hz, 2H, CH2), 2.239 (s, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 175.70 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 164.18 (C), 147.50 (C), 143.35 (C), 137.09 (C), 135.10 (C), 129.86 (C–Ar ×2), 128.82 (C–Ar ×2), 127.58 (C–Ar), 111.35 (C), 59.98 (CH), 53.16 (O–CH3), 35.30 (CH2), 13.72 (CH3). MS (ESI): m/z (M + H) +314.33. Elemental analysis for C16H16N4O3 calculated: C, 61.53; H, 5.16; N, 17.94; O, 15.37. Found: C, 61.51; H, 5.12; N, 17.89; O, 15.34.
O), 164.18 (C), 147.50 (C), 143.35 (C), 137.09 (C), 135.10 (C), 129.86 (C–Ar ×2), 128.82 (C–Ar ×2), 127.58 (C–Ar), 111.35 (C), 59.98 (CH), 53.16 (O–CH3), 35.30 (CH2), 13.72 (CH3). MS (ESI): m/z (M + H) +314.33. Elemental analysis for C16H16N4O3 calculated: C, 61.53; H, 5.16; N, 17.94; O, 15.37. Found: C, 61.51; H, 5.12; N, 17.89; O, 15.34.
Compound 6f. Methyl 3-(4-hydroxyphenyl)-2-(8-methyl-6-oxo-1,6-dihydro-9H-purin-9-yl)propanoate. 1H-NMR (400 MHz, DMSO-d6, ppm): δ 8.142 (s, 1H, CH–Ar), 6.789 (d, J = 8.4 Hz, 2H, CH–Ar), 6.576 (d, J = 8.4 Hz, 2H, CH–Ar), 4.962 (t, J = 4.8 Hz, 1H, CH), 3.724 (s, 3H, O–CH3), 3.094 (dd, J = 4.4, 10.4 Hz, 2H, CH2), 2.211 (s, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 175.71 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 163.64 (C), 156.81 (C), 149.99 (C), 144.34 (C), 138.70 (C), 137.05 (C–Ar), 130.80 (C–Ar ×2), 124.93 (C–Ar ×2), 115.62 (C), 60.25 (CH), 54.42 (O–CH3), 34.56 (CH2), 13.72 (CH3). MS (ESI): m/z (M + H) +329.33. Elemental analysis for C16H16N4O3 calculated: C, 61.53; H, 5.16; N, 17.94; O, 15.37. Found: C, 61.48; H, 5.11; N, 17.89; O, 15.32.
O), 163.64 (C), 156.81 (C), 149.99 (C), 144.34 (C), 138.70 (C), 137.05 (C–Ar), 130.80 (C–Ar ×2), 124.93 (C–Ar ×2), 115.62 (C), 60.25 (CH), 54.42 (O–CH3), 34.56 (CH2), 13.72 (CH3). MS (ESI): m/z (M + H) +329.33. Elemental analysis for C16H16N4O3 calculated: C, 61.53; H, 5.16; N, 17.94; O, 15.37. Found: C, 61.48; H, 5.11; N, 17.89; O, 15.32.
Compound 7a. 2-(8-Methyl-6-oxo-1,6-dihydro-9H-purin-9-yl)acetic acid. 1H-NMR (400 MHz, D2O, ppm): δ 7.944 (s, 1H, CH–Ar), 4.556 (s, 2H, CH2), 2.325 (s, 3H, CH3). 13C-NMR (100 MHz, D2O, ppm): δ 174.45 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 166.79 (C), 152.78 (C), 150.80 (C), 149.80 (C), 121.56 (C), 45.46 (CH2), 12.61 (CH3). MS (ESI): m/z (M + H) +209.18. Elemental analysis for C8H8N4O3 calculated: C, 46.16; H, 3.87; N, 26.91; O, 23.06. Found: C, 46.11; H, 3.82; N, 26.86; O, 23.00.
O), 166.79 (C), 152.78 (C), 150.80 (C), 149.80 (C), 121.56 (C), 45.46 (CH2), 12.61 (CH3). MS (ESI): m/z (M + H) +209.18. Elemental analysis for C8H8N4O3 calculated: C, 46.16; H, 3.87; N, 26.91; O, 23.06. Found: C, 46.11; H, 3.82; N, 26.86; O, 23.00.
Compound 7b. 2-(8-Methyl-6-oxo-1,6-dihydro-9H-purin-9-yl)propanoic acid. 1H-NMR (400 MHz, D2O, ppm): δ 8.326 (s, 1H, CH–Ar), 5.034–4.978 (m, 1H, CH), 2.180 (s, 3H, CH3), 1.386 (s, 3H, CH3). 13C-NMR (100 MHz, MeOD, ppm): δ 169.10 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 160.03 (C), 155.38 (C), 151.92 (C), 137.16 (C), 109.13 (C), 51.58 (CH), 15.11 (CH3), 11.94 (CH3). MS (ESI): m/z (M + H) +223.20. Elemental analysis for C9H10N4O3 calculated: C, 48.65; H, 4.54; N, 25.21; O, 21.60. Found: C, 48.61; H, 4.49; N, 25.17; O, 21.54.
O), 160.03 (C), 155.38 (C), 151.92 (C), 137.16 (C), 109.13 (C), 51.58 (CH), 15.11 (CH3), 11.94 (CH3). MS (ESI): m/z (M + H) +223.20. Elemental analysis for C9H10N4O3 calculated: C, 48.65; H, 4.54; N, 25.21; O, 21.60. Found: C, 48.61; H, 4.49; N, 25.17; O, 21.54.
Compound 7c. 3-Methyl-2-(8-methyl-6-oxo-1,6-dihydro-9H-purin-9-yl)butanoic acid. 1H-NMR (400 MHz, D2O, ppm): δ 7.857 (s, 1H, CH–Ar), 4.401 (d, J = 10.8 Hz, 1H, CH), 2.236–2.202 (m, 1H, CH), 2.087 (s, 3H, CH3), 0.786 (d, J = 6.8 Hz, 3H, CH3), 0.614 (d, J = 6.8 Hz, 3H, CH3). 13C-NMR (100 MHz, D2O, ppm): δ 175.56 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 167.15 (C
O), 167.15 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 152.73 (C), 149.48 (C), 139.90 (C), 108.48 (C), 65.44 (CH), 29.60 (CH), 16.99 (CH3), 16.20 (CH3), 13.29 (CH3). MS (ESI): m/z (M + H) +251.26. Elemental analysis for C11H14N4O3 calculated: C, 52.79; H, 5.64; N, 22.39; O, 19.18. Found: C, 52.73; H, 5.58; N, 22.33; O, 19.13.
O), 152.73 (C), 149.48 (C), 139.90 (C), 108.48 (C), 65.44 (CH), 29.60 (CH), 16.99 (CH3), 16.20 (CH3), 13.29 (CH3). MS (ESI): m/z (M + H) +251.26. Elemental analysis for C11H14N4O3 calculated: C, 52.79; H, 5.64; N, 22.39; O, 19.18. Found: C, 52.73; H, 5.58; N, 22.33; O, 19.13.
Compound 7d. 3-Hydroxy-2-(8-methyl-6-oxo-1,6-dihydro-9H-purin-9-yl)propanoic acid. 1H-NMR (400 MHz, CD3OD, ppm): δ 8.563 (s, 1H, CH–Ar), 5.422–5.404 (m, 2H, CH2), 3.625–3.500 (m, 1H, CH), 1.190 (s, 3H, CH3). 13C-NMR (100 MHz, CD3OD, ppm): δ 165.54 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 155.45 (C), 149.92 (C), 145.68 (C), 142.10 (C), 116.06 (C), 65.92 (CH), 62.67 (CH2), 14.65 (CH3). MS (ESI): m/z (M + H) +239.07. Elemental analysis for C9H10N4O4 calculated: C, 45.38; H, 4.23; N, 23.52; O, 26.87. Found: C, 45.32; H, 4.19; N, 23.47; O, 26.82.
O), 155.45 (C), 149.92 (C), 145.68 (C), 142.10 (C), 116.06 (C), 65.92 (CH), 62.67 (CH2), 14.65 (CH3). MS (ESI): m/z (M + H) +239.07. Elemental analysis for C9H10N4O4 calculated: C, 45.38; H, 4.23; N, 23.52; O, 26.87. Found: C, 45.32; H, 4.19; N, 23.47; O, 26.82.
Compound 7e. 2-(8-Methyl-6-oxo-1,6-dihydro-9H-purin-9-yl)-3-phenylpropanoic acid. 1H-NMR (400 MHz, CD3OD, ppm): δ 8.557 (s, 1H, CH–Ar), 7.183–7.120 (m, 5H, CH–Ar), 4.620 (t, J = 4.8 Hz, 1H, CH), 3.489 (dd, J = 14.4, 13.6 Hz, H, CH2), 2.146 (s, 3H, CH3). 13C-NMR (100 MHz, CD3OD, ppm): δ 174.11 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 166.77 (C), 155.80 (C), 147.79 (C), 137.50 (C), 135.78 (C), 129.15 (C–Ar ×2), 127.98 (C–Ar ×2), 126.54 (C–Ar), 109.04 (C), 66.73 (CH), 35.65 (CH2), 12.28 (CH3). MS (ESI): m/z (M + H) +299.30. Elemental analysis for C15H14N4O3 calculated: C, 60.40; H, 4.73; N, 18.78; O, 16.09. Found: C, 60.35; H, 4.68; N, 18.71; O, 16.01.
O), 166.77 (C), 155.80 (C), 147.79 (C), 137.50 (C), 135.78 (C), 129.15 (C–Ar ×2), 127.98 (C–Ar ×2), 126.54 (C–Ar), 109.04 (C), 66.73 (CH), 35.65 (CH2), 12.28 (CH3). MS (ESI): m/z (M + H) +299.30. Elemental analysis for C15H14N4O3 calculated: C, 60.40; H, 4.73; N, 18.78; O, 16.09. Found: C, 60.35; H, 4.68; N, 18.71; O, 16.01.
Compound 7f. 3-(4-Hydroxyphenyl)-2-(8-methyl-6-oxo-1,6-dihydro-9H-purin-9-yl)propanoic acid. 1H-NMR (400 MHz, D2O, ppm): δ 8.349 (s, 1H, CH–Ar), 6.640 (d, J = 8.0 Hz, 2H, CH–Ar), 6.325 (d, J = 8.0 Hz, 2H, CH–Ar), 5.150–5.145 (m, 1H, CH), 3.225 (dd, J = 4.4, 14.8 Hz, 2H, CH2), 2.014 (s, 3H, CH3). 13C-NMR (100 MHz, CD3OD, ppm): δ 174.77 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 169.08 (C), 151.65 (C), 148.17 (C), 137.93 (C), 129.59 (C–Ar ×2), 128.89 (C–Ar), 120.59 (C), 118.55 (C–Ar ×2), 108.94 (C), 63.40 (CH), 36.14 (CH2), 12.38 (CH3). MS (ESI): m/z (M + H) +315.30. Elemental analysis for C15H14N4O4 calculated: C, 57.32; H, 4.49; N, 17.83; O, 20.36. Found: C, 57.27; H, 4.43; N, 17.77; O, 20.31.
O), 169.08 (C), 151.65 (C), 148.17 (C), 137.93 (C), 129.59 (C–Ar ×2), 128.89 (C–Ar), 120.59 (C), 118.55 (C–Ar ×2), 108.94 (C), 63.40 (CH), 36.14 (CH2), 12.38 (CH3). MS (ESI): m/z (M + H) +315.30. Elemental analysis for C15H14N4O4 calculated: C, 57.32; H, 4.49; N, 17.83; O, 20.36. Found: C, 57.27; H, 4.43; N, 17.77; O, 20.31.
Cell cultures
A 549 (human lung epithelial carcinoma) cell lines were grown in DMEM 1640 medium supplemented with 10% fetal calf serum (FCS); glutamine 0.3 mg mL−1; penicillin 100 U mL−1 and streptomycin 100 mg mL−1. Cell viability was estimated by trypan blue (0.02%) exclusion. All reagents were purchased from Invitrogen (Milan, Italy).
Cell toxicity assays
The cytotoxicity of the compounds was evaluated by the inhibition of MTT reduction into formazan and by the trypan blue staining assay.39 Briefly, in the MTT assay, A549 cells were seeded in 96-well plates at a density of 2 × 104 cells per well in 100 μL of complete DMEM without phenol red for 24 h at 37 °C. Subsequently, cell monolayers were treated or not with increasing concentrations (range 5–80 μg mL−1) of compounds for 24 h at 37 °C. After 24 h, 10 μL of MTT solution (5 mg mL−1) were added to each well for 3–4 h at 37 °C. Each sample was then acidified by adding 0.1 N HCl in isopropanol (100 μL per well) for 30 min under mild agitation to ensure dissolution of all formazan crystals. Absorbance of samples was read at 570 nm, using an automatic plate reader (Multiskan EX, Ascent Software, Thermo Fisher Scientific). Untreated cells were used as control. The 50% cytotoxic concentration (CC50) was defined as the compound concentration required to reduce cell viability by 50% and calculated by regression analysis considering untreated cells as control (100%).
Virus production and infection
Influenza A/Puerto Rico/8/34 H1N1 (PR8) virus was grown in the allantoic cavities of 10 day-old embryonated chicken eggs. After 48 h at 37 °C, the allantoic fluid was harvested, centrifuged at 5000 rpm for 30 min to remove cellular debris, and stored at −80 °C. Virus titration was performed by Tissue Culture Infectious Dose 50% (TCID50%). Confluent monolayers of A549 epithelial cells were challenged for 1 h at 37 °C with PR8 at a multiplicity of infection (MOI) of 0.001 (TCID50%/cell) incubated for 1 h at 37 °C, washed with PBS, and then incubated with medium supplemented with 2% FCS. Mock infection was performed with the same dilution of allantoic fluid from uninfected eggs.40
In cell western (ICW) assay
The ICW assay was performed using the Odyssey Imaging System (LI-COR, Lincoln, NE, USA) as previously described.41,42 Briefly, A549 cells grown in 96-well plates (2 × 104 cells per well) infected with PR8, and treated or not with high concentrations (100, 150 and 200 μg mL−1) of selected compounds were fixed with 4% formaldehyde, washed, permeabilized with 0.1% Triton X-100 and incubated with Odyssey Blocking Buffer for 1 h (LI-COR Biosciences, Lincoln, NE, USA). The cells were then stained at 4 °C overnight with mouse anti HA (Santa Cruz Biotechnology, Germany) diluted in Odyssey Blocking Buffer. Cells were then washed and stained with a fluorochrome-conjugated secondary antibody (fluorescence emission at 800 nm) (LI-COR Biosciences, Lincoln, NE, USA) properly diluted in Odyssey Blocking Buffer. Subsequently, three washes with PBS plus 0.1% Tween 20 were performed and plates were analyzed by the Odyssey infrared imaging system (LI-COR). The relative fluorescence unit (RFU) was expressed as a percentage compared to untreated infected cells (100%). The 50% infectious dose (IC50) was defined as the dose of compound required to reduce viral replication by 50%. The Selectivity Index (SI) of each compound was calculated as the ratio between CC50 and IC50 (SI = CC50/IC50).
Conclusions
Aminomalononitrile inspired prebiotic chemistry furnished an efficient synthetic tool for the synthesis of a large panel of imidazole and purine derivatives decorated with a natural amino acid side-chain motif. These products resemble the first heterocycle derivatives possibly produced by an original multicomponent reaction involving well-recognized prebiotic precursors, such as HCN oligomers and amino acids. The annulation of amino-imidazole-carbonitrile derivatives into the corresponding purine analogues generally reduced the activity against influenza A/Puerto Rico/8/34 H1N1 (PR8) virus, suggesting the key role of the imidazole scaffold. The derivatives bearing a free carboxylic moiety in the amino acid residue (compounds 5 and 7) showed an IC50 value of the same order of magnitude than the corresponding esters (compounds 4 and 6). The comparable antiviral activity between imidazole derivatives bearing a free or a protected carboxyl moieties has been previously reported in the case of HSV.43 Derivatives decorated with the valine and phenylalanine structural motif showed the higher activity in both AIC and APA series, compounds 4b, 4e, 5a, 5e, 6c, 6f and 7a being characterized by the highest IC50 value (Table 3, entries 2, 3, 9, 10, 14, 17, 23). Among the AIC family, the presence of the phenylalanine residue produced the most active derivatives, namely compounds 4e and 5e (IC50 values of 0.39 μM and 0.46 μM), respectively. They also showed the highest selectivity index (2.1 and 7.2, respectively). The valine derivatives 6c and 7c were the most active compounds in the APA series, showing IC50 value of 0.18 μM and 0.59 μM, respectively, and SI value of 1.7 and 1.8, respectively. Compounds 4b, 4e, 5a, 5e, 6c, 6f and 7a showed an antiviral activity against influenza A/Puerto Rico/8/34 H1N1 (PR8) virus higher than standard antiviral agents (oseltamivir phosphate, ribavirin and amantadine) and a selectivity index comparable to ribavirin (5.84) and higher than amantadine.44 In addition, compounds 5a and 7c decreased the expression of viral HA on infected monolayers, suggesting the occurrence of inhibition of the viral budding from the cell, thus preventing the viral release from infected cells and the spreading to other cells. This hypothesis is in accordance with previous data on the capacity of imidazole derivatives to block the release of virions from infected cells.45
The peptide nucleic acid building blocks we synthesized are also relevant to prebiotic research, suggesting novel structural motifs to be considered in pristine molecular evolution processes.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
This work is supported by the Italian Space Agency (ASI) DC-VUM-2017-034 contratto ASI N. 2019-3-U.0, CUP F86C16000000006 “Vita nello spazio – Origine, presenza, persistenza della vita nello spazio, dalle molecole agli estremofili” and MIUR Ministero dell'Istruzione, dell'Università e della Ricerca Italiano, project PRIN 2017, ORIGINALE CHEMIAE in Antiviral Strategy – Origin and Modernization of Multi-Component Chemistry as a Source of Innovative Broad Spectrum Antiviral Strategy, cod. 2017BMK8JR.
References
- R. C. Cioc, E. Ruijtera and R. V. A. Orru, Green Chem., 2014, 16, 2958 RSC.
- B. Shirt-Ediss, S. Murillo-Sànchez and K. Ruiz-Mirazo, Beilstein J. Org. Chem., 2017, 13, 1388–1395 CrossRef CAS PubMed.
- K. Ruiz-Mirazo, C. Briones and A. De La Escosura, Chem. Rev., 2014, 114, 285–366 CrossRef CAS PubMed.
- R. F. Socha, A. H. Weiss and M. M. Sakharov, J. Catal., 1981, 67, 207–217 CrossRef CAS.
- J. Oró, Nature, 1961, 191, 1193–1194 CrossRef PubMed.
- R. Saladino, G. Botta, S. Pino, G. Costanzo and E. Di Mauro, Chem. Soc. Rev., 2012, 41, 5526–5565 RSC.
- L. Botta, B. M. Bizzarri, D. Piccinino, T. Fornaro, J. Robert Brucato and R. Saladino, Eur. Phys. J. Plus, 2017, 132, 317 CrossRef.
- L. Botta, R. Saladino, B. M. Bizzarri, B. Cobucci-Ponzano, R. Iacono, R. Avino, S. Caliro, A. Carandente, F. Lorenzini, A. Tortora, E. Di Mauro and M. Moracci, Adv. Space Res., 2018, 62, 2372–2379 CrossRef CAS.
- R. Saladino, B. M. Bizzarri, L. Botta, J. Šponer, J. E. Šponer, T. Georgelin, M. Jaber, B. Rigaud, M. Kapralov, G. N. Timoshenko, A. Rozanov, E. Krasavin, A. M. Timperio and E. Di Mauro, Sci. Rep., 2017, 7, 14709 CrossRef PubMed.
- B. M. Bizzarri, L. Botta, M. I. Pérez-Valverde, R. Saladino, E. Di Mauro and J. M. García-Ruiz, Chem.–Eur. J., 2018, 24, 8126–8132 CrossRef PubMed.
- R. Saladino, G. Botta, B. M. Bizzarri, E. Di Mauro and J. M. Garcia Ruiz, Biochemistry, 2016, 55, 2806–2811 CrossRef CAS PubMed.
- B. M. Bizzarri, R. Saladino, I. Delfino, J. M. García-Ruiz and E. Di Mauro, Int. J. Mol. Sci., 2021, 22, 1–12 Search PubMed.
- B. M. Bizzarri, P. Manini, V. Lino, M. d'Ischia, M. Kapralov, E. Krasavin, K. Mráziková, J. Šponer, J. E. Šponer, E. Di Mauro and R. Saladino, Chem.–Eur. J., 2020, 26, 14919–14928 CrossRef CAS PubMed.
- P. E. Nielsen, M. Egholm, R. H. Berg and O. Buchardt, Science, 1991, 254, 1497–1500 CrossRef CAS PubMed.
- R. Saladino, C. Crestini, F. Ciciriello, G. Costanzo and E. Di Mauro, Chem. Biodiversity, 2007, 4, 694–720 CrossRef CAS PubMed.
- S. Freeman and J. M. Gardiner, Mol. Biotechnol., 1996, 5, 125–137 CrossRef CAS PubMed.
- B. Ndeboko, O. Hantz, G. J. Lemamy and L. Cova, Biomolecules, 2018, 8, 55 CrossRef PubMed.
- B. Friedland, J.–R. Soc. Health, 1991, 111, 170–171 CAS.
- D. W. Green, G. S. Watson, J. A. Watson, D. J. Lee, J. M. Lee and H. S. Jung, Acta Biomater., 2016, 42, 33–45 CrossRef CAS PubMed.
- F. Raulin, F. Fonsalas and M. Wolny, Origins Life, 1984, 14, 151–156 CrossRef CAS.
- S. Nandi, D. Bhattacharyya and A. Anoop, Chem.–Eur. J., 2018, 24, 4885–4894 CrossRef CAS.
- J. P. Ferris and W. J. Hagan Jr, Tetrahedron, 1984, 40, 1093–1120 CrossRef CAS.
- H. L. Barks, R. Buckley, G. A. Grieves, E. Di Mauro, N. V. Hud and T. M. Orlando, ChemBioChem, 2010, 11, 1240–1243 CrossRef CAS.
- N. Jazi, S. Fozooni and H. Hamidian, J. Chem. Res., 2018, 42, 80–85 CrossRef CAS.
- A. V. Galochkina, K. R. Bollikanda, V. V. Zarubaev, D. G. Tentler, I. N. Lavrenteva, A. V. Slita, N. Chirra and S. Kantevar, Arch. Pharm. Chem. Life Sci., 2019, 352, 1800225 CrossRef PubMed.
- J. Dong, S. Chen, R. Li, W. Cui, H. Jiang, Y. Ling, Z. Yang and W. Hu, Eur. J. Med. Chem., 2016, 108, 605–615 CrossRef CAS PubMed.
- T. M. A. Eldebss, A. M. Farag, M. M. Abdulla and R. K. Arafa, Med. Chem., 2016, 16, 67–83 CAS.
- M. Bollier, F. Klupsch, P. Six, L. Dubuquoy, N. Azaroual, R. Millet and N. Leleu-Chavain, J. Org. Chem., 2018, 83, 422–430 CrossRef CAS PubMed.
- B. M. A. G. Piette and J. G. Heddle, Trends Ecol. Evol., 2020, 35, 397–406 CrossRef PubMed.
- P. K. Martin, H. R. Matthews, H. Rapoport and G. Thyagarajan, J. Org. Chem., 1968, 33, 3758–3761 CrossRef CAS.
- A. A. Watson, J. Org. Chem., 1977, 42, 1610–1612 CrossRef CAS PubMed.
- E. Mohammadi, L. Petera, H. Saeidfirozeh, H. A. Knížek, P. Kubelík, R. Dudžák, M. Krůs, L. Juha, S. Civiš, R. Coulon, O. Malina, J. Ugolotti, V. Ranc, M. Otyepka, J. Šponer, M. Ferus and J. E. Šponer, Chem.–Eur. J., 2020, 26, 12075–12080 CrossRef CAS PubMed.
- M. d'Ischia, P. Manini, M. Moracci, R. Saladino, V. Ball, H. Thissen, R. A. Evans, C. Puzzarini and V. Barone, Int. J. Mol. Sci., 2020, 20, 4079 CrossRef PubMed.
- W. Y. Lou, M. H. Zong, Y. Y. Liu and J. F. Wang, J. Biotechnol., 2006, 125, 64–74 CrossRef CAS PubMed.
- E. I. Al-Afaleq and S. A. Abubshait, Molecules, 2001, 6, 621–638 CrossRef CAS.
- A. A. Aly and S. A. Nassar, Heteroat. Chem., 2004, 15, 2–8 CrossRef CAS.
- E. Deau, Y. Loidreau, P. Marchand, M. R. Nourrisson, N. Loaec, L. Meijer, V. Levacher and T. Besson, Bioorg. Med. Chem. Lett., 2013, 23, 6784–6788 CrossRef CAS PubMed.
- R. Sgarbanti, D. Amatore, I. Celestino, A. T. Palamara and L. Nencioni, Curr. Top. Med. Chem., 2014, 14, 2529–2541 CrossRef CAS PubMed.
- T. J. Rashamuse, Z. Njengele, E. MabelCoyanis, Y. Sayed, S. Mosebi and M. L. Bode, Eur. J. Med. Chem., 2020, 190, 112111 CrossRef.
- A. Di Sotto, S. Di Giacomo, D. Amatore, M. Locatelli, A. Vitalone, C. Toniolo, G. L. Rotino, R. Lo Scalzo, A. T. Palamara, M. E. Marcocci and L. Nencioni, Molecules, 2018, 23, 2066 CrossRef PubMed.
- M. De Angelis, B. Casciaro, A. Genovese, D. Brancaccio, M. E. Mercocci, E. Novellino, A. Carotenuto, A. T. Palamara, M. L. Mangoni and L. Nencioni, FASEB J., 2021, 35, 1–14 CrossRef PubMed.
- C. Zippilli, L. Botta, B. M. Bizzarri, L. Nencioni, M. De Angelis, V. Protto, G. Giorgi, M. C. Baratto, R. Pogni and R. Saladino, Int. J. Mol. Sci., 2021, 22, 1337 CrossRef CAS PubMed.
- S. Davidson, Front. Immunol., 2018, 9, 1946 CrossRef PubMed.
- M. Kim, J. H. Yim, S.-Y. Kim, H. S. Kim, W. G. Lee, S. J. Kim, P.-S. Kang and C.-K. Lee, Antiviral Res., 2012, 93, 253–259 CrossRef CAS PubMed.
- J. Leis, C. H. Luan, J. E. Audia, S. F. Dunne and C. M. Heath, J. Virol., 2021, 95, e00190 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: 1H-NMR and 13C-NMR of 4a–f, 5a–f, 6a–f and 7a–f. See DOI: 10.1039/d1ra05240c |
|
| This journal is © The Royal Society of Chemistry 2021 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *a,
Angelica Fanellia,
Lorenzo Bottaa,
Marta De Angelisb,
Anna Teresa Palamarabc,
Lucia Nencioni*b and
Raffaele Saladino
*a,
Angelica Fanellia,
Lorenzo Bottaa,
Marta De Angelisb,
Anna Teresa Palamarabc,
Lucia Nencioni*b and
Raffaele Saladino a
a













![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n-hexane (2
n-hexane (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 4a–f from 15 to 60% yield.
1) to afford 4a–f from 15 to 60% yield.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (CH3OH) (5
methanol (CH3OH) (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 6a–f in quantitative yield.
1) to afford 6a–f in quantitative yield.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 148.883 (C), 140.104 (C), 118.080 (C), 88.533 (C), 52.912 (O–CH3), 43.907 (CH2), 13.297 (CH3). MS (ESI): m/z (M + H) +195.19. Elemental analysis for C8H10N4O2 calculated: C, 49.48; H, 5.19; N, 28.85; O, 16.48. Found: C, 49.45; H, 5.16; N, 28.81; O, 16.43.
O), 148.883 (C), 140.104 (C), 118.080 (C), 88.533 (C), 52.912 (O–CH3), 43.907 (CH2), 13.297 (CH3). MS (ESI): m/z (M + H) +195.19. Elemental analysis for C8H10N4O2 calculated: C, 49.48; H, 5.19; N, 28.85; O, 16.48. Found: C, 49.45; H, 5.16; N, 28.81; O, 16.43.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) petroleum ether (2
petroleum ether (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H-NMR (400 MHz, CDCl3, ppm): δ 4.897–4.878 (m, 1H, CH), 4.150 (s, 2H, NH2), 3.847 (s, 3H, O–CH3), 2.336 (s, 3H, CH3), 1.761 (d, J = 7.6 Hz, 3H, CH3). 13C-NMR (100 MHz, CDCl3, ppm): δ 170.573 (C
1). 1H-NMR (400 MHz, CDCl3, ppm): δ 4.897–4.878 (m, 1H, CH), 4.150 (s, 2H, NH2), 3.847 (s, 3H, O–CH3), 2.336 (s, 3H, CH3), 1.761 (d, J = 7.6 Hz, 3H, CH3). 13C-NMR (100 MHz, CDCl3, ppm): δ 170.573 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 145.708 (C), 140.664 (C), 115.587 (C), 96.244 (C), 53.373 (CH), 53.297 (O–CH3), 15.965 (CH3), 14.096 (CH3). MS (ESI): m/z (M + H) +209.22. Elemental analysis for C9H12N4O2 calculated: C, 51.92; H, 5.81; N, 26.91; O, 15.37. Found: C, 51.89; H, 5.77; N, 26.87; O, 15.32.
O), 145.708 (C), 140.664 (C), 115.587 (C), 96.244 (C), 53.373 (CH), 53.297 (O–CH3), 15.965 (CH3), 14.096 (CH3). MS (ESI): m/z (M + H) +209.22. Elemental analysis for C9H12N4O2 calculated: C, 51.92; H, 5.81; N, 26.91; O, 15.37. Found: C, 51.89; H, 5.77; N, 26.87; O, 15.32.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n-hexane (2
n-hexane (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H-NMR (400 MHz, DMSO-d6, ppm): δ 6.042 (s, 2H, NH2), 4.577 (d, J = 10.8 Hz, 1H, CH), 3.705 (s, 3H, O–CH3), 2.597–2.553 (m, 1H, CH), 2.123 (s, 3H, CH3), 1.113 (d, J = 6.4 Hz, 3H, CH3), 0.639 (d, J = 6.4 Hz, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 169.805 (C
1). 1H-NMR (400 MHz, DMSO-d6, ppm): δ 6.042 (s, 2H, NH2), 4.577 (d, J = 10.8 Hz, 1H, CH), 3.705 (s, 3H, O–CH3), 2.597–2.553 (m, 1H, CH), 2.123 (s, 3H, CH3), 1.113 (d, J = 6.4 Hz, 3H, CH3), 0.639 (d, J = 6.4 Hz, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 169.805 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 148.935 (C), 139.722 (C), 117.724 (C), 89.925 (C), 61.883 (CH), 53.198 (O–CH3), 28.372 (CH), 20.673 (CH3), 18.860 (CH3), 14.716 (CH3). MS (ESI): m/z (M + H) +238.28. Elemental analysis for C11H16N4O2 calculated: C, 55.92; H, 6.83; N, 23.71; O, 13.54. Found: C, 55.86; H, 6.78; N, 23.65; O, 13.49.
O), 148.935 (C), 139.722 (C), 117.724 (C), 89.925 (C), 61.883 (CH), 53.198 (O–CH3), 28.372 (CH), 20.673 (CH3), 18.860 (CH3), 14.716 (CH3). MS (ESI): m/z (M + H) +238.28. Elemental analysis for C11H16N4O2 calculated: C, 55.92; H, 6.83; N, 23.71; O, 13.54. Found: C, 55.86; H, 6.78; N, 23.65; O, 13.49.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 148.98 (C), 140.56 (C), 117.99 (C), 90.09 (C), 62.85 (CH2), 59.86 (CH), 53.09 (O–CH3), 14.55 (CH3). MS (ESI): m/z (M + H) +225.22. Elemental analysis for C9H12N4O3 calculated: C, 48.21; H, 5.39; N, 24.99; O, 21.41. Found: C, 48.17; H, 5.35; N, 24.94; O, 21.38.
O), 148.98 (C), 140.56 (C), 117.99 (C), 90.09 (C), 62.85 (CH2), 59.86 (CH), 53.09 (O–CH3), 14.55 (CH3). MS (ESI): m/z (M + H) +225.22. Elemental analysis for C9H12N4O3 calculated: C, 48.21; H, 5.39; N, 24.99; O, 21.41. Found: C, 48.17; H, 5.35; N, 24.94; O, 21.38.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n-hexane (3
n-hexane (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H-NMR (400 MHz, CDCl3, ppm): δ 7.283–7.254 (m, 3H, CH–Ar), 6.995–6.973 (m, 2H, CH–Ar), 4.837–4.799 (dd, J = 4.0, 11.2 Hz, 1H, CH2), 4.323 (s, 2H, NH2), 3.877 (s, 3H, O–CH3), 3.569 (t, J = 12.8 Hz, 1H, CH), 3.385–3.340 (dd, J = 4.4, 13.6 Hz, 1H, CH2), 1.809 (s, 3H, CH3). 13C-NMR (100 MHz, CDCl3, ppm): δ 169.934 (C
1). 1H-NMR (400 MHz, CDCl3, ppm): δ 7.283–7.254 (m, 3H, CH–Ar), 6.995–6.973 (m, 2H, CH–Ar), 4.837–4.799 (dd, J = 4.0, 11.2 Hz, 1H, CH2), 4.323 (s, 2H, NH2), 3.877 (s, 3H, O–CH3), 3.569 (t, J = 12.8 Hz, 1H, CH), 3.385–3.340 (dd, J = 4.4, 13.6 Hz, 1H, CH2), 1.809 (s, 3H, CH3). 13C-NMR (100 MHz, CDCl3, ppm): δ 169.934 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 146.094 (C), 141.435 (C), 135.347 (C), 129.080 (C–Ar ×2), 128.757 (C–Ar ×2), 127.768 (C–Ar), 115.833 (C), 95.757 (C), 60.286 (CH), 53.498 (O–CH3), 35.296 (CH2), 13.456 (CH3). MS (ESI): m/z (M + H) +285.32. Elemental analysis for C15H16N4O2 calculated: C, 63.37; H, 5.67; N, 19.71; O, 11.25. Found: C, 63.31; H, 5.62; N, 19.68; O, 11.20.
O), 146.094 (C), 141.435 (C), 135.347 (C), 129.080 (C–Ar ×2), 128.757 (C–Ar ×2), 127.768 (C–Ar), 115.833 (C), 95.757 (C), 60.286 (CH), 53.498 (O–CH3), 35.296 (CH2), 13.456 (CH3). MS (ESI): m/z (M + H) +285.32. Elemental analysis for C15H16N4O2 calculated: C, 63.37; H, 5.67; N, 19.71; O, 11.25. Found: C, 63.31; H, 5.62; N, 19.68; O, 11.20.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) petroleum ether (4
petroleum ether (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H-NMR (400 MHz, DMSO-d6, ppm): δ 9.249 (s, 1H, OH), 6.846 (d, J = 8.4 Hz, 2H, CH–Ar), 6.593 (d, J = 8.4 Hz, 2H, CH–Ar), 5.700 (s, 2H, NH2), 5.199–5.160 (m, 1H, CH), 3.724 (s, 3H, O–CH3), 3.266–3.219 (m, 2H, CH2), 1.794 (s, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 169.44 (C
1). 1H-NMR (400 MHz, DMSO-d6, ppm): δ 9.249 (s, 1H, OH), 6.846 (d, J = 8.4 Hz, 2H, CH–Ar), 6.593 (d, J = 8.4 Hz, 2H, CH–Ar), 5.700 (s, 2H, NH2), 5.199–5.160 (m, 1H, CH), 3.724 (s, 3H, O–CH3), 3.266–3.219 (m, 2H, CH2), 1.794 (s, 3H, CH3). 13C-NMR (100 MHz, DMSO-d6, ppm): δ 169.44 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 156.59 (C–OH), 148.22 (C), 140.24 (C), 130.41 (C–Ar ×2), 126.71 (C–Ar), 117.83 (C), 115.60 (C–Ar ×2), 90.92 (C), 60.22 (CH), 53.26 (O–CH3), 34.19 (CH2), 14.56 (CH3). MS (ESI): m/z (M + H) +301.32. Elemental analysis for C15H16N4O3 calculated: C, 59.99; H, 5.37; N, 18.66; O, 15.98. Found: C, 59.95; H, 5.29; N, 18.61; O, 15.94.
O), 156.59 (C–OH), 148.22 (C), 140.24 (C), 130.41 (C–Ar ×2), 126.71 (C–Ar), 117.83 (C), 115.60 (C–Ar ×2), 90.92 (C), 60.22 (CH), 53.26 (O–CH3), 34.19 (CH2), 14.56 (CH3). MS (ESI): m/z (M + H) +301.32. Elemental analysis for C15H16N4O3 calculated: C, 59.99; H, 5.37; N, 18.66; O, 15.98. Found: C, 59.95; H, 5.29; N, 18.61; O, 15.94.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 144.50 (C), 141.60 (C), 109.61 (C), 95.61 (C), 45.88 (CH2), 12.00 (CH3). MS (ESI): m/z (M + H) +181.17. Elemental analysis for C7H8N4O2 calculated: C, 46.67; H, 4.48; N, 31.10; O, 17.76. Found: C, 46.62; H, 4.43; N, 31.05; O, 17.71.
O), 144.50 (C), 141.60 (C), 109.61 (C), 95.61 (C), 45.88 (CH2), 12.00 (CH3). MS (ESI): m/z (M + H) +181.17. Elemental analysis for C7H8N4O2 calculated: C, 46.67; H, 4.48; N, 31.10; O, 17.76. Found: C, 46.62; H, 4.43; N, 31.05; O, 17.71.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 142.78 (C), 139.26 (C), 117.28 (C), 93.45 (C), 58.05 (CH), 22.48 (CH3), 14.60 (CH3). MS (ESI): m/z (M + H) +195.19. Elemental analysis for C8H10N4O2 calculated: C, 49.48; H, 5.19; N, 28.85; O, 16.48. Found: C, 49.43; H, 5.15; N, 28.79; O, 16.41.
O), 142.78 (C), 139.26 (C), 117.28 (C), 93.45 (C), 58.05 (CH), 22.48 (CH3), 14.60 (CH3). MS (ESI): m/z (M + H) +195.19. Elemental analysis for C8H10N4O2 calculated: C, 49.48; H, 5.19; N, 28.85; O, 16.48. Found: C, 49.43; H, 5.15; N, 28.79; O, 16.41.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 148.33 (C), 141.22 (C), 117.46 (C), 92.58 (C), 67.34 (CH), 29.76 (CH), 16.76 (CH3), 16.17 (CH3), 13.25 (CH3). MS (ESI): m/z (M + H) +223.25. Elemental analysis for C10H14N4O2 calculated: C, 54.04; H, 6.35; N, 25.21; O, 14.40. Found: C, 53.97; H, 6.31; N, 25.17; O, 14.35.
O), 148.33 (C), 141.22 (C), 117.46 (C), 92.58 (C), 67.34 (CH), 29.76 (CH), 16.76 (CH3), 16.17 (CH3), 13.25 (CH3). MS (ESI): m/z (M + H) +223.25. Elemental analysis for C10H14N4O2 calculated: C, 54.04; H, 6.35; N, 25.21; O, 14.40. Found: C, 53.97; H, 6.31; N, 25.17; O, 14.35.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 145.80 (C), 140.76 (C), 116.44 (C), 94.79 (C), 64.43 (CH2), 61.41 (CH), 12.92 (CH3). MS (ESI): m/z (M + H) +211.19. Elemental analysis for C8H10N4O3 calculated: C, 45.71; H, 4.80; N, 26.66; O, 22.83. Found: C, 45.66; H, 4.75; N, 26.61; O, 22.78.
O), 145.80 (C), 140.76 (C), 116.44 (C), 94.79 (C), 64.43 (CH2), 61.41 (CH), 12.92 (CH3). MS (ESI): m/z (M + H) +211.19. Elemental analysis for C8H10N4O3 calculated: C, 45.71; H, 4.80; N, 26.66; O, 22.83. Found: C, 45.66; H, 4.75; N, 26.61; O, 22.78.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 154.94 (C), 139.62 (C), 134.76 (C), 129.24 (C–Ar ×2), 128.38 (C–Ar ×2), 127.08 (C–Ar), 108.66 (C), 95.90 (C), 60.23 (CH), 34.70 (CH2), 12.80 (CH3). MS (ESI): m/z (M + H) +271.29. Elemental analysis for C14H14N4O2 calculated: C, 62.21; H, 5.22; N, 20.73; O, 11.84. Found: C, 62.16; H, 5.28; N, 20.69; O, 11.79.
O), 154.94 (C), 139.62 (C), 134.76 (C), 129.24 (C–Ar ×2), 128.38 (C–Ar ×2), 127.08 (C–Ar), 108.66 (C), 95.90 (C), 60.23 (CH), 34.70 (CH2), 12.80 (CH3). MS (ESI): m/z (M + H) +271.29. Elemental analysis for C14H14N4O2 calculated: C, 62.21; H, 5.22; N, 20.73; O, 11.84. Found: C, 62.16; H, 5.28; N, 20.69; O, 11.79.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 156.01 (C–OH), 147.00 (C), 137.88 (C), 129.61 (C–Ar ×2), 120.47 (C–Ar), 118.54 (C–Ar ×2), 108.95 (C), 95.90 (C), 59.81 (CH), 35.99 (CH2), 12.47 (CH3). MS (ESI): m/z (M + H) +287.29. Elemental analysis for C14H14N4O3 calculated: C, 58.74; H, 4.93; N, 19.57; O, 16.77. Found: C, 58.69; H, 4.88; N, 19.52; O, 16.71.
O), 156.01 (C–OH), 147.00 (C), 137.88 (C), 129.61 (C–Ar ×2), 120.47 (C–Ar), 118.54 (C–Ar ×2), 108.95 (C), 95.90 (C), 59.81 (CH), 35.99 (CH2), 12.47 (CH3). MS (ESI): m/z (M + H) +287.29. Elemental analysis for C14H14N4O3 calculated: C, 58.74; H, 4.93; N, 19.57; O, 16.77. Found: C, 58.69; H, 4.88; N, 19.52; O, 16.71.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 155.96 (C), 151.77 (C), 149.80 (C), 145.45 (C), 122.02 (C), 53.42 (O–CH3), 43.91 (CH2), 12.49 (CH3). MS (ESI): m/z (M + H) +221.20. Elemental analysis for C9H10N4O3 calculated: C, 48.65; H, 4.54; N, 25.21; O, 21.60. Found: C, 48.61; H, 4.48; N, 25.16; O, 21.55.
O), 155.96 (C), 151.77 (C), 149.80 (C), 145.45 (C), 122.02 (C), 53.42 (O–CH3), 43.91 (CH2), 12.49 (CH3). MS (ESI): m/z (M + H) +221.20. Elemental analysis for C9H10N4O3 calculated: C, 48.65; H, 4.54; N, 25.21; O, 21.60. Found: C, 48.61; H, 4.48; N, 25.16; O, 21.55.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 163.66 (C), 156.83 (C), 145.26 (C), 136.91 (C), 111.34 (C), 54.79 (O–CH3), 50.28 (CH), 15.96 (CH3), 13.40 (CH3). MS (ESI): m/z (M + H) +237.23. Elemental analysis for C10H12N4O3 calculated: C, 50.84; H, 5.12; N, 23.72; O, 20.32. Found: C, 50.79; H, 5.06; N, 23.67; O, 20.27.
O), 163.66 (C), 156.83 (C), 145.26 (C), 136.91 (C), 111.34 (C), 54.79 (O–CH3), 50.28 (CH), 15.96 (CH3), 13.40 (CH3). MS (ESI): m/z (M + H) +237.23. Elemental analysis for C10H12N4O3 calculated: C, 50.84; H, 5.12; N, 23.72; O, 20.32. Found: C, 50.79; H, 5.06; N, 23.67; O, 20.27.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 163.69 (C), 145.67 (C), 138.57 (C), 136.98 (C), 111.13 (C), 63.61 (CH), 52.92 (O–CH3), 29.96 (CH), 17.52 (CH3), 16.57 (CH3), 13.88 (CH3). MS (ESI): m/z (M + H) +265.29. Elemental analysis for C12H16N4O3 calculated: C, 54.54; H, 6.10; N, 21.20; O, 18.16. Found: C, 54.49; H, 6.05; N, 21.16; O, 18.11.
O), 163.69 (C), 145.67 (C), 138.57 (C), 136.98 (C), 111.13 (C), 63.61 (CH), 52.92 (O–CH3), 29.96 (CH), 17.52 (CH3), 16.57 (CH3), 13.88 (CH3). MS (ESI): m/z (M + H) +265.29. Elemental analysis for C12H16N4O3 calculated: C, 54.54; H, 6.10; N, 21.20; O, 18.16. Found: C, 54.49; H, 6.05; N, 21.16; O, 18.11.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 162.93 (C), 150.65 (C), 146.34 (C), 143.51 (C), 115.97 (C), 63.38 (CH), 61.55 (CH2), 52.59 (O–CH3), 13.11 (CH3). MS (ESI): m/z (M + H) +253.23. Elemental analysis for C10H12N4O4 calculated: C, 47.62; H, 4.80; N, 22.21; O, 25.37. Found: C, 47.57; H, 4.76; N, 22.18; O, 25.32.
O), 162.93 (C), 150.65 (C), 146.34 (C), 143.51 (C), 115.97 (C), 63.38 (CH), 61.55 (CH2), 52.59 (O–CH3), 13.11 (CH3). MS (ESI): m/z (M + H) +253.23. Elemental analysis for C10H12N4O4 calculated: C, 47.62; H, 4.80; N, 22.21; O, 25.37. Found: C, 47.57; H, 4.76; N, 22.18; O, 25.32.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 164.18 (C), 147.50 (C), 143.35 (C), 137.09 (C), 135.10 (C), 129.86 (C–Ar ×2), 128.82 (C–Ar ×2), 127.58 (C–Ar), 111.35 (C), 59.98 (CH), 53.16 (O–CH3), 35.30 (CH2), 13.72 (CH3). MS (ESI): m/z (M + H) +314.33. Elemental analysis for C16H16N4O3 calculated: C, 61.53; H, 5.16; N, 17.94; O, 15.37. Found: C, 61.51; H, 5.12; N, 17.89; O, 15.34.
O), 164.18 (C), 147.50 (C), 143.35 (C), 137.09 (C), 135.10 (C), 129.86 (C–Ar ×2), 128.82 (C–Ar ×2), 127.58 (C–Ar), 111.35 (C), 59.98 (CH), 53.16 (O–CH3), 35.30 (CH2), 13.72 (CH3). MS (ESI): m/z (M + H) +314.33. Elemental analysis for C16H16N4O3 calculated: C, 61.53; H, 5.16; N, 17.94; O, 15.37. Found: C, 61.51; H, 5.12; N, 17.89; O, 15.34.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 163.64 (C), 156.81 (C), 149.99 (C), 144.34 (C), 138.70 (C), 137.05 (C–Ar), 130.80 (C–Ar ×2), 124.93 (C–Ar ×2), 115.62 (C), 60.25 (CH), 54.42 (O–CH3), 34.56 (CH2), 13.72 (CH3). MS (ESI): m/z (M + H) +329.33. Elemental analysis for C16H16N4O3 calculated: C, 61.53; H, 5.16; N, 17.94; O, 15.37. Found: C, 61.48; H, 5.11; N, 17.89; O, 15.32.
O), 163.64 (C), 156.81 (C), 149.99 (C), 144.34 (C), 138.70 (C), 137.05 (C–Ar), 130.80 (C–Ar ×2), 124.93 (C–Ar ×2), 115.62 (C), 60.25 (CH), 54.42 (O–CH3), 34.56 (CH2), 13.72 (CH3). MS (ESI): m/z (M + H) +329.33. Elemental analysis for C16H16N4O3 calculated: C, 61.53; H, 5.16; N, 17.94; O, 15.37. Found: C, 61.48; H, 5.11; N, 17.89; O, 15.32.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 166.79 (C), 152.78 (C), 150.80 (C), 149.80 (C), 121.56 (C), 45.46 (CH2), 12.61 (CH3). MS (ESI): m/z (M + H) +209.18. Elemental analysis for C8H8N4O3 calculated: C, 46.16; H, 3.87; N, 26.91; O, 23.06. Found: C, 46.11; H, 3.82; N, 26.86; O, 23.00.
O), 166.79 (C), 152.78 (C), 150.80 (C), 149.80 (C), 121.56 (C), 45.46 (CH2), 12.61 (CH3). MS (ESI): m/z (M + H) +209.18. Elemental analysis for C8H8N4O3 calculated: C, 46.16; H, 3.87; N, 26.91; O, 23.06. Found: C, 46.11; H, 3.82; N, 26.86; O, 23.00.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 160.03 (C), 155.38 (C), 151.92 (C), 137.16 (C), 109.13 (C), 51.58 (CH), 15.11 (CH3), 11.94 (CH3). MS (ESI): m/z (M + H) +223.20. Elemental analysis for C9H10N4O3 calculated: C, 48.65; H, 4.54; N, 25.21; O, 21.60. Found: C, 48.61; H, 4.49; N, 25.17; O, 21.54.
O), 160.03 (C), 155.38 (C), 151.92 (C), 137.16 (C), 109.13 (C), 51.58 (CH), 15.11 (CH3), 11.94 (CH3). MS (ESI): m/z (M + H) +223.20. Elemental analysis for C9H10N4O3 calculated: C, 48.65; H, 4.54; N, 25.21; O, 21.60. Found: C, 48.61; H, 4.49; N, 25.17; O, 21.54.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 167.15 (C
O), 167.15 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 152.73 (C), 149.48 (C), 139.90 (C), 108.48 (C), 65.44 (CH), 29.60 (CH), 16.99 (CH3), 16.20 (CH3), 13.29 (CH3). MS (ESI): m/z (M + H) +251.26. Elemental analysis for C11H14N4O3 calculated: C, 52.79; H, 5.64; N, 22.39; O, 19.18. Found: C, 52.73; H, 5.58; N, 22.33; O, 19.13.
O), 152.73 (C), 149.48 (C), 139.90 (C), 108.48 (C), 65.44 (CH), 29.60 (CH), 16.99 (CH3), 16.20 (CH3), 13.29 (CH3). MS (ESI): m/z (M + H) +251.26. Elemental analysis for C11H14N4O3 calculated: C, 52.79; H, 5.64; N, 22.39; O, 19.18. Found: C, 52.73; H, 5.58; N, 22.33; O, 19.13.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 155.45 (C), 149.92 (C), 145.68 (C), 142.10 (C), 116.06 (C), 65.92 (CH), 62.67 (CH2), 14.65 (CH3). MS (ESI): m/z (M + H) +239.07. Elemental analysis for C9H10N4O4 calculated: C, 45.38; H, 4.23; N, 23.52; O, 26.87. Found: C, 45.32; H, 4.19; N, 23.47; O, 26.82.
O), 155.45 (C), 149.92 (C), 145.68 (C), 142.10 (C), 116.06 (C), 65.92 (CH), 62.67 (CH2), 14.65 (CH3). MS (ESI): m/z (M + H) +239.07. Elemental analysis for C9H10N4O4 calculated: C, 45.38; H, 4.23; N, 23.52; O, 26.87. Found: C, 45.32; H, 4.19; N, 23.47; O, 26.82.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 166.77 (C), 155.80 (C), 147.79 (C), 137.50 (C), 135.78 (C), 129.15 (C–Ar ×2), 127.98 (C–Ar ×2), 126.54 (C–Ar), 109.04 (C), 66.73 (CH), 35.65 (CH2), 12.28 (CH3). MS (ESI): m/z (M + H) +299.30. Elemental analysis for C15H14N4O3 calculated: C, 60.40; H, 4.73; N, 18.78; O, 16.09. Found: C, 60.35; H, 4.68; N, 18.71; O, 16.01.
O), 166.77 (C), 155.80 (C), 147.79 (C), 137.50 (C), 135.78 (C), 129.15 (C–Ar ×2), 127.98 (C–Ar ×2), 126.54 (C–Ar), 109.04 (C), 66.73 (CH), 35.65 (CH2), 12.28 (CH3). MS (ESI): m/z (M + H) +299.30. Elemental analysis for C15H14N4O3 calculated: C, 60.40; H, 4.73; N, 18.78; O, 16.09. Found: C, 60.35; H, 4.68; N, 18.71; O, 16.01.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 169.08 (C), 151.65 (C), 148.17 (C), 137.93 (C), 129.59 (C–Ar ×2), 128.89 (C–Ar), 120.59 (C), 118.55 (C–Ar ×2), 108.94 (C), 63.40 (CH), 36.14 (CH2), 12.38 (CH3). MS (ESI): m/z (M + H) +315.30. Elemental analysis for C15H14N4O4 calculated: C, 57.32; H, 4.49; N, 17.83; O, 20.36. Found: C, 57.27; H, 4.43; N, 17.77; O, 20.31.
O), 169.08 (C), 151.65 (C), 148.17 (C), 137.93 (C), 129.59 (C–Ar ×2), 128.89 (C–Ar), 120.59 (C), 118.55 (C–Ar ×2), 108.94 (C), 63.40 (CH), 36.14 (CH2), 12.38 (CH3). MS (ESI): m/z (M + H) +315.30. Elemental analysis for C15H14N4O4 calculated: C, 57.32; H, 4.49; N, 17.83; O, 20.36. Found: C, 57.27; H, 4.43; N, 17.77; O, 20.31.
