Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microbial diversity in full-scale water supply systems through sequencing technology: a review
Wei Zhouab,
Weiying Li *ab,
Jiping Chena,
Yu Zhoua,
Zhongqing Weic and
Longcong Gongd
*ab,
Jiping Chena,
Yu Zhoua,
Zhongqing Weic and
Longcong Gongd
aCollege of Environmental Science and Engineering, Tongji University, Shanghai 200092, China. E-mail: 123lwyktz@tongji.edu.cn
bState Key Laboratory of Pollution Control and Resource Reuse, Tongji University, Shanghai 200092, China
cFuzhou Water Affairs Investment Development Co., Ltd., Fuzhou 350000, Fujian, China
dFuzhou Water Co., Ltd., Fuzhou 350000, Fujian, China
First published on 22nd July 2021
Abstract
The prevalence of microorganisms in full-scale water supply systems raises concerns about their pathogenicity and threats to public health. Clean tap water is essential for public health safety. The conditions of the water treatment process from the source water to tap water, including source water quality, water treatment processes, the drinking water distribution system (DWDS), and building water supply systems (BWSSs) in buildings, greatly influence the bacterial community in tap water. Given the importance of drinking water biosafety, the study of microbial diversity from source water to tap water is essential. With the development of molecular biology methods and bioinformatics in recent years, sequencing technology has been applied to study bacterial communities in full-scale water supply systems. In this paper, changes in the bacterial community and the influence of each treatment stage on microbial diversity in full-scale water supply systems are classified and analyzed. Microbial traceability analysis and control are discussed, and suggestions for future drinking water biosafety research and its prospects are proposed.
Introduction
Water security is not only an ecological and environmental issue, but also an economic, social, and political issue that is directly related to national security. Safe drinking water is essential to public health and an integral part of effective policies to protect health.1 However, as of 2017, billions of people worldwide still do not have access to safe drinking water and basic health services. According to the World Health Organization (WHO), 80% of human diseases and 50% of child deaths worldwide are related to drinking water quality. With the outbreak of COVID-19, the safety of microorganisms in the environment, especially in water, has become a greater concern.2–4 Unlike chemical pollution, microbial pollution is proliferative, secondary, and infectious. The explosive proliferation of microorganisms can result in deterioration of water quality, and the presence of odor or toxins,5,6 and induce secondary pollution. Water-mediated pathogenic microorganisms can be transmitted through diet, aerosols, and contact, endangering human health.Culture-based methods are one of the most widely used traditional analytical approaches to evaluate the microbiological quantity of drinking water. However, due to the overlooked of some bacteria (e.g., viable but non-culturable (VBNC) bacteria), the culture-based method leads to an underestimation of the microbial density and diversity in drinking water.7 As a result, nucleic acid-based approaches have been widely applied in recent investigations of drinking water distribution system (DWDS) microbial communities.8,9 These culture-independent methods, such as sequencing technology, include internal transcribed spacer (ITS) fingerprints, terminal restriction fragment length polymorphisms (T-RFLP), 16S rRNA gene surveys, and metagenomics surveys,10 which could not only detect low concentrations of microorganism (including VBNC), but also obtain microbial diversity information, providing a good technical support for the study of microbial fate.
Given the advantages, sequencing technology, especially high-throughput sequencing (HTS), is widely used to analyze microbial diversity in drinking water to obtain a more comprehensive understanding of bacterial ecology. This review paper summarize the findings of microbial community analysis in full-scale water supply system through sequence technology, especially from the perspective of biological safety of the tap water through distribution system, focusing on (i) the development of sequencing technologies and influence on the study the microorganism in water supply system; (ii) the microbial diversity and environmental impact on full-scale water supply systems using sequencing technology; (iii) evaluation of microbial safety in water source, water treatment process as well as the drinking water distribution system (DWDS); and (iv) proposed biosafety assurance measures for a full-scale water supply system. The goals of this review are to understand application of sequencing technology in the study of drinking water microbial communities, analyze the possible causes of microorganism safety problems in full-scale water supply systems, guide operational practices to obtain safe drinking water, and enhance future research on the drinking water microbiome.
Development of sequencing technology and its contribution to drinking water investigation
Development of sequencing technology
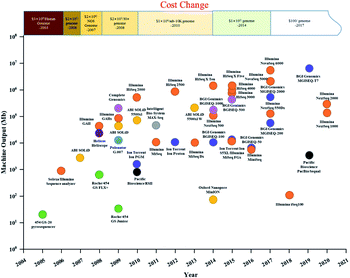
Gene sequencing technology has developed in the last 50 years as a result of the pioneering Sanger and Coulson chain termination method. With the high cost and low throughput of first-generation sequencing technology,11 continuous technological development and improvement yielded Roche's 454 technology, Illumina's Solexa technology, and ABI's Solid technology. The comparison of four generation sequencing technologies was list in Table 1. Compared with first-generation Sanger sequencing, they offered high throughput12 and fast sequencing, greatly reducing sequencing cost and expanding the scale of genomics research.13 The timeline and comparison of commercial HTS instruments and costs since 2003 are shown in Fig. 1. After the introduction of the Genome Sequencer 20 System by 454 Life Sciences in 2005, and the Genome Analyzer II by Illumina/Solexa in 2006, high-throughput sequencing companies were emerging, providing a solid foundation for the development of high-throughput sequencing and price reduction. However, second-generation sequencing technology was still costly, with a short-read length. Since 2008, single-molecule real-time (SMRT, PacBio) sequencing technology and the Heliscope (Helicos Biosciences) genetic analysis system have been developed, known as third-generation sequencing. In NGS method, DNA is broken into short pieces, amplified, and then sequenced. Third generation technologies do not break down or amplify the DNA: they directly sequence a single DNA molecule. Fourth-generation sequencing technology (e.g., Nanopore sequencing technology by Oxford Nanopore Technologies) was invented in 2014.14 However, third- and fourth-generation sequencing technologies have relatively lower accuracy and have not been widely used as NGS. Currently, NGS technology is still the predominant sequencing technology in the market. The launch of Illumina's NovaSeq 6000 in 2017 brought the cost of sequencing under $100 per human genome, promoting widespread use of HTS in recent years in medicine, health, and environmental fields (Fig. 1).| First-generation sequencing technology | Next-generation sequencing technology | Third-generation sequencing technology | Fourth-generation sequencing technology | |
|---|---|---|---|---|
| Characteristics | Dideoxy chain termination method | Sequencing by synthesis | Single-molecule sequencing | Nanopore sequencing |
| Read length | ∼1000 bp | 50–300 bp | 8–12 kb | ∼100 kb |
| Throughput | Low | High | High | High |
| Instrument time | Long | Short | Short | Short |
| Relative cost | High | Relatively low | Low | Low |
| Advantage | Long read length and high accuracy | High throughput, accuracy, speed, and output | Long read length, high throughput, and high speed | Long read length, high throughput, low cost, high speed, simpleness on sample preparation and analysis |
| Disadvantage | Low throughput and long instrument time | Short read length | Relatively low accuracy | Relatively low accuracy |
| Represented platforms | ABI | Illumina | Pacbio | Nanopore |
Sequencing technology application in water supply system
With the development of sequencing technology, especially high-throughput technology in recent years, analysis of drinking water microorganisms in relation to human health has been widely conducted.15 As the analysis method has gradually shifted from traditional analytical approaches to sequencing analysis methods, the research and focus on the whole process of microorganisms in drinking water has also changed from the original quantity and species to the present diversity, transformation, and function. Searching related papers on the topic of “Water” and “Bacterial community” in Web of Science, the number of publications has reached 39540 (as of July 1, 2021), among which the number of annual increased papers reached more than 1000 since 2007. Searching related papers on the Web of Science with the topic of “Drinking Water” and “Bacterial Community”, the article number has reached 2708 (as of July 1, 2021), among which the growth rate of the increased papers number was largely improved from 2006 (Fig. 2).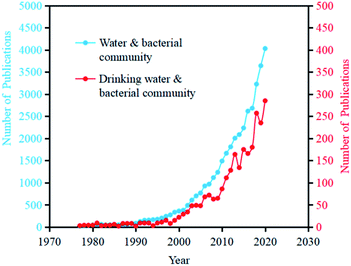 | ||
| Fig. 2 The annual increasing number of publications with the topics of “water & bacterial community” and “drinking water & bacterial community” in Web of Science (as of July 1st, 2021). | ||
Here in this paper, the changes in microbial diversity from source water to tap water were categorized based on reported research which using sequencing technologies, as well as the health risks associated with these changes.
Impact on microbial diversity in full-scale water supply system
Water source effects
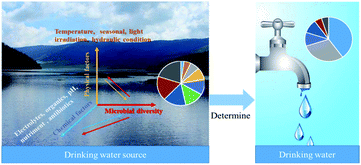
The microbial diversity of water sources detected by sequencing technology were categorized in Table 2. Microbial diversity in drinking water sources is influenced by environmental factors, including chemical factors (electrolytes, organics, pH, nutrients, antibiotics) and physical factors (temperature, seasonality, light irradiation). The microbial diversity in the source water also affects the chemical and physical characteristics of the water. In conventional treatment, the source water microbial community is important because it is the source, and the tap water microbial community is the sink (Fig. 3). In studying the dynamic changes of microorganisms in water sources, physical, chemical, and biological properties must be considered together for systematic analysis to evaluate the microorganism diversity more comprehensively.
| Factors | Impacts on drinking water microbiome | Ref. |
|---|---|---|
| Biological effects | ✓ Different water sources have different microbial community compositions, resulting in different bacterial communities in the final tap water | 16–18 and 21–25 |
| ✓ The dominant microbial composition may be similar in different water sources, but the abundances may vary | ||
| Chemical and physical effects | ✓ Temperature, seasonality seasonal, pH, electrolyte type, salinity, dissolved particles, dissolved oxygen (DO), C/N ratio, total nitrogen (TN), total phosphorus (TP), and COD have been verified to influence the composition of the microbial community in drinking water | 19, 22 and 25–36 |
| ✓ Actinobacteria negatively correlated with phosphorus, sulfate, dissolved particles, and chloride levels. Proteobacteria positively correlated with sulfate, dissolved particles, chloride, dissolved oxygen, and nitrite levels | ||
| ✓ The bacterial diversity was positively correlated with CODMn, turbidity, and pH | ||
| ✓ The bacterial diversity in water source was higher in wet season than in dry season | ||
| Organics effects | ✓ Organic nutrients (such as assimilable organic carbon) or DBPs precursors of organic matter have positive effect on the bacteria proliferation | 37 and 39–43 |
| ✓ Some pharmaceutical and personal care products (such as antibiotics, and environmental endocrine disruptor) have negative effect on the bacterial diversity, however, they may pose a great threat to drinking water safety | ||
| Unconventional water sources effects | ✓ The presence of one or more fecal indicators, and potential bacterial and protozoan pathogens were detected in rainwater, giving suggestion to that it may not be suitable for drinking | 17–21 and 45–47 |
| ✓ Disinfection was recommended whenever possible |
Drinking water treatment processes effects
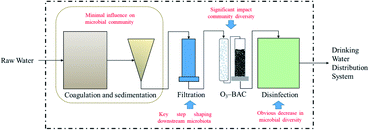
Drinking water treatment is the key to preventing waterborne diseases and their spread. There is a potential relationship between bacterial community composition and the emergence of opportunistic pathogens;48 problems encountered in drinking water treatment plants or water distribution may lead to the proliferation of conditioned pathogenic bacteria (Mycobacterium, Pseudomonas aeruginosa, Legionella pneumophila, etc.).49–51 Currently, conventional water treatment processes (coagulation–flocculation, sedimentation, filtration, and disinfection) are widely used to purify drinking water in China.52 In recent years, with the deterioration of source water quality, especially from an increase in organic matter, advanced treatment technologies such as ozone–biological activated carbon (O3–BAC) and membrane treatment have been applied. Understanding changes in the microbial community during treatment is vital for the management of DWTSs. Usually, O3–BAC and disinfection processes are regarded as the primary units influencing the microbial density and diversity; other units also have some influence.53 A great change in the proportions of Actinobacteria, Proteobacteria, and Firmicutes during the treatment process was detected by Hou et al., the proportion of Actinobacteria decreased sharply, and the proportions of Proteobacteria and Firmicutes increased and predominated in treated water.54 During drinking water treatment processes, the microbial activity and bacterial diversity showed obvious spatial differences; the bacterial community changed significantly after chlorination disinfection, indicating that the disinfection process affected the bacterial community. In addition, the bacterial community structure of the finished water was like that of the biofilm on the GAC, indicating that the application of biological treatment technology can significantly change the microbial community composition inherited from the source.Drinking water treatment processes effects on its microbiome was counted in Table 3. Generally, drinking water treatment processes have a significant impact on the microbial diversity in tap water. Microbial changes, whether in the traditional processes of coagulation and sedimentation or in subsequent filtration and O3–BAC, are adjusted in the disinfection process, resulting in (i) the culturable bacteria transfer from predominantly Gram-negative to predominantly Gram-positive;93 (ii) alpha- and betaproteobacteria are dominant in water; (iii) chlorine-resistant bacteria (e.g., VBNC and ARB) may be hidden dangers in subsequent DWDSs. The influence of each unit in the DWTP on the microbial community is shown in Fig. 4. Coagulation and sedimentation have minimal influence on the community; filtration is the key step shaping downstream microbiota. The O3–BAC and disinfection processes have the strongest effect in changing the microbial community.
| Treatment processes | Impacts on drinking water microbiome | Ref. |
|---|---|---|
| Coagulation and sedimentation | ✓ Early studies suggested that they have minimal influence on the microbial community | 54–56, 67 and 68 |
| ✓ With the develop of analytical approach, the results show that they are important for the removal of bacteria in source water | ||
| ✓ The removal of microorganisms from source water by coagulation and clarification mostly refers to microorganisms that are easily adsorbed on suspended particles and colloids | ||
| Filtration | ✓ Filtration is the key step shaping downstream microbiota through removing incoming particles and seeding outflow with microorganisms sloughed from filters | 57–61, 69 and 70 |
| ✓ The filtration process can significantly reduce suspended substances such as bacteria and viruses, further affecting the microbial diversity | ||
| ✓ Various biological processes can occur in filters | ||
| O3–BAC | ✓ Ozonation increased taxonomic diversity but decreased functional diversity of the bacterial communities in the BAC filters | 71–77 |
| ✓ With the removal of organic matters in DWTP, the leakage of bacteria which could be have been seeding of distribution system, becomes an important issue in this unit | ||
| ✓ The influent water quality, oxidative pretreatment, empty bed contact time, and backwashing frequency can affect the redox environment of the system, influencing the microbial diversity of the effluent | ||
| ✓ O3–BAC effect on assimilable organic carbon (AOC) removal in three DWTPs and AOC increased after O3 treatment, and BAC could remove most AOC from water, which may be attributed to the microbial community differences | ||
| Disinfection | ✓ Different disinfection type and dosage might result in different bacterial populations | 78–91 |
| ✓ Generally, after disinfection, alpha- and beta-proteobacteria were dominant in chlorinated water. Betaproteobacteria was more abundant after chloramine disinfection than the other two processes | ||
| ✓ Although disinfection process could inactivate most bacteria, there are still some chlorine-resistant bacteria existed in finished water, leading to the formation of biofilms in drinking water distribution systems and thus affecting the biosafety of residential water | ||
| ✓ The molecular mechanism of chlorine resistance is attributed to glutathione synthesis | ||
| ✓ Much attentions have been paid on the new approached of disinfection |
Influence in DWDS
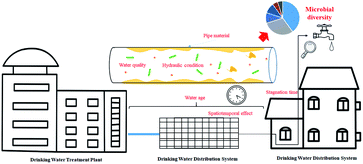
Mathieu et al.102 have reviewed the bugs systematically found in drinking water distribution systems all over the world (bacteria, viruses, yeasts, fungi, protozoa, microcrustaceans, rotifers, and oligochaete worms), here, we will analysis and discuss several factors which influence the microbial diversity in DWDS, especially the fate of biofilm on pipes according to the results from HTS. Based on the microbial diversity analysis of finished water and tap water, a diverse core microbiome was shared between the two locations; however, the microbial community was changed in the DWDS,103 which was attributed to the shedding of biofilms (the environmental reservoirs for pathogenic microorganisms) from the inner wall of the pipe, posing a potential threat to human health.104 Microbial regrowth with spatiotemporal variation is a major concern in distribution, as the physicochemical and nutritional conditions provided by pipe walls are very different from those found during treatment. Recent studies have identified the microbial community and dominant species associated with many factors in the DWDS. These factors include pipe materials, hydraulic conditioning, spatiotemporal effects, and the quality of the treated water (Fig. 5).| Materials | Impacts on biofilms bacterial community | Ref. |
|---|---|---|
| PVC and cast ion | ✓ Hyphomicrobia was the most dominant bacteria identified in the PVC | 105 |
| ✓ Corrosion associated bacteria was the most dominant bacteria identified cast-iron biofilms | ||
| ✓ Bacterial colonization on the material surfaces was selective | ||
| HDPE, PEX and PVC | ✓ Coupon material did not have a significant impact on biomass levels or composition of the biofilm communities in the chloraminated reactors | 80 and 106 |
| ✓ The biological diversity of different metal pipes was significantly different due to the metal precipitation problem | ||
| ✓ A higher biological diversity was observed in biofilms on metallic material than that on plastic materials | ||
| ✓ The most extensive biofilm was found in pipes of HDPE material | ||
| ✓ The most numerous quantities of bacterial was found in pipes of PEX surface | ||
| EPDM and PEX | ✓ The biofilm populations on EPDM were higher than those on PEX | 107 |
| Copper | ✓ Copper could significantly reduce the microbial diversity downstream | 108 and 109 |
| ✓ Effect of copper surface on Legionella pneumophila biofilm formation in drinking water | ||
| ✓ Copper could inactivate Lactobacillus pneumophilus. In biofilms | ||
| UPVC and copper | ✓ Significant differences between bacterial and eukaryotic member in biofilm on UPVC and Cu | 110 |
| Epoxying iron, PVC, and cement | ✓ Free chlorine was most stable in the presence of PVC while chloramine was most stable in the presence of cement | 111 and 112 |
| ✓ The influence of pipe material became apparent at water ages corresponding to low disinfectant residual | ||
| ✓ Each target microbe appeared to display a distinct response to disinfectant type, pipe materials, water age, and their interactions | ||
| EPDM and PEX | ✓ Total cell counts and HPC values were highest on EPDM followed by the plastic materials and copper | 113 |
| ✓ P. aeruginosa and L. pneumophila became incorporated into drinking water biofilms on EPDM and PEX | ||
| ✓ Copper biofilms were colonized only by L. pneumophila in low culturable numbers | ||
| Copper and PEX | ✓ Pipe material seemed to affect mycobacteria occurrence, and bacterial communities with MWT in copper but not in PEX pipes | 114 |
| Plastic and stainless steel | ✓ The microbiome of biofilms formed on stainless steel and plastics was quite different | 115 |
| ✓ Metallic materials facilitate the formation of higher diversity biofilms | ||
| Copper (CU), chlorinated poly vinyl chloride (CP), polybutylene (PB), polyethylene (PE), stainless steel (SS), steel coated with zinc (ST) | ✓ Steel pipes (SS and ST) had the highest biofilm formation potential (BFP) and CU showed the lowest BFP | 116 |
| ✓ The BFP of CP in drinking water and mixed water were comparable to those of CU | ||
| ✓ PB and PE showed relatively high BFP |
In addition, treated drinking water quality, including temperature,132,144,145 suspended solids,35,146 electrolytes,28,125,147–149 disinfectants,149 and organic matter28 also influence microbial diversity in tap water. Sun et al.28 reported that pH and COD were positively correlated with the relative abundance of Proteobacteria and Firmicutes. Ma et al.148 reported that bacterial richness and diversity were positively related to SO42−, Cl−, and HCO3− in the water supply, and negatively related to pH value. Chemical reactions other than microbial processes play a major role in the release of iron during the transition period of the water supply. Moreover, the role of residual chlorine in water quality cannot be underestimated. The disinfectant changes the bacterial community structure of the pipeline biofilm, and affects the water quality and the remodeling of the corrosion scale, further changing the kinetics of the corrosion process.149
Prospects of microbial diversity analysis in drinking water biosafety
With the development of sequencing technology, in-depth characterization and evaluation has been conducted by researchers on microbial communities in DWDSs and BWSSs. Although microbiological safety assurance technology is relatively good, harmful bacteria such as pathogenic bacteria and ARB may still be detected in drinking water, posing a threat to public health.150–154 It has been verified that every stage from the source to the tap has some influence on microbial diversity. Thus, researchers have conducted traceability analyses of microorganisms in tap water or estimated the impact on microbial conditions in drinking water based on existing water quality conditions to determine if emergency treatment methods are necessary. Marshall et al.155 investigated the genotype similarities and geographic relationships of bacterial communities between humans and drinking water. The results indicated that drinking water may be a source of human Mycobacterium lentiflavum infection. Liu et al.156 used the Bayesian “source tracing” method to determine the proportional contributions of source water, treatment water, and the distribution system in shaping the bacterial community in faucet water based on bacterial community fingerprints. The results showed that the source water had no obvious contribution to the bacterial communities of tap water and water in the distribution system. Loose sediments and biofilms show significant effects on phytoplankton and particle-related bacteria in faucet water, which are position dependent and subject to hydraulic changes. In addition, sequencing technology has been used to assess the safety of rural drinking water systems. The rich genetic footprint of pathogens in water samples from many reports suggests that the bacteria can be transmitted to humans. Thus, the importance of disinfection of raw water must be clearly communicated to rural communities to ensure the safe use of water. Studying the microbial community structure and its influence on drinking water is critical.With the continuous improvement of sequencing technology in depth, accuracy, and economy, how and why the microbial diversity changes in the whole process of drinking water will be clearer. Future research may focus on the impact of new pollutants, traceability analysis and source control, as well as rapid detection and intelligent feedback.
Conclusion
This paper summarizes and clarifies the biological sequencing technologies applied in research on drinking water microbial communities, including the source drinking water quality, the treatment process, and the distribution system supply conditions, and indicates that all three steps can affect the tap water microbial community. A significant correlation was observed between the microbial populations in the source water and tap water, and the abundance of bacteria was largely affected by the treatment process and the distribution system condition. Thus, the microbiological safety assurance of drinking water must start from the source. The treatment process must be improved, the pipeline network route material must be carefully selected, and drinking water management should be strengthened from the factory to the client, to block the source (water protection), decrease the concentration (optimization of disinfection during DWTP), and control the flow (reduce growth in the DWDS). These mechanisms that need to be explored require the development of cheaper and more accurate biological sequencing technologies.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (51979194). We also thank the research on water quality stability characteristics and countermeasures of the Fuzhou Water Supply System (Project No. 20203000) from Fuzhou Water Group Co. Ltd.References
- W. H. Organization, Guidelines for Drinking-water Quality, 4th edn, 2011 Search PubMed.
- S. Khadse, N. Gharami and T. Sapate, Int. J. Tech. Res. Sci., 2020, 5, 35–41 CrossRef.
- M. Keulertz, M. Mulligan and J. A. Allan, Water International, 2020, 45, 430–434 CrossRef.
- M. J. Neal, Water International, 2020, 45, 435–440 CrossRef.
- L. Shang, M. Feng, X. Xu, F. Liu, F. Ke and W. Li, Toxins, 2018, 10, 26 CrossRef PubMed.
- P. Lebaron, B. Cournoyer, K. Lemarchand, S. Nazaret and P. Servais, Environmental Microbiology: Fundamentals and Applications, 2015, pp. 619–658 Search PubMed.
- D. G. Lee, S. J. Park and S. J. Kim, J. Microbiol. Biotechnol., 2007, 17, 1558 Search PubMed.
- E. I. Prest, F. Hammes, M. C. Van Loosdrecht and J. S. Vrouwenvelder, Front. Microbiol., 2016, 7, 1–24 Search PubMed.
- J. F. Petrosino, H. Sarah, L. R. Ann, R. A. Gibbs and V. James, Clin. Chem., 2009, 856 CrossRef CAS PubMed.
- Y. Zhang and W.-T. Liu, Crit. Rev. Environ. Sci. Technol., 2019, 49, 1188–1235 CrossRef CAS.
- Y. Luo, X. Liao, F. X. Wu and J. Wang, Curr. Bioinf., 2020, 15, 2–16 CrossRef CAS.
- E. L. Van Dijk, H. Auger, Y. Jaszczyszyn and C. Thermes, Trends Genet., 2014, 30, 418–426 CrossRef CAS PubMed.
- J. J. Salk, M. W. Schmitt and L. A. Loeb, Nat. Rev. Genet., 2018, 19, 269–285 CrossRef CAS PubMed.
- J. A. Reuter, D. V. Spacek and M. P. Snyder, Mol. Cell, 2015, 58, 586–597 CrossRef CAS PubMed.
- J. Vierheilig, D. Savio, R. E. Ley, R. L. Mach and G. H. Reischer, Water Sci. Technol., 2015, 72, 1962–1972 CrossRef CAS PubMed.
- L. Kahlisch, K. Henne, L. Groebe, J. Draheim, M. G. Höfle and I. Brettar, Water Sci. Technol., 2010, 61, 9–14 CrossRef CAS PubMed.
- A. P. Mucha, M. T. S. D. Vasconcelos and A. A. Bordalo, Estuarine, Coastal Shelf Sci., 2004, 59, 663–673 CrossRef CAS.
- D. Boucher, L. Jardillier and D. Debroas, FEMS Microbiol. Ecol., 2010, 55, 79–97 CrossRef PubMed.
- W. Li, F. Wang, J. Zhang, Y. Qiao, C. Xu, Y. Liu, L. Qian, W. Li and B. Dong, Sci. Total Environ., 2016, 544, 499–506 CrossRef CAS PubMed.
- X. Guo, J. Li, F. Yang, J. Yang and D. Yin, Sci. Total Environ., 2014, 493, 626–631 CrossRef CAS PubMed.
- V. Gomez-Alvarez, B. W. Humrighouse, R. P. Revetta and J. W. Santo Domingo, J. Water Health, 2015, 13, 140–151 CrossRef PubMed.
- T. Jiang, S. Sun, Y. Chen, Y. Qian and D. An, Sci. Total Environ., 2021, 769, 144698 CrossRef CAS PubMed.
- D. A. Pearce, C. J. Gast, B. Lawley and J. C. Ellis-Evans, FEMS Microbiol. Ecol., 2010, 45, 59–70 CrossRef.
- K. Henne, L. Kahlisch, I. Brettar and M. G. Hofle, Appl. Environ. Microbiol., 2012, 78, 3530–3538 CrossRef CAS PubMed.
- W. Sun, C. Xia, M. Xu, J. Guo and G. Sun, Front. Microbiol., 2017, 8, 1644 CrossRef PubMed.
- M. Kaevska, P. Videnska, K. Sedlar and I. Slana, SpringerPlus, 2016, 5, 1–8 CrossRef CAS PubMed.
- M. Moitra and L. G. Leff, Hydrobiologia, 2015, 747, 201–215 CrossRef CAS.
- H. Sun, B. Shi, Y. Bai and D. Wang, Sci. Total Environ., 2014, 472, 99–107 CrossRef CAS PubMed.
- D. R. Joshi, Y. Zhang, Y. Gao, Y. Liu and M. Yang, Water Res., 2017, 121, 338 CrossRef CAS PubMed.
- H. Zhang, J. Jia, S. Chen, T. Huang, Y. Wang, Z. Zhao, J. Feng, H. Hao, S. Li and X. Ma, Int. J. Environ. Res. Public Health, 2018, 15, 361 CrossRef PubMed.
- M. Kaestli, D. O'Michelle, A. Rose, J. R. Webb, M. Mayo, B. J. Currie and K. Gibb, PLoS Neglected Trop. Dis., 2019, 13, e0007672 CrossRef CAS PubMed.
- L. Zhang, G. Gao, X. Tang and K. Shao, Can. J. Microbiol., 2014, 60, 319–326 CrossRef CAS PubMed.
- Y. Zhang, S. Oh and W. T. Liu, Environ. Microbiol., 2017, 19, 3163–3174 CrossRef CAS PubMed.
- G. Roeselers, J. Coolen, P. W. van der Wielen, M. C. Jaspers, A. Atsma, B. de Graaf and F. Schuren, Environ. Microbiol., 2015, 17, 2505–2514 CrossRef PubMed.
- A. Nescerecka, T. Juhna and F. Hammes, Water Res., 2018, 135, 11–21 CrossRef CAS PubMed.
- Q. Wu, X. Zhao, Q. Tian, L. Wang and X. Zhao, IOP Conference Series: Earth and Environmental Science, 2018, 113, 012144 CrossRef.
- P. Servais, P. Laurent and D. Gatel, Appl. Mech. Mater., 1995, 781, 329–332 Search PubMed.
- F. L. Rosario-Ortiz, S. A. Snyder and I. H. Suffet, Water Res., 2007, 41, 4115–4128 CrossRef CAS PubMed.
- E. J. Rosi-Marshall, D. W. Kincaid, H. A. Bechtold, T. V. Royer, M. Rojas and J. J. Kelly, Ecological Applications, 2013, 23, 583–593 CrossRef PubMed.
- L. Proia, G. Lupini, V. Osorio, S. Perez, D. Barcelo, T. Schwartz, S. Amalfitano, S. Fazi, A. M. Romani and S. Sabater, Chemosphere, 2013, 92, 1126–1135 CrossRef CAS PubMed.
- D. Li, R. Qi, M. Yang, Y. Zhang and T. Yu, Water Res., 2011, 45, 6063–6073 CrossRef CAS PubMed.
- E. K. Radwan, M. B. Ibrahim, A. Adel and M. Farouk, Environ. Sci. Pollut. Res., 2020, 27, 1776–1788 CrossRef CAS PubMed.
- K. Yuan, S. Xiao, X. Jiang, L. Yang, B. Chen, T. Luan, L. Lin and N. F. Y. Tam, Mar. Pollut. Bull., 2017, 122, 122–128 CrossRef CAS PubMed.
- W. J. Deng, N. Li and G. G. Ying, Environ. Geochem. Health, 2018, 40, 1–13 CrossRef PubMed.
- W. Ahmed, L. Hodgers, J. P. S. Sidhu and S. Toze, Appl. Environ. Microbiol., 2012, 78, 219–226 CrossRef CAS PubMed.
- F. Alves, T. Köchling, J. Luz, S. M. Santos and S. Gavazza, J. Water Health, 2014, 12, 513–525 CrossRef PubMed.
- A. Belila, J. El-Chakhtoura, N. Otaibi, G. Muyzer, G. Gonzalez-Gil, P. E. Saikaly, M. C. M. van Loosdrecht and J. S. Vrouwenvelder, Water Res., 2016, 94, 62–72 CrossRef CAS PubMed.
- P. Ji, W. J. Rhoads, M. A. Edwards and A. Pruden, ISME J., 2017, 11, 1318 CrossRef PubMed.
- J. Lu, I. Struewing, S. Yelton and N. Ashbolt, J. Appl. Microbiol., 2015, 119, 278–288 CrossRef CAS PubMed.
- N. J. Ashbolt, Curr. Environ. Health Rep., 2015, 2, 95–106 CrossRef CAS PubMed.
- Y. Zhang, S. Oh and W. Liu, Environ. Microbiol., 2017, 3163–3174 CrossRef CAS PubMed.
- S. Feng, X. J. Zhang, C. Chen and Z. Y. Yang, China Water & Wastewater, 2012, 28, 16–19 Search PubMed.
- Q. Li, S. Yu, L. Li, G. Liu, Z. Gu, M. Liu, Z. Liu, Y. Ye, Q. Xia and L. Ren, Front. Microbiol., 2017, 8, 2465 CrossRef PubMed.
- L. Hou, Q. Zhou, Q. Wu, Q. Gu and J. Zhang, Sci. Total Environ., 2017, 625, 449–459 CrossRef PubMed.
- J. B. Poitelon, M. Joyeux, B. Welte, J. P. Duguet, E. Prestel and M. S. DuBow, J. Ind. Microbiol. Biotechnol., 2010, 37, 117–128 CrossRef CAS PubMed.
- W. Lin, Z. Yu, H. Zhang and I. P. Thompson, Water Res., 2014, 52, 218–230 CrossRef CAS PubMed.
- J. L. Shaw, P. Monis, R. Fabris, L. Ho, K. Braun, M. Drikas and A. Cooper, Chemosphere, 2014, 117, 185–192 CrossRef CAS PubMed.
- C. N. Albers, L. Ellegaard-Jensen, C. B. Harder, S. Rosendahl, B. E. Knudsen, F. Ekelund and J. Aamand, Environ. Sci. Technol., 2015, 49, 839–846 CrossRef CAS PubMed.
- Y. Bai, R. Liu, J. Liang and J. Qu, PLoS One, 2013, 8, e61011 CrossRef CAS PubMed.
- S. Oh, F. Hammes and W.-T. Liu, Water Res., 2018, 128, 278–285 CrossRef CAS PubMed.
- C. O. Lee, R. Boe-Hansen, S. Musovic, B. Smets, H. J. Albrechtsen and P. Binning, Water Res., 2014, 64, 226–236 CrossRef CAS PubMed.
- K. Tatari, S. Musovic, A. Guelay, A. Dechesne, H. J. Albrechtsen and B. F. Smets, Water Res., 2017, 127, 239–248 CrossRef CAS PubMed.
- S. Feng, S. Xie, X. Zhang, Z. Yang, D. Wei, X. Liao, Y. Liu and C. Chao, J. Environ. Sci., 2012, 1587–1593 CrossRef CAS.
- T. M. LaPara, K. Hope Wilkinson, J. M. Strait, R. M. Hozalski, M. J. Sadowksy and M. J. Hamilton, Appl. Environ. Microbiol., 2015, 81, 6864–6872 CrossRef CAS PubMed.
- D. Zhang, Biomed. Environ. Sci., 2011, 24, 122–131 Search PubMed.
- C. Liu, C. I. Olivares, A. J. Pinto, C. V. Lauderdale, J. Brown, M. Selbes and T. Karanfil, Water Res., 2017, 124, 630–653 CrossRef CAS PubMed.
- D. N. Zeng, Z. Y. Fan and C. Liang, World J. Microbiol. Biotechnol., 2013, 29, 1573–1584 CrossRef PubMed.
- W. Lin, Z. Yu, H. Zhang and I. P. Thompson, Water Res., 2014, 52, 218–230 CrossRef CAS PubMed.
- C. N. Albers, L. Ellegaard-Jensen, C. B. Harder, S. Rosendahl, B. E. Knudsen, F. Ekelund and J. Aamand, Environ. Sci. Technol., 2015, 49, 839–846 CrossRef CAS PubMed.
- C. O. Lee, R. Boe-Hansen, S. Musovic, B. Smets, H. J. Albrechtsen and P. Binning, Water Res., 2014, 64, 226–236 CrossRef CAS PubMed.
- I. X. Zhu, T. Getting and D. Bruce, J. - Am. Water Works Assoc., 2010, 102, 67–77 CrossRef CAS.
- S. Velten, M. Boiler, O. Koester, J. Helbing, H. U. Weilenmann and F. Hammes, Water Res., 2011, 45, 6347–6354 CrossRef CAS PubMed.
- S. Soonglerdsongpha, I. Kasuga, F. Kurisu and H. Furumai, Sustainable Environ. Res., 2011, 21, 59–64 CAS.
- X. Liao, C. Chen, Z. Wang, R. Wan, C. H. Chang, X. Zhang and S. Xie, Process Biochem., 2013, 48, 312–316 CrossRef CAS.
- K. Lautenschlager, C. Hwang, F. Ling, W. T. Liu, N. Boon, O. Koester, T. Egli and F. Hammes, Water Res., 2014, 62, 40–52 CrossRef CAS PubMed.
- X. Liao, C. Chen, Z. Wang, R. Wan and S. Xie, Process Biochem., 2013, 48, 703–707 CrossRef CAS.
- O. E. Kaarela, H. A. Haerkki, M. R. T. Palmroth and T. A. Tuhkanen, Environ. Technol., 2015, 36, 681–692 CrossRef CAS PubMed.
- X. Bai, F. Wu, B. Zhou and X. Zhi, J. Water Health, 2010, 8, 593–600 CrossRef CAS PubMed.
- R. S. Roeder, J. Lenz, P. Tarne, J. Gebel, M. Exner and U. Szewzyk, Int. J. Hyg. Environ. Health, 2010, 213, 183–189 CrossRef CAS PubMed.
- S. Aggarwal, C. K. Gomez-Smith, Y. Jeon, T. M. LaPara, M. B. Waak and R. M. Hozalski, Environ. Sci. Technol., 2018, 52, 13077–13088 CrossRef CAS PubMed.
- X. Li, H. Wang, Y. Zhang, C. Hu and M. Yang, Int. Biodeterior. Biodegrad., 2014, 96, 71–79 CrossRef.
- Y. Zhu, H. Wang, X. Li, C. Hu, M. Yang and J. Qu, Water Res., 2014, 60, 174–181 CrossRef CAS PubMed.
- J. J. Kelly, N. Minalt, A. Culotti, M. Pryor and A. Packman, PLoS One, 2014, 9, e98542 CrossRef PubMed.
- Z. G. Homonnay, G. Török, J. Makk, A. Brumbauer, E. Major, K. Márialigeti and E. Tóth, J. Basic Microbiol., 2014, 54, 729–738 CrossRef CAS PubMed.
- Z. Mi, Y. Dai, S. Xie, C. Chen and X. Zhang, J. Environ. Sci., 2015, 37, 200–205 CrossRef CAS PubMed.
- M. M. Williams, J. W. S. Domingo, M. C. Meckes, C. A. Kelty and H. S. Rochon, J. Appl. Microbiol., 2010, 96, 954–964 CrossRef PubMed.
- W. Ding, W. Jin, S. Cao, X. Zhou, C. Wang, Q. Jiang, H. Huang, R. Tu, S.-F. Han and Q. Wang, Water Res., 2019, 160, 339–349 CrossRef CAS PubMed.
- W. Zhou, L. Fu, L. Zhao, X. Xu, W. Li, M. Wen and Q. Wu, ACS Appl. Mater. Interfaces, 2021, 13, 10878–10890 CrossRef CAS PubMed.
- Y. Ahmed, J. Lu, Z. Yuan, P. L. Bond and J. Guo, Water Res., 2020, 179, 115878 CrossRef CAS PubMed.
- P. Xiong and J. Hu, Water Res., 2013, 47, 4547–4555 CrossRef CAS PubMed.
- Y. Chao, L. Ma, Y. Yang, F. Ju, X. X. Zhang, W. M. Wu and T. Zhang, Rep, 2013, 3, 3550 Search PubMed.
- W. Qi, W. Li, J. Zhang, X. Wu, J. Zhang and W. Zhang, Front. Environ. Sci. Eng., 2018, 13, 15 CrossRef.
- I. Vaz-Moreira, C. Egas, O. C. Nunes and C. M. Manaia, FEMS Microbiol. Ecol., 2013, 83, 361–374 CrossRef CAS PubMed.
- S. Potgieter, A. Pinto, M. Sigudu, H. Du Preez, E. Ncube and S. Venter, Water Res., 2018, 139, 406 CrossRef CAS PubMed.
- N. Kotlarz, N. Rockey, T. M. Olson, S. J. Haig, L. Sanford, J. J. LiPuma and L. Raskin, Environ. Sci. Technol., 2018, 52, 2618–2628 CrossRef CAS PubMed.
- Y. Zhu, H. Wang, X. Li, C. Hu, M. Yang and J. Qu, Water Res., 2014, 60, 174–181 CrossRef CAS PubMed.
- C. Wang, N. Moore, K. Bircher, S. Andrews and R. Hofmann, Water Res., 2019, 161, 448–458 CrossRef CAS PubMed.
- S. Min, Z. Hao, X. Bei and J. Bao, Environ. Sci. Pollut. Res., 2018, 25, 1–11 Search PubMed.
- X. Qian, Y. Ren and S. Yu, Chem. Eng. J., 2017, 330, 1203–1210 CrossRef.
- F. Zeng, S. Cao, W. Jin, X. Zhou, W. Ding, R. Tu, S. F. Han, C. Wang, Q. Jiang and H. Huang, J. Cleaner Prod., 2020, 243, 118666 CrossRef CAS.
- X. Ao, Z. Chen, S. Li, C. Li, Z. Lu and W. Sun, J. Environ. Sci., 2020, 87, 398–410 CrossRef PubMed.
- L. Mathieu, T. Paris and J. C. Block, The Structure and Function of Aquatic Microbial Communities, 2019, vol. 7, pp. 261–311 Search PubMed.
- J. El-Chakhtoura, E. Prest, P. Saikaly, M. Van Loosdrecht, F. Hammes and H. Vrouwenvelder, Water Res., 2015, 74, 180–190 CrossRef CAS PubMed.
- J. Wingender and H. C. Flemming, Int. J. Hyg. Environ. Health, 2011, 214, 417–423 CrossRef PubMed.
- R. Liu, J. Zhu, Z. Yu, D. R. Joshi, H. Zhang, W. Lin and M. Yang, J. Environ. Sci., 2014, 26, 865–874 CrossRef CAS.
- A. Rożej, A. Cydzik-Kwiatkowska, B. Kowalska and D. Kowalski, World J. Microbiol. Biotechnol., 2015, 31, 37–47 CrossRef PubMed.
- R. S. Roeder, K. Heeg, P. Tarne, J. K. Benolken, G. Schaule, B. Bendinger, H. C. Flemming and U. Szewzyk, Water Practice & Technology, 2010, 5, wpt2010082 Search PubMed.
- J. Lu, H. Y. Buse, V. Gomez-Alvarez, I. Struewing and N. J. Ashbolt, J. Appl. Microbiol., 2014, 117, 905–918 CrossRef CAS PubMed.
- M. S. Gião, S. A. Wilks and C. W. Keevil, BioMetals, 2015, 28, 329–339 CrossRef PubMed.
- H. Y. Buse, J. Lu, X. Lu, X. Mou and N. J. Ashbolt, FEMS Microbiol. Ecol., 2014, 88, 280–295 CrossRef CAS PubMed.
- S. Masters, H. Wang, A. Pruden and M. A. Edwards, Water Res., 2015, 68, 140–149 CrossRef CAS PubMed.
- H. Wang, S. Masters, Y. Hong, J. Stallings, J. O. r. Falkinham, M. A. Edwards and A. Pruden, Environ. Sci. Technol., 2012, 46, 11566–11574 CrossRef CAS PubMed.
- M. M. Moritz, H. C. Flemming and J. Wingender, Int. J. Hyg. Environ. Health, 2010, 213, 190–197 CrossRef CAS PubMed.
- J. Inkinen, B. Jayaprakash, M. Ahonen, T. Pitkänen, R. Mäkinen, A. Pursiainen, J. Santo Domingo, H. Salonen, M. Elk and M. Keinänen-Toivola, J. Appl. Microbiol., 2018, 124, 611–624 CrossRef CAS PubMed.
- Y. Chao, Y. Mao, Z. Wang and T. Zhang, Sci. Rep., 2015, 5, 10044 CrossRef CAS PubMed.
- J. Yu, D. Kim and T. Lee, Water Sci. Technol., 2010, 61, 163 CrossRef CAS PubMed.
- H. J. Jang, Y. J. Choi and J. O. Ka, J. Microbiol. Biotechnol., 2011, 21, 115–123 CrossRef CAS PubMed.
- Y. Shi, A. Babatunde, B. Bockelmann-Evans and G. Webster, Environ. Sci.: Water Res. Technol., 2019, 5, 977–992 RSC.
- B. Han, R. Chen, B. Shi, W. Xu and Y. Zhuang, J. Water Supply: Res. Technol.--AQUA, 2018, 67, 12–21 CrossRef.
- I. Douterelo, R. L. Sharpe and J. B. Boxall, Water Res., 2013, 47, 503–516 CrossRef CAS PubMed.
- K. Fish, A. M. Osborn and J. B. Boxall, Sci. Total Environ., 2017, 593–594, 571–580 CrossRef CAS PubMed.
- E. Tsagkari and W. T. Sloan, Bioprocess Biosyst. Eng., 2018, 41, 1–14 CrossRef PubMed.
- I. Douterelo, R. Sharpe and J. Boxall, J. Appl. Microbiol., 2014, 117, 286–301 CrossRef CAS PubMed.
- K. Lautenschlager, C. Hwang, W.-T. Liu, N. Boon, O. Koester, H. Vrouwenvelder, T. Egli and F. Hammes, Water Res., 2013, 47, 3015–3025 CrossRef CAS PubMed.
- P. Ji, J. Parks, M. A. Edwards and A. Pruden, PLoS One, 2015, 10, e0141087 CrossRef PubMed.
- F. Ling, R. Whitaker, M. W. Lechevallier and W.-T. Liu, ISME J., 2018, 12, 1520–1531 CrossRef PubMed.
- C. Hwang, F. Ling, G. L. Andersen, M. W. Lechevallier and W. T. Liu, Appl. Environ. Microbiol., 2012, 78, 7856–7865 CrossRef CAS PubMed.
- A. J. Pinto, J. Schroeder, M. Lunn, W. Sloan and L. Raskin, mBio, 2014, 5, e01135-14 CrossRef PubMed.
- F. Wang, W. Li, Y. Li, J. Zhang, J. Chen, W. Zhang and X. Wu, Front. Environ. Sci. Eng., 2018, 12, 6 CrossRef.
- S. J. Haig, N. Kotlarz, J. J. Lipuma, L. Raskin and M. J. Bailey, mbio, 2018, 9, e02354-17 CrossRef PubMed.
- C. Hwang, F. Ling, G. L. Andersen, M. W. Lechevallier and W. T. Liu, Appl. Environ. Microbiol., 2012, 78, 7856–7865 CrossRef CAS PubMed.
- P. Ji, W. J. Rhoads, M. A. Edwards and A. Pruden, ISME J., 2017, 11, 1318–1330 CrossRef PubMed.
- V. H. Alcocer-Yamanaka, V. G. Tzatchkov and F. I. Arreguin-Cortes, Water Resources Management, 2012, 26, 1779–1792 CrossRef.
- N. B. Hallam, J. R. West, C. F. Forster and J. Simms, Water Res., 2001, 35, 4063–4071 CrossRef CAS PubMed.
- S. L. Zhou, T. A. Mahon, A. Walton and J. Lewis, J. Hydrol., 2000, 236, 153–164 CrossRef.
- C. Nguyen, C. Elfland and M. Edwards, Water Res., 2012, 46, 611–621 CrossRef CAS PubMed.
- W. J. Rhoads, A. Pruden and M. A. Edwards, Environ. Sci.: Water Res. Technol., 2016, 2, 164–173 RSC.
- K. Lautenschlager, N. Boon, Y. Wang, T. Egli and F. Hammes, Water Res., 2010, 44, 4868–4877 CrossRef CAS PubMed.
- X. Chen, Y. Wang, W. Li, X. Zhao and Z. Ding, Environ. Res., 2020, 188, 109715 CrossRef CAS PubMed.
- P. Y. Hong, C. Hwang, F. Ling, G. L. Andersen and W.-T. Liu, Appl. Environ. Microbiol., 2010, 76, 5631–5635 CrossRef CAS PubMed.
- R. Liu, Z. Yu, H. Zhang, M. Yang, B. Shi and X. Liu, Can. J. Microbiol., 2012, 58, 261–270 CrossRef CAS PubMed.
- Q. M. Bautista-de Los Santos, J. L. Schroeder, O. Blakemore, J. Moses, M. Haffey, W. Sloan and A. J. Pinto, Water Res., 2016, 90, 216–224 CrossRef CAS PubMed.
- Y. Perrin, D. Bouchon, V. Delafont, L. Moulin and Y. Héchard, Water Res., 2019, 149, 375–385 CrossRef CAS PubMed.
- F. Ling, C. Hwang, M. W. Lechevallier, G. L. Andersen and W. Liu, ISME J., 2015, 10, 582–595 CrossRef PubMed.
- J. Inkinen, B. Jayaprakash, J. W. Santo Domingo, M. M. Keinnen-Toivola, H. Ryu and T. Pitkänen, J. Appl. Microbiol., 2016, 120, 1723–1738 CrossRef CAS PubMed.
- G. Liu, G. L. Bakker, S. Li, J. H. G. Vreeburg, J. Q. J. C. Verberk, G. J. Medema, W. T. Liu and J. C. V. Dijk, Environ. Sci. Technol., 2014, 48, 5467–5476 CrossRef CAS PubMed.
- Z. Dwidjosiswojo, J. Richard, M. M. Moritz, E. Dopp, H. C. Flemming and J. Wingender, Int. J. Hyg. Environ. Health, 2011, 214, 485–492 CrossRef CAS PubMed.
- X. Ma, G. Zhang, G. Li, Y. Wan, H. Sun, H. Wang and B. Shi, Environ. Sci.: Water Res. Technol., 2018, 4, 644–653 RSC.
- H. Zhang, Y. Tian, M. Kang, C. Chen, Y. Song and H. Li, Chemosphere, 2019, 215, 62–73 CrossRef CAS PubMed.
- D. Corsaro, G. S. Pages, V. Catalan, J. F. Loret and G. Greub, Int. J. Hyg. Environ. Health, 2010, 213, 158–166 CrossRef CAS PubMed.
- K. Huang, X. X. Zhang, P. Shi, B. Wu and H. Ren, Ecotoxicol. Environ. Saf., 2014, 109, 15–21 CrossRef CAS PubMed.
- A. Karkey, T. Jombart, A. W. Walker, C. N. Thompson, A. Torres, S. Dongol, N. T. V. Thieu, D. P. Thanh, D. T. T. Ngoc and P. V. Vinh, PLoS Neglected Trop. Dis., 2016, 10, e0004346 CrossRef PubMed.
- V. Delafont, F. Mougari, E. Cambau, M. Joyeux, D. Bouchon, Y. Héchard and L. Moulin, Environ. Sci. Technol., 2014, 48, 11872–11882 CrossRef CAS PubMed.
- F. Saleem, A. Mustafa, J. A. Kori, M. S. Hussain and M. Kamran Azim, Microb. Ecol., 2018, 76, 899–910 CrossRef CAS PubMed.
- H. M. Marshall, R. Carter, M. J. Torbey, S. Minion, C. Tolson, H. E. Sidjabat, F. Huygens, M. Hargreaves and R. M. Thomson, Emerging Infect. Dis., 2011, 17, 395–402 CrossRef PubMed.
- G. Liu, Y. Zhang, E. Van der Mark, A. Magic-Knezev, A. Pinto, B. Van Den Bogert, W.-T. Liu, W. Van Der Meer and G. Medema, Water Res., 2018, 138, 86–96 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |