Open Access Article
Open Access ArticleArchitecture of a multi-channel and easy-to-make microfluidic paper-based colorimetric device (μPCD) towards selective and sensitive recognition of uric acid by AuNPs: an innovative portable tool for the rapid and low-cost identification of clinically relevant biomolecules†
Fatemeh Farshchiab,
Arezoo Saadatic,
Mohammad Hasanzadeh *bd and
Farzad Seidi
*bd and
Farzad Seidi *a
*a
aJiangsu Co-Innovation Center for Efficient Processing and Utilization of Forest Resources, International Innovation Center for Forest Chemicals and Materials, Nanjing Forestry University, Nanjing 210037, China. E-mail: f_seidi@njfu.edu.cn
bPharmaceutical Analysis Research Center, Tabriz University of Medical Sciences, Tabriz, Iran. E-mail: hasanzadehm@tbzmed.ac.ir
cDrug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
dNutrition Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
First published on 10th August 2021
Abstract
Uric acid (UA) is the end product of purine metabolism. Uric acid is usually excreted in the urine, but its abnormal increase and toxic amount can lead to diseases such as gout, hyperuricemia, Lesch–Nyhan syndrome, and cardiovascular disease. On the other hand, UA reduction can lead to neurodegenerative diseases such as sarcoma, glioblastoma, Hodgkin, and etc. Therefore, rapid identification of UA is of great importance. In this work, a simple, portable, inexpensive, and fast microfluidic paper-based colorimetric sensor based on the color change in the presence of UA by using AuNPs was developed. The results can be easily identified with naked eye and further confirmed by UV-vis spectrophotometry. In this method, iron pattern and fiberglass paper were used to construct diagnostic areas and hydrophilic microfluidic channels. We greatly reduced the preparation time of this pattern using a magnet (about three minutes). In this work, four types of nanoparticles with different lower limit of quantification (LLOQ) were used. Linear range of 10−6 to 10−3 M and LLOQ of 10−6 M were obtained for the determination of uric acid using AuNPs–CysA as optical probe. Also, by AuNPs as optical probe a linear range of 10−4 to 10−2 M and the obtained LLOQ was 10−4 M. Finally, by AuNFs as optical probe linear range from 10−6 to 10−2 M and 5 × 10−5 to 10−2 M along with LLOQ of 10−6 and 5 × 10−5 M, respectively. The designed system successfully studied in human urine samples.
1. Introduction
Uric acid (UA) is an organic compound of the elements carbon, oxygen, nitrogen and hydrogen. UA is the end product of purine nucleoside metabolism. Uric acid is mainly produced in the liver and intestines and is excreted through the kidneys (two thirds) and intestines (one third).1,2 Elevated uric acid can be a sign of diseases such as cancer, alcohol consumption, overeating meat, obesity, or high blood triglycerides. Gout is a complication of elevated uric acid caused by the deposition of uric acid crystals in the joints of the body, especially the joints of the legs.3 Measurement of uric acid is very important in clinical trials. Diagnosis of uric acid is of particular importance for the diagnosis of syndromes as well as its monitoring in patients undergoing chemotherapy or radiation therapy.4,5 The standard concentration of UA in urine is 1.4 to 4.4 mM and in serum 0.24 to 0.52 mM. Uric acid has usually excreted in the urine, but its abnormal increase can lead to diseases such as gout, hyperuricemia, Lesch–Nyhan syndrome, and cardiovascular disease.6 Many of these conditions depend on the diet and lifestyle of patients.7 Therefore, early awareness of excessive uric acid highlights the demand for a cheap and simple system to prevent recurrence by changing the diet. On the other hand, reducing uric acid can lead to neurodegenerative diseases such as sarcoma, glioblastoma, Hodgkin, etc. Therefore, rapid identification of UA is of great importance.8–10 Common methods for identifying UA were based on the uricase spectrometer, which is the enzymatic oxidation of uric acid by oxygen to produce carbon dioxide, allantoin, and hydrogen peroxide.11 Other common methods include chromatography and mass spectrometry.12 These methods have drawbacks such as the high cost and length of sample preparation.13 Also, one of the major drawbacks of these methods is the use of blood serum, which prevents rapid analysis. On the other hand, they require skilled technicians to do the job and require expensive tools.9,14 Therefore, one of the urgent needs in the diagnostic field is the design and construction of devices with the study of biochemical processes in the human body for rapid and timely detection of related diseases.15 A biosensor is a device that uses specific biochemical reactions, usually by specific enzymes, tissues or cells, to electrically detect the chemical elements of the substance, optical, or thermal detection.16 These sensors have designed to react with only a specific substance. The result of this reaction is the messages that a microprocessor can analyze.17 Sensors due to the unique features such as excellent accuracy and sensitivity have been widely used in UA detection.18 Techniques and methods used in the detection of UA include electrochemical methods,19,20 fluorometry,21 chemiluminescence,22 surface Raman spectroscopy and colorimetry.23 One of the best methods is colorimetric methods, which in addition to being simple, are also very cheap and can be detect by naked eye.24 In recent years, several colorimetric biosensors have designed for UA detection.23,25–27 Recently, the integration of advanced biosensors on microfluidic substrates has received much attention.28 Microfluidics system is made up of channels with the size of tens to hundreds of micrometers that can analyze small amounts of liquids. Microfluidics has an excellent structure that allows them to be combined with biosensors and increase their application and capability.29 These sensors can also be made on paper.30,31 Point of care (POC) tests can provide physicians with important and good information, and nowadays these tests are one of the promising points for the use of biosensors.32 This system can be a good tool in the rapid and timely diagnosis of diseases, but the design and construction of new tools face serious challenges.33 One of the major challenges in microfluid design is its small and portable design and the use of a cheap and accessible substrate without damaging the environment. Nowadays, paper-based sensors have attracted the attention of researchers due to their low cost, ease of movement and use, and great biocompatibility.34 These sensors are a great launching pad for the simultaneous detection of multiple analytes.35 Due to its hydrophilic nature, paper transfers liquids through its porous structure.36 Various methods have used to create and construct a specific pattern that has a hydrophobic and hydrophilic region, such as laser therapy, wax printing, screen printing, and poly dimethyl siloxane (PDMS).37,38 One of the best, simplest and cheapest ways to design a pattern and make hydrophobic microchannels on paper is to use liquid paraffin.39 Although there is a lot of studies in the use of wax printing technology in the detection of UA, which has good resistance to most chemical compounds,40 paraffin is inexpensive and more accessible, so paraffin is a better option.41 Paper-based microfluidic analyzers are a new generation of microfluidics that is very simple with high flexibility and can be very suitable as a disposable diagnostic kit.42,43 So far, many analytical techniques have been used in paper sensors, fluorescence, electrochemical detection, electrical conductivity, and colorimetry.44 Martinez et al. proposed a paper-based biosensor using ELISA, which used micro zone paper for measurement.45 Chen et al. designed a microfluidic paper-based colorimetric biosensor that could be detected by the naked eye.46 This technique can be employed in smartphones that have high analysis speed. These methods have complexities such as high sample preparation time and cost and low portability. Among these methods, colorimetric detection had higher sensitivity and selectivity in the manufacture of diagnostic kits, and their commercialization is more cost-effective.47 Some colorimetric methods use single nanoparticles (gold, platinum, silver, etc.). These nanoparticles have excellent physicochemical properties that can improve the performance of biosensors.48 Among nanoparticles, gold nanoparticles (AuNPs) have high optical properties and biocompatibility.49 The target molecule causes the accumulation of AuNPs and due to the accumulation of analyte with this nanoparticle, discoloration occurs.50,51 As the particle size increases, their catalytic properties decrease, reducing the surface-to-volume ratio.52In this work, we designed a simple, portable, inexpensive and fast microfluidic paper-based colorimetric biosensor based on the color change in the presence of target UA. The results can be easily identified with the naked eye and also proved by UV spectroscopy. Recently, UV-based AuNPs spectroscopy methods for sensitive and selective detection of target species in complex matrices have attracted a lot of attention.53,54 AuNPs have a small size, high electron density and excellent catalytic performance, which are the most useful optical properties of fluorescence quenching and surface plasmon intensification (SPR).55 According to previous studies, AuNPs can be very useful in identifying uric acid. Additionally, AuNPs are non-toxic and stable against the light.56 The AuNPs also have peroxidase-like properties that can catalyze TMB (3,3′,5,5′-tetramethylbenzidine) reduction in the presence of H2O2 and increase the sensitivity of the resulting dye.57,58 These nanoparticles have high stability, high surface area and adjustable catalytic activity.59 Uric acid changes color through the accumulation of nanoparticles.60 Also, liquid paraffin and an iron pattern and fiberglass and TLC papers used to make hydrophilic channels. For easy transfer of the designed pattern on the surface of the paper, paraffin-free paper and paraffin-immersed paper placed in the middle of the magnet and the hot iron pattern. By pressing between the iron and magnet pattern, microfluidic hydro-fluid channels created. This method speeds up pattern making (three minutes) on paper and reduces hand pressure. The low requirements of the stamp tool and the disposable of the microfluidic paper-based device make it possible to use for commercial application in clinical samples analysis based on POC. The designed system is very cheap, simple and portable.
The main innovation of this research work is the simultaneous use of several types of gold nanoparticles and showing their strengths and weaknesses. It is important to point out that, there is no report concerning the effect of size, morphology and other physico-chemical properties of AuNPs on the detection of UA in real samples. So, in this study various type of AuNPs were used for the colorimetric detection of UA in real samples. It is important to point out that, there is no comparative study for the evaluation of various type of AuNPs on the detection of UA by TMB/H2O2 reduction process till now. On the other hand, there is no report on the use of paper based microfluid modified by AuNPs for the detection and determination of UA in real samples. Finally, one of the main strengths of this work is the optimization of time and production of very fast and inexpensive paper based microfluidic tool for the on-site detection of UA in biomedical samples using POC.
2. Experimental
2.1. Chemical and reagents
3,3′,5,5′-Tetramethylbenzidine (TMB), hydrogen peroxide, sodium hydroxide, hydrogen chloride, chloroauric acid, cysteamine (CysA), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), potassium carbonate, silver nitrate, sodium citrate, sodium borohydride, cetyltrimethylammonium bromide (CTAB), ascorbic acid. TLC paper, fiberglass paper, white paraffin candle.2.2. Instrumentation
The optical examination was performed by UV-VIS spectrophotometer Shimadzu UV-1800 with a resolution of 1 nm. Energy scattering spectroscopy (EDS) was employed to study chemical elements. High-resolution field emission scanning electron microscope (FE-SEM) Hitachi SU8020, Czech with a 3 kV effectual voltage was employed for considering the morphology of electrode surface. Dynamic light scattering (DLS) for zeta potential and size distribution of synthesized nanoparticles was conducted by the zeta potential instrument Malvern Instruments Ltd (Zetasizer Ver. 7.11, MAL1032660, England). AFM was achieved by Nanosurf (AG Gräubernstrasse 12, 4410 Liestal, Switzerland) device.2.3. Stock solution preparation
A mixture of TMB and methanol was prepared at 0.02 M H2O2 (in a volume ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and added with synthesized gold nanoparticles (in a volume ratio of 1
1) and added with synthesized gold nanoparticles (in a volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The color of the final solution oxidized to bluish-green. For the control solution, NaOH (0.01 N) has prepared with HCl (0.01 N), which treated with the combination. Saturated solution of UA (0.06 M) was prepared in 0.01 (N) NaOH at room temperature. Different concentrations of UA was also prepared using deionized water for further investigation.
1). The color of the final solution oxidized to bluish-green. For the control solution, NaOH (0.01 N) has prepared with HCl (0.01 N), which treated with the combination. Saturated solution of UA (0.06 M) was prepared in 0.01 (N) NaOH at room temperature. Different concentrations of UA was also prepared using deionized water for further investigation.
2.4. Universal process for detection of UA
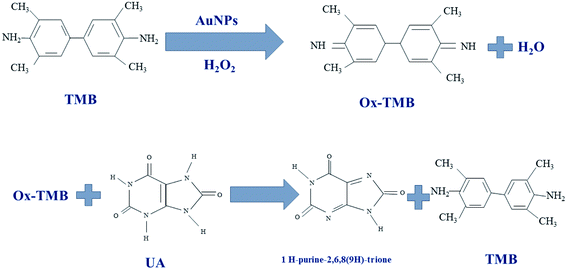
In this work, different gold nanoparticles with different morphologies were used to detect UA and the color redox reactions of TMB used for UA analysis. The TMB solution is initially colorless and oxidizes to bluish-green in the presence of H2O2. The gold nanoparticles exhibits peroxidase-like catalytic activity and their color is changed upon mixing with TMB in the presence of H2O2. After adding UA to the oxidized TMB solution, the TMB was reduced and the colorless solution produced again. Color changes due to TMB redox reaction investigated by UV-Vis spectrophotometry in different solutions (Scheme 1).2.5. Preparation of paper-based microfluidic device
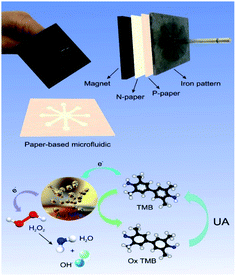
An inexpensive portable microfluidic device was designed and built for colorimetric analysis to examine color changes on paper as well as the effect of concentration on paper. For this purpose, an iron pattern was designed using CorelDRAW software. This pattern has 8 branches that create 8 hydrophilic circular regions that have microfluidic channels 10 mm long and 30 mm wide. These channels end in a central area that has a diameter of 10 mm. The overall size of this pattern is 4 cm. Fiberglass and TLC papers were used to create the pattern. Melted paraffin was used to create a hydrophilic environment and transfer the pattern to the paper. Creating a pattern on paper required a lot of pressure and a long time, so for the first time, a very strong magnet was used to prepare the paper bed in the shortest time (about 3 minutes). In the first stage, the metal pattern is placed at a temperature of 150 °C for 2 minutes, in the second stage, the candle is placed at a temperature of 90 °C until it melts completely and the paper is immersed in it for 30 seconds, in the third stage after drying complete paraffin paper, paraffin-free paper is placed on the magnet and on it is placed paraffin paper and on it is a hot iron pattern and after 15 seconds we separate it. This allows the candle to penetrate the paper texture and create hydrophilic channels on the paper that are also hydrophobic around it. This system was used to detect uric acid and capillary levels in ducts using a variety of gold nanoparticles (Fig. S1 (see ESI†)) (Scheme 2).2.6. Synthesis of nanoparticles (NPs)
Ag-seeds synthesis. In the first step, 156 μL of AgNO3 (0.0163 M) and 250 μL of 0.005 sodium citrate (Na3C6H5O7) were added in 10 ccs of deionized water, respectively. Then 400 μL of cold NaBH4 (0.04 M) added to the solution. The yellow color obtained at a distance. This solution kept in a dark place for 2 hours.
Ag growth solution. To prepare gold nano-stars, 0.364 g of CTAB was added to deionized water and stirred at 30 °C to obtain a clear solution, then the temperature turned off and 60 μL of AgNO3, 100 μL of 50 mM HAuCl4 solution and 100 μL of 0.08 M ascorbic acid solution added at intervals of 1 minute. After 20 seconds of stirring, 50 μL of silver seeds synthesized in the previous step added. The prepared solution was mixed for 15 minutes and then keep at room temperature for 24 hours. The color of the synthesized solution first turns sky blue and then gradually turns brown. After 24 hours, crystals are formed that can be placed at a temperature of 30 degrees and then centrifuged for 15 minutes at 3000 rpm and 5 minutes at 2500 rpm, respectively. The colorless supernatant is separated and used.
3. Characterization
DLS detection from aggregation analysis provides several advantages and limitations and processes higher sensitivity than conventional colorimeter readings. DLS method, zeta potential analysis used for zeta potential and nanocomposite size distribution.61 Also, FE-SEM has been widely used to identify functional nanocomposites in biosensors.62 FE-SEM was used in the present work to study the morphology of the synthesized AuNPs. EDX spectroscopy is an analytical method used to study the chemical properties and initial analysis of a sample.63 Each element has a unique atomic structure, and this principle is important in EDX, as well as the interaction of a sample and the X-ray excitation source. TEM is a microscopic technique in which an electron beam is transmitted through a sample to create an image.64 The image is made of the interaction of the sample and the electrons when the beam is transmitted through the sample. The image is then focused and magnified on an imaging device such as a photographic film layer, a fluorescent screen or a sensor such as a spark plug with a charger. TEM is capable of imaging with significantly higher resolution than light microscopes and is used in several fields of science, such as materials science and nanotechnology. AFM is suitable for observing the structural changes of complex nanostructures in solution and can be said to be the only imaging technique that makes it possible to observe biological compounds without labelling in three-dimensional resolution and at high molecular speed.653.1. Characterization of AuNPs–CysA
As shown in Fig. S2 (see ESI†), FE-SEM imaging was used to evaluate the morphology of AuNPs–CysA. As it turned out, the spherical nanoparticles were evenly distributed. Preliminary EDX analysis has also used to examine the elements. As shown in Fig. S3 (see ESI†), the synthesis was successful, and S also demonstrate the successful binding of cysteamine to AuNP. DLS was used to measure the particle size distribution and charge. The various magnifications obtained from TEM were shown in Fig. S4 (see ESI†). As shown in Fig. S5 (see ESI†), the particle distribution range of nanoparticles is 10–20 nm. Also, after combining AuNPs–CysA and uric acid DLS was performed. According to Fig. S4C,† as it is clear, the size of nanoparticles has increased to more than 1000 nm. According to the results, it is clear that the interaction of uric acid and nanoprobes has been successfully performed. The zeta potential is negative (−25 mV). Saeb et al. have shown that particles with zeta potential values greater than 30 mV or negative more than −30 mV have good stability.66 Also, if the amount of zeta potential increases, the aggregation decreases and the size of the nanoparticles become larger.67 Also, the AFM technique was used for 3D imaging. As shown in Fig. S6 (see ESI†). Spherical and white particles of AuNPs are clearly visible on the surface, so this method is a good way to measure the size of nanoparticles in all three dimensions compared to other methods.3.2. Characterization of AuNFs
According to Fig. S7 (see ESI†), in different synthesis conditions, i.e. different pH, AuNFs have the same structure. In this study, the EDC method also used to determine the percentage and frequency of different elements, which was shown in Fig. S8 (see ESI†). Also, to stabilize the successful synthesis and morphology of nanoparticles, TEM imaging was used at different magnifications, as shown in Fig. S9 (see ESI†), the size of AuNFs is about 40 nm. Based on the obtained results from the DLS technique, although the synthesized AuNFs have the same structure at different pHs, their size was different, as shown in Fig. S10A and B (see ESI†), when the pH was 6.15 the average size of AuNFs was 100 nm while at pH 4.19 the nanoparticle size is 10 nm. It is also shown in Fig. S11C and D† these nanoparticles, after combining with uric acid, are about 100 times larger in size, confirming the interaction of uric acid and nanoparticles. According to Fig. S10E and F (see ESI†), the zeta potential of AuNFs at pH 6.15 was 23.9 mV and at pH 4.19 was 31.9 mV. Therefore, pH plays an important role in the size and zeta potential of nanoparticles. The AFM pictures of AuNFs also show in Fig. S11 (see ESI†). At pH 6.15, the AuNFs provided better images and were spread evenly across the surface, possibly due to the stability of the AuNFs at different pHs.3.3. Characterization of GNSs
Fig. S12 (see ESI†) shows the FE-SEM images of star-shaped AuNPs that have an average size of 100 to 60 nm at different magnifications. Also, in TEM imaging, nanoparticles are observed in a completely stellar shape (Fig. S13 (see ESI†)). For the study of smaller nanoparticles, we also used the DLS technique, which shows the micro to the nanoscale, because since the DLS method depends on the interaction of light with particles, it can be used to evaluate the particle size distribution, especially in the range of 2–500 nm. The particle size distribution showed that the average particle size is 20 nm, and after combining with uric acid, the size of nanoparticles has increased to 430 nm (Fig. S14B (see ESI†)). The zeta potential obtained is 42.6 mV, which indicates that this NPs have good stability. Also, the AFM technique was used for 3D imaging. As shown in Fig. S15 (see ESI†). AuNPs are a white spot peak and are well visible in the figure, so it can be said that this method is a good way to measure the size of nanoparticles in all three dimensions compared to other methods.4. Results and discussion
4.1. UV-Vis spectroscopic measurements
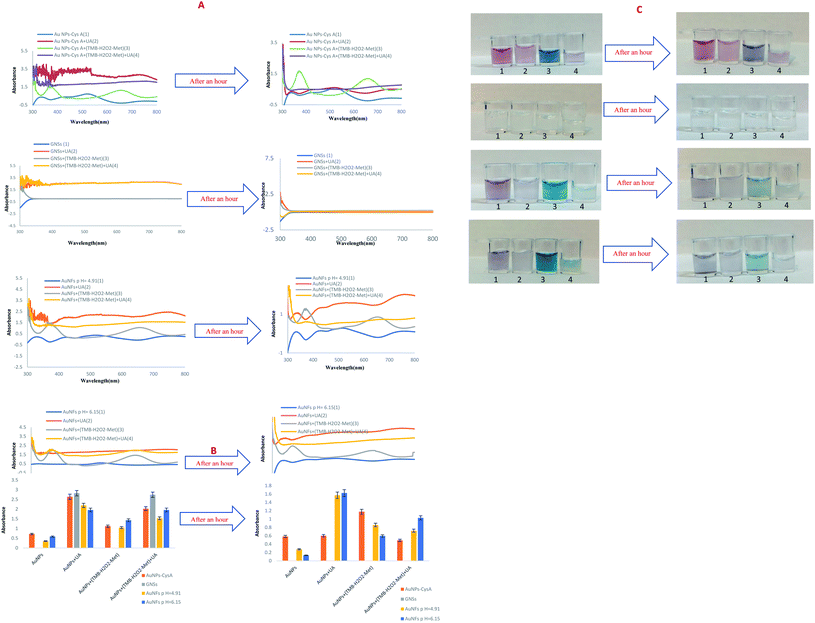
Initially, the UV-Vis method was used to monitor color change for detection in the liquid phase. The UV-Vis sub-forums show the synthesized NPs, the bluish-green TMB solution and the UA solutions. No. 1 contains synthesized NPs, no. 2 is a combination of a mixture of UA and 50 μL of synthesized AuNPs, no. 3 contains 50 μL of bluish-green solution from TMB oxidation and 50 μL of synthesized NPs, no. 4 is a combination of no. 3 and a mixture of NaOH and UA.4.2. Effect of concentration of UA
The synthesized AuNPs examined in the presence of (AuNPs + TMB + H2O2 + methanol) solution and different concentrations of UA (0.000001 to 0.01 M) and the prepared solutions used to measure the UV-Vis spectrum. In these experiments, a colorimetric method performed to detect UA, using the peroxidase-like catalytic activity of AuNPs. As shown in Fig. S16–S19 (see ESI†), as the color intensity of the solution decreases, the absorption intensity also decreases, which is directly visible to the naked eye. There is also a linear relationship between UV-Vis absorption and uric acid concentration. The regression equation has also plotted for each of the nanoparticles, which listed separately in Table S1 (see ESI†) of the range of uric acid and LLOQ.As shown in Table 1, although, AuNFs (pH = 4.91) and AuNPs–CysA have the best LLOQ, which indicates a successful design of colorimetric assay, AuNFs (pH = 4.91) has better performance and linear range (10−6 to 10−2 M). The LLOQ of colorimetric biosensor based on AuNFs at pH = 6.15 for detection of UA was 5 × 10−5 M and the linear range was from 5 × 10−5 to 10−2 M. GNSs has not good presentation and their UV-Vis absorption was near zero. These nanoparticle is not suitable for the preparation of diagnostic kits, because it does not show any color change and other nanoparticles have better performance. Concentrations less than 10−4 M could not be detected and the adsorption intensity was zero at low concentrations. As shown in the images above, the intensity of adsorption also decreases with decreasing concentration. So, in a nutshell, we can say that AuNFs (pH = 4.91) is the most successful nanoparticle in this study (Table 1).
| Analytical technique | Reaction system | Linear range (M) | LLOQ–LOD (M) | Ref. |
|---|---|---|---|---|
| Electrochemical | Nafion/uricase/ferrocene/GCE | 0.5–600 × 10−6 | 230 × 10−12 | 74 |
| Electrochemical | Uricase/Au-rGO/ITO | 50–800 × 10−6 | 7.32 ± 0.21 × 10−6 | 75 |
| Fluorescence/colorimetric | N-CQDs/Ag TNPs-H2O2 | 0.1–45 × 10−6 | 0.05 × 10−6 | 76 |
| Electrochemical | UOx/Fc/Cu2O/GCE | 0.01–1 × 10−3 | 0.0596 × 10−6 | 77 |
| Colorimetry | TMB/g-C3N4/uricase | 10–100 × 10−6 | 8.9 × 10−6 | 78 |
| Chemiluminescence | TCPO–H2O2–rubrene | 10–1000 × 10−6 | 5.0 × 10−6 | 79 |
| Fluorescence/colorimetric | AuNPs–CysA/TMB–H2O2 | 10−3 to 10−6 | 10−6 | This work |
| GNSs/TMB–H2O2 | 10−2 to 10−4 | 10−4 | ||
| AuNFs pH = 4.91/TMB–H2O2 | 10−2 to 10−6 | 10−6 | ||
| AuNFs pH = 6.15/TMB–H2O2 | 10−2 to 5 × 10−5 | 5 × 10−5 |
According to Table 1, the designed system has a high diagnostic limit and is much simpler and cheaper compared to other studies, its detection and preparation time is much lower than other proposed systems. One of the most important features of the designed sensor is its portability, which can be used as a simple and inexpensive diagnostic kit for easier use by patients. Based on the obtained results, the dynamic recognition range is wider than these literature values. So, it is expected the prepared sensor paves a novel way for subsequent works in biological studies, and environmental safety test.
4.3. UV-Vis spectroscopic measurements in real sample
To evaluate the effect of uric acid concentration in the real sample, a healthy urine sample used without any pretreatment. First, urine samples combined with known concentrations of UA. In addition, UV-Vis spectrophotometry performed to calculate its improvement and calculated according to the response current with a standard calibration curve. Except for AuNPs–CysA, which decreases with decreasing adsorption intensity, the rest of the nanoparticles increase with decreasing adsorption intensity, depending on the type of reaction (Fig. S20–S23 (see ESI†)).5. Paper-based colorimetric detection of uric acid
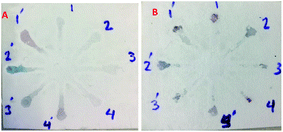
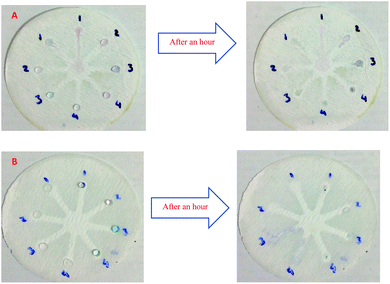
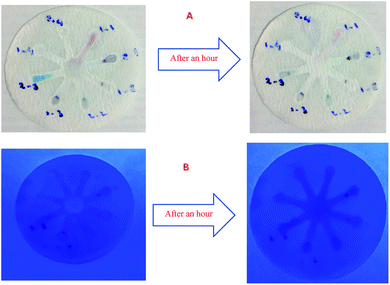
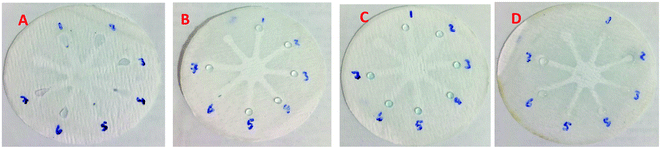
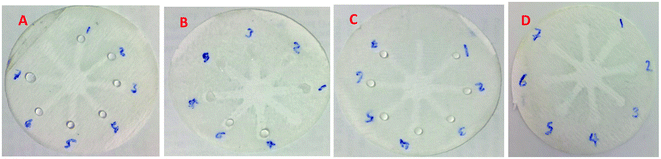
Although UV-Vis spectrophotometric results are very suitable for UA detection, their use is limited and can only be used in laboratories. Paper-based microfluidics can be used to apply and fabricate the kit as a tool to detect and determine the effect of UA concentration change. In recent years various sensors have been designed to increase the efficiency and recognition of UA. Microfluidic paper-based sensors have been designed with unique features. Recently, with the rapid development of microfluidics in various fields such as chemistry and life sciences, their integration with sensors promises a new way of identifying different analytes. They have made significant strides in this area.80 Also, they have great potential in identifying and analyzing various analytes such as UA, which we hope can become a commercial product in the future. Paper-based microfluidic analyzers are a new generation of microfluidics that are very simple and highly flexible. Paper-based microfluidics can be used to apply and fabricate kits as a tool to identify and determine the effect of UA concentration changes in laboratories.30 The prepared paper-based template has micro-channels that have a capillary flow and the liquid can pass through it easily. So, it has a pump that makes it an attractive layer for the detection of UA. On the other hand, paper is very cheap, available, and portable. All this adds to the benefits of the designed biosensor. The prepared microfluidic paper-based biosensor was made in a small size, which has better advantages compared to the old sensors, such as low material consumption and easy sample transfer. Older systems require complex and multi-stage tools that have limited use in some areas. The designed microfluid can analyze several samples simultaneously. In this work, to evaluate the performance of papers, TLC and fiberglass paper were used, the results showed that fiberglass paper has less friction due to better flow. So this type of substrate used to investigate the detection of UA and its determination (Fig. 5 and 6) which recorded one hour later fully confirm this claim. Also, to evaluate the effect of UA concentration in the urine sample on the prepared microfluidic substrate, the prepared concentrations placed on the sensing zones of the prepared paper. The results showed that proposed platform can be used as a colorimetric kit for clinical analysis of UA. To check the capillary power of the microchannel, 10 μL of UA placed in the center of the prepared microfluidic paper-based sensor and the compounds containing nanoparticles and dye solution were placed in the assay area (Fig. 3). According to the results, the channels have good pumping power. Hence, low friction and flow in microchannels are appropriate, and over time, reaction systems and uric acid flow into each other inside microfluid channels (Fig. 4). Therefore, this technique can be used to design and fabricate microfluidic paper-based colorimetric sensor to detect glucose, amino acids, and so on (Fig. 2).6. Conclusion
In this study, we developed a facile, portable, inexpensive and fast microfluidic paper-based colorimetric sensor based on the color change in the presence of target UA. The results can be easily identified with the naked eye. In this research work, different type of AuNPs with various morphologies were used to detection of UA and the color change in the redox reaction of TMB/H2O2 was used for the analysis UA in real samples. Color changes resulting from the redox reaction were investigated by UV-Vis spectrophotometry in various solutions. The AuNPs which used in this work have high stability, high surface area and adjustable catalytic activity. Also, liquid paraffin and an iron pattern and fiberglass and TLC papers were used to make hydrophilic channels. The employed AuNPs have different lower limit of quantification (LLOQ). AuNPs–CysA has a linear range of 10−6 to 10−3 M and the obtained LLOQ was 10−6 M. GNSs have a linear range of 10−4 to 10−2 M and the obtained LLOQ was 10−4 M. AuNFs at pHs of 4.91 and 6.15 have a linear range from 10−6 to 10−2 M, and 10−2 to 5 × 10−5 M, LLOQ 10−6 and 5 × 10−5 M, respectively. The AuNFs with pH = 4.91 showed the best results. Also, GNSs have not shown suitable results. So, it is not a suitable nanoparticle for the preparation of diagnostic kits. The designed system exhibited excellent performance in detection of UA in real samples such as human urine samples. Therefore, this system has high ability to be applied as a powerful kit in clinical laboratories and has great features to be commercialized.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge Tabriz University of Medical Sciences for instrumental supporting of this research.References
- G. Glantzounis, E. Tsimoyiannis, A. Kappas and D. Galaris, Curr. Pharm. Des., 2005, 11, 4145–4151 CrossRef CAS PubMed.
- Y. Y. Sautin and R. J. Johnson, Nucleosides, Nucleotides Nucleic Acids, 2008, 27, 608–619 CrossRef CAS.
- D.-H. Kang, T. Nakagawa, L. Feng, S. Watanabe, L. Han, M. Mazzali, L. Truong, R. Harris and R. J. Johnson, J. Am. Soc. Nephrol., 2002, 13, 2888–2897 CrossRef CAS PubMed.
- N. M. Elsayed, J. M. Nakashima and E. M. Postlethwait, Arch. Biochem. Biophys., 1993, 302, 228–232 CrossRef CAS.
- R. Archibald, Clin. Chem., 1957, 3, 102–105 CrossRef CAS.
- A. Leiba, S. Vinker, D. Dinour, E. J. Holtzman and M. Shani, J. Am. Soc. Hypertens., 2015, 9, 600–609 CrossRef CAS PubMed.
- H. K. Choi and G. Curhan, Curr. Opin. Rheumatol., 2005, 17, 341–345 CrossRef PubMed.
- R. El Ridi and H. Tallima, J. Adv. Res., 2017, 8, 487–493 CrossRef CAS.
- A. Virdis, S. Masi, E. Casiglia, V. Tikhonoff, A. F. Cicero, A. Ungar, G. Rivasi, M. Salvetti, C. M. Barbagallo and M. Bombelli, Hypertension, 2020, 75, 302–308 CrossRef CAS.
- C. King, M. A. Lanaspa, T. Jensen, D. R. Tolan, L. G. Sánchez-Lozada and R. J. Johnson, Uric Acid in Chronic Kidney Disease, 2018, vol. 192, pp. 88–102 Search PubMed.
- K. N. Glazebrook, L. S. Guimarães, N. S. Murthy, D. F. Black, T. Bongartz, N. J. Manek, S. Leng, J. G. Fletcher and C. H. McCollough, Radiology, 2011, 261, 516–524 CrossRef.
- L. Xu, Y. Shi, S. Zhuang and N. Liu, Oncotarget, 2017, 8, 100852 CrossRef PubMed.
- I. Ramazzina, C. Folli, A. Secchi, R. Berni and R. Percudani, Nat. Chem. Biol., 2006, 2, 144–148 CrossRef CAS.
- J. Dawson, T. Quinn and M. Walters, Curr. Med. Chem., 2007, 14, 1879–1886 CrossRef CAS PubMed.
- M. L. Muiesan, M. Salvetti, A. Virdis, S. Masi, E. Casiglia, V. Tikhonoff, C. M. Barbagallo, M. Bombelli, A. F. Cicero and M. Cirillo, J. Hypertens., 2021, 39, 62–69 CrossRef CAS PubMed.
- C. Ziegler and W. Göpel, Curr. Opin. Chem. Biol., 1998, 2, 585–591 CrossRef CAS.
- S. Vigneshvar, C. Sudhakumari, B. Senthilkumaran and H. Prakash, Frontiers in Bioengineering and Biotechnology, 2016, 4, 11 CrossRef CAS PubMed.
- L. Farzin, M. Shamsipur, L. Samandari and S. Sheibani, Microchim. Acta, 2018, 185, 1–25 CrossRef.
- W. Shi, J. Li, J. Wu, Q. Wei, C. Chen, N. Bao, C. Yu and H. Gu, Anal. Bioanal. Chem., 2020, 412, 7275–7283 CrossRef CAS.
- X.-J. Huang, H.-S. Im, O. Yarimaga, J.-H. Kim, D.-H. Lee, H.-S. Kim and Y.-K. Choi, J. Phys. Chem. B, 2006, 110, 21850–21856 CrossRef CAS.
- Y. Liu, H. Li, B. Guo, L. Wei, B. Chen and Y. Zhang, Biosens. Bioelectron., 2017, 91, 734–740 CrossRef CAS.
- T. Khajvand, M. J. Chaichi and A. H. Colagar, Food Chem., 2015, 173, 514–520 CrossRef CAS PubMed.
- H.-F. Lu, J.-Y. Li, M.-M. Zhang, D. Wu and Q.-L. Zhang, Sens. Actuators, B, 2017, 244, 77–83 CrossRef CAS.
- N. Alizadeh, S. Ghasemi, A. Salimi, T.-K. Sham and R. Hallaj, Colloids Surf., B, 2020, 195, 111228 CrossRef CAS.
- A. Badoei-Dalfard, N. Sohrabi, Z. Karami and G. Sargazi, Biosens. Bioelectron., 2019, 141, 111420 CrossRef CAS PubMed.
- Y. He, F. Qi, X. Niu, W. Zhang, X. Zhang and J. Pan, Anal. Chim. Acta, 2018, 1021, 113–120 CrossRef CAS PubMed.
- K. Khachornsakkul and W. Dungchai, RSC Adv., 2020, 10, 24463–24471 RSC.
- N.-T. Nguyen, S. T. Wereley and S. A. M. Shaegh, Fundamentals and applications of microfluidics, Artech House, 2019 Search PubMed.
- G. M. Whitesides, Nature, 2006, 442, 368–373 CrossRef CAS PubMed.
- X. Li, D. R. Ballerini and W. Shen, Biomicrofluidics, 2012, 6, 011301 CrossRef.
- C. Carrell, A. Kava, M. Nguyen, R. Menger, Z. Munshi, Z. Call, M. Nussbaum and C. Henry, Microelectron. Eng., 2019, 206, 45–54 CrossRef CAS.
- S. K. Sia and L. J. Kricka, Lab Chip, 2008, 8, 1982–1983 RSC.
- S. Sharma, J. Zapatero-Rodríguez, P. Estrela and R. O'Kennedy, Biosensors, 2015, 5, 577–601 CrossRef PubMed.
- J. Sun, Y. Xianyu and X. Jiang, Chem. Soc. Rev., 2014, 43, 6239–6253 RSC.
- F. Farshchi, A. Saadati and M. Hasanzadeh, Anal. Methods, 2020, 12, 4759–4768 RSC.
- B. Liu, D. Du, X. Hua, X. Y. Yu and Y. Lin, Electroanalysis, 2014, 26, 1214–1223 CrossRef CAS.
- Y. He, Y. Wu, J. z. Fu, Q. Gao and J. j. Qiu, Electroanalysis, 2016, 28, 1658–1678 CrossRef CAS.
- G. Luka, A. Ahmadi, H. Najjaran, E. Alocilja, M. DeRosa, K. Wolthers, A. Malki, H. Aziz, A. Althani and M. Hoorfar, Sensors, 2015, 15, 30011–30031 CrossRef CAS.
- P. de Tarso Garcia, T. M. G. Cardoso, C. D. Garcia, E. Carrilho and W. K. T. Coltro, RSC Adv., 2014, 4, 37637–37644 RSC.
- E. Carrilho, A. W. Martinez and G. M. Whitesides, Anal. Chem., 2009, 81, 7091–7095 CrossRef CAS.
- M. Lehto, Paraffin Actuators in Microfluidic Systems, PhD thesis, Acta Universitatis Upsaliensis, 2007.
- L. Y. Yeo, H. C. Chang, P. P. Chan and J. R. Friend, Small, 2011, 7, 12–48 CrossRef CAS PubMed.
- D. J. Guckenberger, E. Berthier and D. J. Beebe, J. Lab. Autom., 2015, 20, 146–153 CrossRef PubMed.
- B. Li, L. Yu, J. Qi, L. Fu, P. Zhang and L. Chen, Anal. Chem., 2017, 89, 5707–5712 CrossRef CAS.
- A. W. Martinez, S. T. Phillips, G. M. Whitesides and E. Carrilho, Anal. Chem., 2010, 82, 3–10 CrossRef CAS PubMed.
- X. Chen, J. Chen, F. Wang, X. Xiang, M. Luo, X. Ji and Z. He, Biosens. Bioelectron., 2012, 35, 363–368 CrossRef CAS.
- S. H. Baek, C. Park, J. Jeon and S. Park, Biosensors, 2020, 10, 187 CrossRef CAS.
- C.-Y. Hou, L.-M. Fu, W.-J. Ju and P.-Y. Wu, Chem. Eng. J., 2020, 398, 125573 CrossRef CAS.
- Y. Zhang, Y.-L. Li, S.-H. Cui, C.-Y. Wen, P. Li, J.-F. Yu, S.-M. Tang and J.-B. Zeng, J. Anal. Test., 2021, 5, 11–18 CrossRef.
- W. Zhao, J. C. Lam, W. Chiuman, M. A. Brook and Y. Li, Small, 2008, 4, 810–816 CrossRef CAS.
- Q. H. Nguyen and M. I. Kim, TrAC, Trends Anal. Chem., 2020, 116038 CrossRef CAS.
- A. Jafari, A. Emami and B. Ashtari, Mater. Sci. Eng., C, 2021, 121, 111810 CrossRef CAS.
- K. Gerelbaatar, A. Tsogoo, R. Dashzeveg, N. Tsedev and E. O. Ganbold, Solid State Phenom., 2018, 271, 76–84 Search PubMed.
- Y. Wang and Y. Ni, Talanta, 2014, 119, 320–330 CrossRef CAS PubMed.
- N. Chen, H. Liu, Y. Zhang, Z. Zhou, W. Fan, G. Yu, Z. Shen and A. Wu, Sens. Actuators, B, 2018, 255, 3093–3101 CrossRef CAS.
- X. Sun, R. Liu, Q. Liu, Q. Fei, G. Feng, H. Shan and Y. Huan, Sens. Actuators, B, 2018, 260, 998–1003 CrossRef CAS.
- A. Kumar, A. Hens, R. K. Arun, M. Chatterjee, K. Mahato, K. Layek and N. Chanda, Analyst, 2015, 140, 1817–1821 RSC.
- S. Kumar, P. Bhushan and S. Bhattacharya, Anal. Methods, 2016, 8, 6965–6973 RSC.
- B. S. Takale, M. Bao and Y. Yamamoto, Org. Biomol. Chem., 2014, 12, 2005–2027 RSC.
- V. Vinoth, J. J. Wu and S. Anandan, Anal. Methods, 2016, 8, 4379–4390 RSC.
- S. Bhattacharjee, J. Controlled Release, 2016, 235, 337–351 CrossRef CAS.
- M. Havrdova, K. Polakova, J. Skopalik, M. Vujtek, A. Mokdad, M. Homolkova, J. Tucek, J. Nebesarova and R. Zboril, Micron, 2014, 67, 149–154 CrossRef CAS PubMed.
- R. White and A. Owens, J. Forensic Sci., 1987, 32, 1595–1603 CAS.
- R. Egerton, P. Li and M. Malac, Micron, 2004, 35, 399–409 CrossRef CAS PubMed.
- C. M. Hoo, N. Starostin, P. West and M. L. Mecartney, J. Nanopart. Res., 2008, 10, 89–96 CrossRef CAS.
- A. Saeb, A. S. Alshammari, H. Al-Brahim and K. A. Al-Rubeaan, Sci. World J., 2014, 2014, 704708 Search PubMed.
- F. Farshchi, A. Saadati, N. Fathi, M. Hasanzadeh and M. Samiei, Anal. Methods, 2021, 13, 1286–1294 RSC.
- V. V. Apyari, V. V. Arkhipova, A. I. Isachenko, P. A. Volkov, S. G. Dmitrienko and I. I. Torocheshnikova, Sens. Actuators, B, 2018, 260, 953–961 CrossRef CAS.
- M. R. Kateshiya, G. George, J. V. Rohit, N. I. Malek and S. K. Kailasa, Microchem. J., 2020, 158, 105212 CrossRef CAS.
- L. M. Fischer, M. Tenje, A. R. Heiskanen and N. Masuda, Microelectron. Eng., 2009, 86, 1282–1285 CrossRef CAS.
- W. Patungwasa and J. H. Hodak, Mater. Chem. Phys., 2008, 108, 45–54 CrossRef CAS.
- R. Cao-Milán and L. M. Liz-Marzán, Expert Opin. Drug Delivery, 2014, 11, 741–752 CrossRef.
- A. Mobed, F. Kohansal, A. Ahmadalipour, M. Hasanzadeh and F. Zargari, Anal. Methods, 2021, 13, 311–321 RSC.
- T. Ghosh, P. Sarkar and A. P. Turner, Bioelectrochemistry, 2015, 102, 1–9 CrossRef CAS.
- S. Verma, J. Choudhary, K. P. Singh, P. Chandra and S. P. Singh, Int. J. Biol. Macromol., 2019, 130, 333–341 CrossRef CAS PubMed.
- Y. Wang, Y. Yang, W. Liu, F. Ding, Q. Zhao, P. Zou, X. Wang and H. Rao, Microchim. Acta, 2018, 185, 1–9 CrossRef.
- Q. Yan, N. Zhi, L. Yang, G. Xu, Q. Feng, Q. Zhang and S. Sun, Sci. Rep., 2020, 10, 1–10 CrossRef.
- Q. Lu, J. Deng, Y. Hou, H. Wang, H. Li and Y. Zhang, Chem. Commun., 2015, 51, 12251–12253 RSC.
- D. Yao, A. G. Vlessidis and N. P. Evmiridis, Anal. Chim. Acta, 2003, 478, 23–30 CrossRef CAS.
- T. A. Burinaru, M. Avram, A. Avram, C. Marculescu, B. Tincu, V. Ţucureanu, A. Matei and M. Militaru, ACS Comb. Sci., 2018, 20, 107–126 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra04764g |
| This journal is © The Royal Society of Chemistry 2021 |