 Open Access Article
Open Access ArticleChemical characterization of red cells from the black sea urchin Arbacia lixula by X-ray photoelectron spectroscopy†
Patrizia Pagliara a,
Daniela Chirizzi
a,
Daniela Chirizzi b and
Maria Rachele Guascito
b and
Maria Rachele Guascito *a
*a
aDipartimento di Scienze e Tecnologie Biologiche e Ambientali, Università del Salento, S.P. Lecce-Monteroni, Lecce, Italy. E-mail: maria.rachele.guascito@unisalento.it
bIstituto Zooprofilattico Sperimentale della Puglia e della Basilicata (IZS_PB), Via Manfredonia 20, Foggia, Italy
First published on 9th August 2021
Abstract
Red spherula cells (RSC) from sea urchin coelomic fluid have attracted great interest for their specific and intriguing properties, such as for example antimicrobial activities and immune response, that probably tie in with their red characteristic pigments. Although to date different studies have been reported aimed to chemically characterize their pigments extracted from the cells, few data are available about the chemical characterization of the cell surface. In this work, a systematic chemical characterization of the RSC surface by X-ray photoelectron spectroscopy (XPS) analysis is described. The results were compared with data on colorless cells from the same coelomic fluid sample. Our observations evidenced that the two cell types were characterized by the presence of different chemical functional groups. In particular, the colorless cells are dominated by the presence of alkyl, alcohol, amide, and carboxyl groups in accordance with other similar cell types, enriched in Na+ and Cl− ions. Traces of elements like S (sulphonates) and P (phosphates) are also present. On the other hand, the RSC in addition to the alkyl groups show a reduction in the content of amide groups, accompanied by the anomalous presence of keto-enolic groups that probably can be associated with the presence of quinones/hydro-quinones from red pigments. A chemical enrichment in elements such as Cl− and Mg2+ and sulphate groups (–R–O–SO3−), as well as the presence of sulphides and phosphates traces, is evident. The absence of carbonate groups is also observed in both cell populations, confirming the absence of sodium and magnesium carbonate salts. No traces of toxic elements (i.e., heavy metals) have been revealed.
1. Introduction
Among marine invertebrates, echinoderms are of particular interest for their phylogenetic position. Moreover, some species are also interesting from an economic point of view, being consumed as food, and even considered a delicacy in some regions. Other species have attracted attention because they represent possible sources of potential pharmacological compounds.1 As for other invertebrates, sea urchins possess a circulatory medium (i.e., coelomic fluid) which contains proteins, metabolites, and a heterogeneous population of cells (coelomocytes) that are involved in immune defense.2 Proteins and metabolites mediate humoral immunity and play important roles in defense against infections.2 On the other hand, coelomocytes are the main effector of the defense response3 carrying out functions similar to those of the vertebrate blood cells. Among coelomocytes, we can distinguish different cell types that present diverse morphologies and functions, including phagocytosis, encapsulation, clotting, cytotoxicity, wound healing among others.3 Some types of coelomocytes can be found in all classes, while others have been considered specific to certain echinoderm classes. Red spherula cells (RSC) are a cell type generally found among the class of Echinoidea (commonly known as sea urchins) and characterized by the presence of structurally related pigments, compounds mainly based on juglone, anthraquinone and β-carotene.4 RSC appear colored red due to the abundant presence of echinochrome A,5 the 1,4-napthoquinone pigment, stored within their cytoplasmic vesicles (or granules).6,7 Echinochrome A is degranulated in presence of bacteria8 and has antimicrobial properties against both Gram-positive and Gram-negative bacteria.7,9,10 Furthermore, RSC accumulation has been observed around injuries and sites of infection11,12 suggesting that these cells and echinochrome A play a role in the immune response in adult sea urchins. Literature data on echinoderms red cells refer to the pigment present inside the cells. Recently, Hira and Coworker (2020),13 by developing a new isolation method obtained a pure population of red cells from the sea urchin Strongylocentrotus droebachiensis. This allowed them to profile the naphthoquinone content in the red cells and to identify sulphated derivatives of different spinochrome and spinochrome dimers.13Currently, some information is available on the pigment contained in the RSC, while little is known about the chemical characteristic of the cell surface that is involved in the recognition of foreign material as well as in most cell defense activities. Knowing the chemical characteristics of the surface can help to better understand which mechanisms are activated in the interactions between the RSCs and all that is non-self and consequently which are the components responsible for triggering the recognition process that leads the cell to activate a series of reactions that allow the neutralization of the danger.
Cell surface can be characterized from a chemical point of view by XPS technique that allows avoiding pretreatment that could damage the cells. In fact, this technique allows to analyze, also in terms of chemical speciation, all the chemical elements present in traces, except for H and He, in different types of solid-state samples as thin layers, both inorganic and organic nature, in atomic percentage concentration (at%) not less than 0.01–0.1% depending on the specific detected element. Specifically, the probing depth of XPS is ∼10 nm, which makes it suitable to analyze of the near-surface region of materials. A cell is typically a few micrometers thick. Therefore, in cells characterization XPS could not study the cell cytoplasm but can represent a powerful method to explore cell surface (the membrane, less than 10 nm thick, and the proteins embedded in it).14 To date, this analytical technique has been widely used to study bio-organic systems materials as for example microorganisms and their related systems, as biofilms, extracellular polymeric substances, and bio-adhesions.15 More recently, analysis by nano-ESCA (Electron Spectroscopy for Chemical Analysis) of biological samples, such as blood cells (i.e., neutrophil and platelets) immobilized on suitable surfaces, have been reported for chemical and morphological cell studies, showing that these typologies of bio-organic sample can be analyzed in a condition of ultra-high-vacuum (UHV).16 As an example, in a successful study, the chemical differences between the membrane and the cytoplasm of dehydrated neuronal cells immobilized on gold surface have been reported.14 However, to our knowledge it does not appear that XPS studies has been applied specifically and systematically to characterize purely biological systems, as coelomocytes from sea urchins, bio-adsorbed on a coverslip and de-hydrated in N2 atmosphere. The aim of the present work was to analyze, for the first time, the RSC from the black sea urchin Arbacia lixula using XPS. The data were then compared with the properties of colorless cells isolated from the same coelomic fluid to highlight macroscopic chemical differences between the two cell populations, including for example the observation of physiological and toxic elements (i.e., heavy metals)17 and to understand if the different role played by the two cell types is due to intrinsic and characteristic chemical properties of each of them.
2. Material and methods
2.1. Sea urchin sampling and red cells isolation
Adult specimens of Arbacia lixula were collected in Porto Cesareo (Northern Ionian Sea, Apulia, Italy) by SCUBA diving, at a depth of 5–10 m.The sea urchin coelomic fluid was harvested directly in an isosmotic anticoagulant solution (20 mM Tris, 0.5 M NaCl, 70 mM EDTA, pH 7.5) (ISO-EDTA) by cutting the peristomal membrane. After centrifugation (900 × g for 10 min at 4 °C), the pellet, containing coelomocytes was washed in ISO-EDTA and resuspended at 5 × 106 cells per mL in ISO-EDTA. Coelomocytes number was calculated by a hemocytometer chamber and cell vitality evaluated by Trypan blue exclusion test. Four milliliters of coelomocytes suspension were layered on the top of an iodixanol (Optiprep; Nycomed Oslo, Norway) discontinuous density gradient prepared as follow: one milliliter of 10, 20, 30 and 70% (v/v) iodixinol stock solutions in 0.5 M NaCl containing 10 mM EDTA were layered into a 15 mL centrifuge tube. The gradient was then centrifuged in a swing-out rotor (800 × g for 30 min at 7 °C). The resulting cell populations on the top of 10, 20, 30 and 70% iodixanol layer, referred to as bands P1, P2, P3 and P4 (Fig. 1b), respectively were collected. Each cell population was gently removed, washed with ISO-EDTA and identified according to Smith 1991.18
2.2. XPS measurements
An AXIS ULTRA DLD (Kratos Analytical) was employed for XPS analysis. The spectrometer was equipped with an Al Kα (1486.6 eV) monochromatic source (line width 0.2 eV), operating at 10 kV and 15 mA (150 W) and configured to provide X-ray spot size of approx. 1 mm × 2 mm on the sample surface. The measured pressure in the analysis chamber was in average 1.0 × 10−9 torr during spectra acquisition. Survey spectra were collected typically in a double scan, in fixed analyzer transmission (FAT) mode at a pass energy of 160 eV with acquisition time of 241 s, using a step size of 1 eV over the energy range of 0–1400 eV. High-resolution (HR) regions (C 1s, O 1s, N 1s, S 2p, P 2p, Ca 2p, K 2p, Mg 2s, and Cl 2p) were acquired in FAT mode at a pass energy of 20 eV, with different acquisition time, depending on the energy window selected for the single/multiple elements analysis and on the sweeps number, decided according to the quality of the signal. Typically, the acquisition time for C 1s and O 1s photo-peaks was 10 min and for N 1s + Ca 2p photo-peaks was 1 h, using a step size of 0.1 eV. The analyzed area, obtained by selecting the “hybrid lens mode”, in all experiments was 700 μm × 300 μm and represents a portion of the X-ray spot area on sample surface. During the data acquisition a careful charging effect correction has been made by using the electron flood gun with which the AXIS spectrometers is equipped19 during all the XPS experiments. In detail, to prevent surface charge build-up, charge neutralization was used with a filament current, filament bias voltage, and charge balance voltage of 2.0 A, 1.3 V and 3.6 V, respectively. Resolution for insulator polyethylene terephthalate (PET) standard specifications is guaranteed on the ester component peak (O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –O). The full width at half maximum (FWHM) of related C 1s at large area mode (hybrid lens mode) is 1.0 eV at pass energy 20 eV, and 1.3 eV at pass energy 160 eV. The PET is used as standard to guarantee neutralizer performance. Resolution for conductor Ag specifications is guaranteed on the Ag 3d5/2 peak. The FWHM of the Ag 3d5/2 signal in our experimental conditions, large area mode acquisition (hybrid lens mode), is 0.47 eV at pass energy 20 eV, and 0.73 eV at pass energy 160 eV.
–O). The full width at half maximum (FWHM) of related C 1s at large area mode (hybrid lens mode) is 1.0 eV at pass energy 20 eV, and 1.3 eV at pass energy 160 eV. The PET is used as standard to guarantee neutralizer performance. Resolution for conductor Ag specifications is guaranteed on the Ag 3d5/2 peak. The FWHM of the Ag 3d5/2 signal in our experimental conditions, large area mode acquisition (hybrid lens mode), is 0.47 eV at pass energy 20 eV, and 0.73 eV at pass energy 160 eV.
The binding energy scale of HR spectra was corrected for charging with respect to the C 1s component assigned to aliphatic carbon at 285.0 eV of adsorbed organic molecules onto sample surfaces as an ultra-high vacuum (UHV) contaminant, in accordance with ISO 19318:2004.20 The peaks assignment refers to literature data, NIST standard reference database21 and experimental standards (i.e., MgCO3, dopamine and polydopamine). The oxygen chemical attributions were confirmed by using the modified Auger parameter α′, an internal parameter not affected from charging effects.22 Difference in α′ experimental data refer to different chemical state of the same element in different samples22 (Wagner plot – NIST database).21 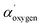 values have been calculated by using the binding energy of the O 1s core line and the kinetic energy of the respective O (KLL) Auger peak obtained on the same survey scan. Details about Auger parameter α′ calculation have been reported in ESI file and Table S1.†
values have been calculated by using the binding energy of the O 1s core line and the kinetic energy of the respective O (KLL) Auger peak obtained on the same survey scan. Details about Auger parameter α′ calculation have been reported in ESI file and Table S1.†
The software New Googly (homemade program, kindly provided by Salvi and Castle) and fully described in Salvi and Castle, (1998),23 has been used for data analysis and peak fitting to elaborate the XPS high-resolution peak regions. The program uses an iterative non-linear least-squares fitting process to modify the peak parameters until the deviation between the synthesized peak envelope and the experimental XPS spectrum is minimized. This program uses a self-consistent Shirley-type background officially recognized by the International Organization for Standardization (designation: E995-16), which allows background correction, as well as curve-fitting of photoelectronic peaks. The software consents to associate the background with the Gaussian/Lorentzian (Gw/Lw) peak shapes. The Gw/Lw function, known as the Voigt function, describes the basic shape of the XPS peak recorded in a spectrometer. In fact, the uncertainty principle gives the initial energy level a Lorentzian energy distribution modified by instrumental and other factors (such as phonon broadening) to give a Gaussian contribution. The synthetic peak contains a mixture of Gaussian and Lorentzian components (Gw/Lw mix), to approximate the shape of the photo-lines after transmission through the analyzer, to which has been added an energy-loss background using the Shirley algorithm,24 thus creating the characteristic photoelectron shape. No peak tail was used. The parameters which can be varied are the Gw/Lw mix, the position of each peak, their FWHM and the amount of background rise across the peak. The goodness of the fit is determined by the value of the chi-squared value (χ2) parameter available from the program. The fit parameters used in this investigation to completely describe the peak shapes are reported in Tables S2 and S3,† as obtained on both sample cell typologies, respectively. Any of these peak parameters can be individually fixed to a preset value or left free to vary but for an effective use it is important to minimize the number of free variables. In detail, for the given core line envelope (i.e., C 1s peaks components) we have fitted the spectral features with a Voigt function constrained to the same Lw/Gw mix ratio associated to the appropriated FWHM as obtained by optimizing the fit parameters. Lw/Gw mix ratio can be different for different elements as far as the FWHM.25 Peak areas as obtained from peak fitting, were converted to atomic percent composition (at%) using the suitable sensitivity factors (SF) to guarantee the correct elemental mass balance.26,27 All reported atomic percentage compositions were representative of the mean values obtained on three different spots of the analyzed colorless and red cells samples, with the relative standard errors that represent the variability observed between these three different randomly selected sample spots on the colorless and red cells. XPS elemental composition allowed to identify and to quantify the elements present in the analyzed samples.
2.3. Red and colorless cells preparation for XPS analysis
P1 (colorless cells) and P4 (red cells) samples were washed in isotonic solution (20 mM Tris, 0.5 M NaCl, pH 7.5) and 60 μL of the cell suspension were coated on a coverslip. Cells were incubated for 1 h in a humid chamber at room temperature to allow their adhesion on the coverslip. Before XPS analysis, samples were N2-dried, washed with distilled water and re-dried by air. In this way a dessicate and well compact bio-adsorbed sample was obtained. As a control a coverslip surface treated as cell samples was also acquired, to exclude chemical interferences from cell support materials. After, all samples were fixed with a double conductive tape mounted on the same sample holder and introduced in the pre-analysis chamber under nitrogen flow. After 12 h, at constant pressure of almost 10−8 torr, samples were transferred in the analysis chamber. In this way, any cell not closely bound to the substrate was removed as far as the presence of residual moisture is reduced. Two spots randomly selected respectively on white and red cells samples, were analyzed in succession alongside the same day. These analyses were replicated for two successive days on two other different pair's points randomly selected for both cells. Between two successive analyses on the same sample, a time of at least 24 hours elapses without the sample being irradiated by X-rays. In this way, it was possible to compare results related to both cell populations acquired at different grade of dehydration and contamination on different spots. The original C 1s, O 1s and N 1s peak signals as acquired in triplicate (three independent sample spots) on both analyzed cells populations were compared as shown in ESI file (Fig. S1 and S2†). Peaks energies and shapes of the same peak signal are relatively stable changing the analysis sample spot position on the same cells populations' sample.To check the lifetime of the sample under irradiation we have monitored the C 1s signals that were stable for an analysis time maximum of 4–5 h under X-ray beam. Typically, after this time the C 1s shows a decrement of the asymmetries around 288 eV. This data gives account of a certain degradation grade, however not associated to the cell destruction. This can be appreciated in Fig. S5 (ESI file†) that shows for comparison the C 1s peaks acquired respectively at the beginning and at the end of the complete XPS analysis of the red cells sample spot. Therefore, the reported data represent the average obtained on three independently selected spots on each of the cell populations analyzed in sequence, with care to avoid analyzing area spot affected by X-rays in previous acquisition. The homogeneity and compactness of cell deposited layers were evident by the absence in the XPS spectra of any significant signals of Si 2p and Si 2s peaks due to the silicon coming up from the support (coverslip).
3. Results and discussion
3.1. Cell populations of Arbacia lixula
Sea urchin coelomic fluid host several circulating cells with an important role in defense mechanisms. Due to the nutritional, immunological and homeostatic status of the individual sea urchin, the proportions of each type of coelomocyte in the coelomic fluid vary considerably both inter- and intra-individual.3 However, in the black sea urchin A. lixula, coelomic fluid is characterized by the presence of a significant number of red cells. In our study, we have separated the different cell types as showed in Fig. 1a by a density gradient and identified four different cell populations, each featured of an enrichment of a particular cell type.Cell population P1 was mainly composed of amoebocytes (92 ± 2.3%), that are uncolored circulating cells (Fig. 1c). The P2 and P3 cell populations present mainly vibratile cells (73.5 ± 2.7%) and colorless spherulocytes (85.3 ± 5.1%) respectively. In P4 cell population (Fig. 1d), red cells were dominant (95.2 ± 7.4%). As shown in Fig. 1a, c and d, the reported protocol used to separate colorless and red cells is suitable to obtain intact whole cells and simultaneously completely remove suspended cell debris. Rare red cells were present in P2 and P3 populations, but completely absent in P1 population that was chosen as a comparison.
The presence of red cells in the sea urchin coelomic fluid is intriguing for several reasons. As an example, in some sea urchin species, the red cells are involved in the inflammation29 and in the immune response,11,12 presenting a bactericidal activity.7–9 In addition, when their number increases, they are considered a biomarker of stress.28 Red cells are also able to migrate towards wounds, infections, and tissue grafts.6,12,30 Furthermore, considering the role of surface in recognize non-self material, we consider important to deepen knowledge about its chemical composition, also to characterize the nature of bonds that RSCs establish in cell–cell or cell–material interaction.
3.2. Red and colorless cells chemical composition by XPS
XPS has been performed to evaluate differences on the chemical composition of colorless and red cells. An initial estimation of differences in chemical composition between the two cell populations analyzed was obtained by comparing their respectively survey spectra (Fig. 2). These spectra show the photoelectron peak signals of the characteristic chemical elements with associated relevant Auger (A) peaks. As expected, the most intense signals for both cell populations were the carbon (C 1s) and oxygen (O 1s) peaks. Relevant Auger C (KLL) most intense components are centered at binding energy 1225.3 eV (261.3 eV in kinetic energy) and 1225.0 eV (261.6 eV in kinetic energy) according to reported literature positions31 for both white and red cells, respectively. Instead the Auger O (KLL) components are centered at 976.6 eV (510.0 eV in kinetic energy) and 976.0 eV (510.6 eV kinetic energy)21 for white and red cells, respectively. The nitrogen N 1s peak signal was also present in both samples. In addition different elements like Na, Cl and Mg have been detected. Among these last elements Mg (Mg 2s, Mg 2p, Mg 1s and Mg(A)) peaks are dominant on RSC samples (Fig. 2 and inset, lines B) while Na (Na 1s, Na 2s Na 2p and Na(A)) peaks are dominant on colorless cells (Fig. 2 and inset, lines A). Chlorine ions are present on both cell samples. The absence in the spectra of all analyzed samples of detectable silicon peaks (Si 2p and Si 2s) from the glass surface used as support (Fig. 2 inset), confirms the uniformity of the cellular layer distribution sufficient to avoid significant interferences of silicon signal. In addition, the absence of iodine peak signals (i.e. I 3d) confirms the effectiveness of washing with water in removing the iodixanol. | ||
| Fig. 2 Survey XPS spectra for colorless (A) and red (B) cells samples. Staked curves have been obtained by shifting red cells original counts along the Y-axis of a constant value of 700000 cps. As obtained original data can be appreciated in Fig. S4.† | ||
To obtain the surface atomic% composition (at%) of the colorless and red cell populations, high resolution (HR) regions of all peaks of interest have been acquired and results reported in Fig. 3. The composition of colorless and red cells, in terms of C, O and N total content was relatively different, considering that the RSC showed a lower carbon and nitrogen content than the colorless one (55% vs. 64% carbon and 4.7 vs. 5.9 nitrogen, respectively) and an enrichment in oxygen content (20.5% vs. 18%), confirmed by the (Otot + Ntot)/Ctot ratios that is 0.37 for colorless cells and 0.46 for red. Instead the ratio between (CO + CN)/Ctot, CO and CN representing the carbon bonded to oxygen and nitrogen respectively (i.e. all components with BE higher than 285 eV), is ∼0.40 for both cells. This value of ∼0.40 smaller than 0.46, for red cells suggests the presence of oxygen in excess not associated to carbon. Based on this data a residual moisture could not be excluded. However, considering the presence of sulphate groups (at% 1.2) the oxygen excess can be associated principally to this species. Finally, the content in elements like Na+, Mg2+ and Cl− ions between the two different cell populations was different. Colorless cells were enriched in Na+ and Cl− ions (6.4% and 4.5% respectively), while red cells in Mg2+ and Cl− ions (6.7% and 10.7% respectively).
Based on these data, it was evident that the quantitative detectable sodium peak signal (Na 1s) related to Na+ ions was present only in the colorless cell samples, while the magnesium, present only in traces in the colorless cell samples, was dominant on the red one. In both samples the chlorine content seems correlated to the Mg2+ and Na+ positive ions components, respectively. Finally sulfureous and phosphorous species were present as traces in all analysed samples and are quantifiable only if acquired at HR relevant spectral region (i.e. S 2p and S 2s, P 2p and P 2s).
3.3. XPS chemical speciation of colorless and red cells populations
To better chemically characterize the cells composition, speciation of elements has been obtained by using an appropriate procedure of curve fitting of acquired high resolution peak signals for C 1s, N 1s, O 1s, Cl 2p, Na 1s, Mg 1s, P 2p, S 2p and K 2p peaks for both colorless and red cells. Results in terms of chemical shift and relevant at% different chemical components composition have been reported in Table 1.| Elements | Assignment | Colorless cells BE (eV) at (%) | Red cells BE (eV) at (%) |
|---|---|---|---|
| C 1s | ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –C/ –C/![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C |
285.0 (39.5 ± 4.6) | 285.0 (33.3 ± 0.8) |
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) α–Cqu α–Cqu![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
— | 285.9 (7.7 ± 0.4) | |
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –N/ –N/![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –N–C –N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/ O/![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –O–H(C) –O–H(C) |
286.4 (15.5 ± 0.8) | — | |
C–O–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/N– O/N–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O–N O–N |
— | 286.9 (8.5 ± 0.2) | |
C–N–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/ O/![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/O– O/O–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –O –O |
288.0 (6.5 ± 0.5) | 288.3 (2.8 ± 0.2) | |
![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OO(H)/C (π–π* satellite) OO(H)/C (π–π* satellite) |
289.0 (2.3 ± 0.3) | 289.3 (3.3 ± 0.3) | |
| N 1s | C–![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) /C– /C–![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) –C –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
399.8 (5.4 ± 0.6) | — |
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) –C –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
— | 400.5 (3.2 ± 0.3) | |
O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C– C–![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) –C –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
— | 401.5 (1.3 ± 0.4) | |
C–![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) H3+ H3+ |
401.8 (0.3*) | — | |
R4![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) + + |
402.7 (0.1*) | 403.0 (0.2*) | |
| O 1s | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) /N–C /N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) /C /C![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) O(H) O(H) |
531.4 (9 ± 1) | — |
C–![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) –H(C)/ –H(C)/![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) –C–O/ –C–O/![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) –C –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/Cqu O/Cqu![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) |
532.7 (10 ± 2) | 532.5 (14 ± 1) | |
CAr–![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) H H |
— | 533.6 (7 ± 1) | |
| Cl 2p | Cl− | 198.8 (4.5 ± 2) | 199.0 (10.7 ± 0.4) |
| Na 1 s | Na+ | 1071.6 (6.5 ± 2) | 1072.5 (0.4 ± 0.1) |
| Na (K1L1L23) | Na+ | 535.8 | |
| Mg 1s | Mg2+ | 1304.5 (0.36*) | 1304.8 (6.7 ± 0.2) |
| P 2p | PO43− | 133.0 (0.40*) | 135.1 (0.2*) |
| S 2p | C–![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) |
— | 163.9 (0.04*) |
C–![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) O3− O3− |
168.2 (0.29*) | — | |
C–O–![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) O3− O3− |
— | 169.8 (1.2 ± 0.1) | |
| K 2p | K+ | 292.8 (0.08*) | — |
3.4. Carbon functional groups
In Fig. 4 the typical peak fit of C 1s signals of colorless (panel A) and red (panel B) cells have been reported. The original data has been reported in triplicate in Fig. S1 and S2,† respectively. The fit of K 2p component was also done when potassium element has been detected. As evident, the C 1s region at low binding energy can be fitted with a dominant component at 285.0 eV, attributed to aliphatic (sp3)/aromatic (sp2) organic carbon (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –C/
–C/![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C),32 for both samples. The evident asymmetries at higher binding energies have been fitted with three different C 1s components for colorless cell samples, while four components were needed for the red one. Also the relative at% distribution was quite different between different analysed cells. Used fitting parameters were reported in Tables S2 and S3.† To quantify C 1s signal components in colorless cells, carbon was classified into four different typologies (Fig. 4 A): (1) the already commented dominant aliphatic/aromatic carbon component (285.0 eV); (2) the amine/alcohol/ether not resolved carbon components (
C),32 for both samples. The evident asymmetries at higher binding energies have been fitted with three different C 1s components for colorless cell samples, while four components were needed for the red one. Also the relative at% distribution was quite different between different analysed cells. Used fitting parameters were reported in Tables S2 and S3.† To quantify C 1s signal components in colorless cells, carbon was classified into four different typologies (Fig. 4 A): (1) the already commented dominant aliphatic/aromatic carbon component (285.0 eV); (2) the amine/alcohol/ether not resolved carbon components (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –N/–
–N/–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –N–C
–N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/
O/![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –O–H(C), 286.4 eV),15,33 (3) carbonyl, amide and (hemi)-acetal carbon (
–O–H(C), 286.4 eV),15,33 (3) carbonyl, amide and (hemi)-acetal carbon (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/C–N–
O/C–N–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/O–
O/O–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –O, 288.0 eV);15 (4) carboxyl carbon (
–O, 288.0 eV);15 (4) carboxyl carbon (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OO(H), 289.0 eV).15,34 Instead, in red cells, the carbon was now classified in five components (Fig. 4, panel B), the first one at 285.0 eV being aliphatic/aromatic carbon (1). The second component (2) at 285.9 eV, characteristic of red cells could be tentatively attributed to the presence of α-carbon in carbon double bonded with oxygen in quinone-like ring (here reported as
OO(H), 289.0 eV).15,34 Instead, in red cells, the carbon was now classified in five components (Fig. 4, panel B), the first one at 285.0 eV being aliphatic/aromatic carbon (1). The second component (2) at 285.9 eV, characteristic of red cells could be tentatively attributed to the presence of α-carbon in carbon double bonded with oxygen in quinone-like ring (here reported as ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) α–Cqu
α–Cqu![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O).35 The third component (3), that could be attributed to amide/alcohol carbon, shows a significant positive chemical shift with respect to the same component in colorless cells (286.9 eV vs. 286.4 eV). Probably the signal was due to the overlap of alcohol carbon with ester (C–O–
O).35 The third component (3), that could be attributed to amide/alcohol carbon, shows a significant positive chemical shift with respect to the same component in colorless cells (286.9 eV vs. 286.4 eV). Probably the signal was due to the overlap of alcohol carbon with ester (C–O–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and/or diamide (N–
O) and/or diamide (N–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) O–N) carbon.15 The absence of amide carbon is coherent with the absence of the related nitrogen peak signals of peptidic carbonyl group (see oxygen and nitrogen discussion reported below). The attribution of the forth (4) component at 288.3 eV, is the same as for colorless cells: carbonyl (
O–N) carbon.15 The absence of amide carbon is coherent with the absence of the related nitrogen peak signals of peptidic carbonyl group (see oxygen and nitrogen discussion reported below). The attribution of the forth (4) component at 288.3 eV, is the same as for colorless cells: carbonyl (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and (hemi)-acetal carbon (O–
O) and (hemi)-acetal carbon (O–![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –O) contribution, even if a drastic decrement in (
–O) contribution, even if a drastic decrement in (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) component was observed (2.8 at% vs. 6.5 at%). The fifth (5) component at 289.3 eV, has been attributed to the π–π* satellite transitions associated to the aromaticity36 of naphthoquinone pigments. This is also in according to the discussion in the attribution of O 1s, where no detectable carboxyl oxygen groups were observed. This data, also taking into account the loss in amide nitrogen content (3.2 at% vs. 0.0 at%), confirms the reduced peptidic content of the red cells surface in comparison with the colorless one.
O) component was observed (2.8 at% vs. 6.5 at%). The fifth (5) component at 289.3 eV, has been attributed to the π–π* satellite transitions associated to the aromaticity36 of naphthoquinone pigments. This is also in according to the discussion in the attribution of O 1s, where no detectable carboxyl oxygen groups were observed. This data, also taking into account the loss in amide nitrogen content (3.2 at% vs. 0.0 at%), confirms the reduced peptidic content of the red cells surface in comparison with the colorless one.
 | ||
| Fig. 4 XPS high-resolution region of C 1s (panels A and B), N 1s (panels C and D) and O 1s (panels E and F) for colorless and red cells, respectively. | ||
3.5. Nitrogen functional groups
Fig. 4 (panel C) shows N 1s peak signals, relevant to colorless cells. The original data has been reported in triplicate in Fig. S1.† According with these peak shapes, N 1s peak signals were fitted with three different nitrogen components: a dominant amine/amide nitrogen component (C–![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) /C–
/C–![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) –C
–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, 399.8 eV),15 a protonated α-amine nitrogen component (C–
O, 399.8 eV),15 a protonated α-amine nitrogen component (C–![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) H3+, 401.8 eV),28 and a quaternary ammonium component (R4–N+, 402.7).15,21 This result indicates that nitrogen was present in the bioorganic matrix, prevalently as amine/amide groups, most likely originating from amino acids in peptidic membrane components, where only a small percentage of the constituent amino acids is present in stable zwitterionic forms.34 Regarding red cells, as evidenced in Fig. S2,† the three replicates show a shift in binding energy, of ∼0.5 eV if compared with colorless cells. Particularly, the N 1s signal related the second spot also shows an important asymmetry at the highest binding energies. However, all three nitrogen peaks were fitted with three components (Fig. 4, panel D). The most intense and well resolved peak is at 400.5 eV. The second one at 401.5 eV is more variable in intensity between three spots. However, all three peaks were fitted by using components with the same FWHM (compare as example Fig. 4, panel D and Fig. S3†). Differences in the asymmetries of these peaks can be related to the variability in nitrogen composition of different spots. Peak fitting parameters were reported in Table S3.† The observed chemical shift with respect to the typical amine/amide binding energies values reported on colorless cells, as well as on other types of analyzed cell membranes, are difficult to be assigned based on the available reference literature.15 However, tentatively they can be assigned to nitrogen in heterocyclic compounds, as pyrimidine and/or purine bases35 (here reported as C
H3+, 401.8 eV),28 and a quaternary ammonium component (R4–N+, 402.7).15,21 This result indicates that nitrogen was present in the bioorganic matrix, prevalently as amine/amide groups, most likely originating from amino acids in peptidic membrane components, where only a small percentage of the constituent amino acids is present in stable zwitterionic forms.34 Regarding red cells, as evidenced in Fig. S2,† the three replicates show a shift in binding energy, of ∼0.5 eV if compared with colorless cells. Particularly, the N 1s signal related the second spot also shows an important asymmetry at the highest binding energies. However, all three nitrogen peaks were fitted with three components (Fig. 4, panel D). The most intense and well resolved peak is at 400.5 eV. The second one at 401.5 eV is more variable in intensity between three spots. However, all three peaks were fitted by using components with the same FWHM (compare as example Fig. 4, panel D and Fig. S3†). Differences in the asymmetries of these peaks can be related to the variability in nitrogen composition of different spots. Peak fitting parameters were reported in Table S3.† The observed chemical shift with respect to the typical amine/amide binding energies values reported on colorless cells, as well as on other types of analyzed cell membranes, are difficult to be assigned based on the available reference literature.15 However, tentatively they can be assigned to nitrogen in heterocyclic compounds, as pyrimidine and/or purine bases35 (here reported as C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) –C
–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and O
O and O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–
C–![[N with combining low line]](https://www.rsc.org/images/entities/char_004e_0332.gif) –C
–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). These results are in agreement with a reduced content in detectable proteic component in the red cell surface compared to colorless cell population where the nitrogen components are prevalently amine/amide.
O). These results are in agreement with a reduced content in detectable proteic component in the red cell surface compared to colorless cell population where the nitrogen components are prevalently amine/amide.
3.6. Oxygen functional groups
Actually also the oxygen peak signal shapes show marked differences between colorless and red samples. This can be observed in original data reported in triplicate in Fig. S1 and S2.† According, all characteristic O 1s peak signals from colorless cells, have been fitted with two principal components, where the first at 531.4 eV (Fig. 4, panel E) originates from oxygen involved in a double bond with carbon in carbonyl, amide and carboxyl groups (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) /N–C
/N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) /C
/C![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) OH), and the second at 532.7 eV is attributed to oxygen involved in the single bond in alcohol/acetal and carboxyl groups (C
OH), and the second at 532.7 eV is attributed to oxygen involved in the single bond in alcohol/acetal and carboxyl groups (C![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) H/
H/![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) –C–
–C–![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) /CO
/CO![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) H). A contribution coming from (phospho)-ester and sulfonate groups, can be also considered, according to phosphorus and sulphur detected elements. These attributions, together with carbon and nitrogen functional groups designations, are consistent with polysaccharides or peptides/proteins structurally present in external cellular membrane and already highlighted in previous XPS cells surface characterizations.15,34,37,38 The characteristic averaged oxygen ratio content (
H). A contribution coming from (phospho)-ester and sulfonate groups, can be also considered, according to phosphorus and sulphur detected elements. These attributions, together with carbon and nitrogen functional groups designations, are consistent with polysaccharides or peptides/proteins structurally present in external cellular membrane and already highlighted in previous XPS cells surface characterizations.15,34,37,38 The characteristic averaged oxygen ratio content (![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) –C/
–C/![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) obtained from the fitting is typically near 1
C) obtained from the fitting is typically near 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. A third weak component found at higher binding energy (535.8 eV), was attributed to Na (K1L1L23) Auger peak21 coupled with relevant Na 1s peak signal at 1071.6 eV. The used fitting parameters were reported in Table S1.†
1. A third weak component found at higher binding energy (535.8 eV), was attributed to Na (K1L1L23) Auger peak21 coupled with relevant Na 1s peak signal at 1071.6 eV. The used fitting parameters were reported in Table S1.†
In RSC samples, the typical O 1s peak fit shape is quite different from colorless cells (Fig. 4, panel F). In fact, the O 1s asymmetric peak has been fitted by using two components, where the most intense component is at 532.5 eV while a second new one component is observed at 533.6 eV with an oxygen ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. In addition the absence of detectable oxygen double bonded to carbon (C
1. In addition the absence of detectable oxygen double bonded to carbon (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) ) component at 531.4 eV, characteristic of colorless cells, confirms the absence of prevalent peptidic bond on red cell membrane surface. Indeed, based on the binding energy values, the peak at 532.5 eV is attributed principally to alcohol oxygen single bonded (C–
) component at 531.4 eV, characteristic of colorless cells, confirms the absence of prevalent peptidic bond on red cell membrane surface. Indeed, based on the binding energy values, the peak at 532.5 eV is attributed principally to alcohol oxygen single bonded (C–![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) H), with a possible contribution from quinoid oxygen double bonded to carbon (here reported as Cqu
H), with a possible contribution from quinoid oxygen double bonded to carbon (here reported as Cqu![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) ). The typical component at highest binding energy 533.6 eV has been attributed to oxygen single bonded to arylic carbon (CAr–
). The typical component at highest binding energy 533.6 eV has been attributed to oxygen single bonded to arylic carbon (CAr–![[O with combining low line]](https://www.rsc.org/images/entities/char_004f_0332.gif) H). These attributions compatible with naphthoquinone pigments presence, considering the high level of discordance data reported in literature, particularly for O 1s at highest binding energy (533.0–534.0 eV), have been confirmed by comparing XPS values experimentally observed for dopamine powder and polidopamine films, used as references, for quinole and quinone oxygen structures binding energy (data not shown). In details, dopamine O 1s peak was dominated from a component at 533.4 eV attributed to hydroquinone oxygen typic of dopamine aromatic ring. Polidopamine O 1s peak, was fitted with two components: the awaited hydroquinone oxygen component at 533.4 eV, and the component at 532.5, attributed to quinone present in polidopamine characteristic aromatic ring. The component at 532.5 eV is also compatible with oxygen carbon double bonded in imide groups, according to carbon and nitrogen attributions. To confirm the chemical shifts attributed to O 1s peak components in red cells population respect to white cells, we have monitored the modified Auger parameter α′ for oxygen, as an internal parameter not affected from charging effects.20–22 In detail,
H). These attributions compatible with naphthoquinone pigments presence, considering the high level of discordance data reported in literature, particularly for O 1s at highest binding energy (533.0–534.0 eV), have been confirmed by comparing XPS values experimentally observed for dopamine powder and polidopamine films, used as references, for quinole and quinone oxygen structures binding energy (data not shown). In details, dopamine O 1s peak was dominated from a component at 533.4 eV attributed to hydroquinone oxygen typic of dopamine aromatic ring. Polidopamine O 1s peak, was fitted with two components: the awaited hydroquinone oxygen component at 533.4 eV, and the component at 532.5, attributed to quinone present in polidopamine characteristic aromatic ring. The component at 532.5 eV is also compatible with oxygen carbon double bonded in imide groups, according to carbon and nitrogen attributions. To confirm the chemical shifts attributed to O 1s peak components in red cells population respect to white cells, we have monitored the modified Auger parameter α′ for oxygen, as an internal parameter not affected from charging effects.20–22 In detail,  = 1041.7 ± 0.5 eV value obtained for colorless cells is within the values range 1041.5–1042.5 eV, attributed to oxygen involved in double bond with carbon in carbonyl and carboxyl groups and/or in single bond in alcohol/acetal and carboxyl groups, according to Wagner plot (NIST standard reference database).21 Instead, red cells
= 1041.7 ± 0.5 eV value obtained for colorless cells is within the values range 1041.5–1042.5 eV, attributed to oxygen involved in double bond with carbon in carbonyl and carboxyl groups and/or in single bond in alcohol/acetal and carboxyl groups, according to Wagner plot (NIST standard reference database).21 Instead, red cells  = 1042.9 ± 0.3 is near to the experimental value measured on the dopamine used as standard (
= 1042.9 ± 0.3 is near to the experimental value measured on the dopamine used as standard ( = 1042.8 eV) confirming the chemical oxygen attributions.
= 1042.8 eV) confirming the chemical oxygen attributions.
The different behavior between red and colorless cells can be explained by assuming that different chemical species are distributed on related cellular membranes. Principally, the O 1s peak components in RSC could be attributed to the presence of napthoquinones (i.e. echinochrome A), that are responsible of their red distinct coloration (packaged within cytoplasmic vesicles or granules).6 Moreover the colorless cells are characterized, from oxygen expected functional groups compatible with protein, phospholipide and (lipo)-polysaccharides molecules, according to the cellular membrane surface composition in the external depth of 5–10 nm detectable by XPS analysis on similar cells.14,16,38
3.7. Chlorine, sodium and magnesium ions
As stated before, both cell populations are characterized from a quantitative presence of chlorine species. Typical Cl 2p peak signals have been reported in Fig. 5 (panels A and B). Relevant peak fit consisted of a spin–split doublet, Cl 2p1/2 and Cl 2p3/2 of 1.65 eV energy splitting and a relative intensity ratio of about 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. Based on Cl 2p3/2 binding energies (198.8–199.0 eV), these signals have been attributed to Cl− ions.19 However, it is important to point out that the peak fit shapes differ in FWHM between red and colorless cells. In detail, the best fit of colorless cells Cl 2p peak was obtained with FWHM 1.2 eV, otherwise a FWHM of 1.5 eV, was necessary to fit the Cl 2p peak shape for red cells samples. This behavior can suggest that Cl− ions chlorine species present on both samples are not equivalent because a chemically different environment. In fact, the colorless cells are rich in Na+ counter-ions, (Fig. 5, panel C and Table 1)14,21, with Cl−/Na+ ratio of 0.7 that is quite dissimilar to 1.2 reported value for Cl− and Na+ content in seawater.39 Instead, red cells are enriched in Mg2+ ions (Fig. 5, panel D, Table 1) with Cl−/Mg2+atomic ratio of 1.4. In both cases, an excess in positive ions, respectively Na+ and Mg2+, content has been observed, considering NaCl and MgCl2 pure salts stoichiometry. The excess in positive charge, probably is offset by carboxylate, phosphate and sulphone/sulphate non protonate functional groups that can be present on the external membrane.14 This data is also confirmed by the total absence of MgCO3 and Na2CO3. No C 1s peaks are observed at 290.1 and 290.3 eV (see Table 1) as assigned to carbonate groups C 1s experimentally acquired respectively on MgCO3 and Na2CO3 standard powder samples (Fig. S6†). These data confirm that Na+ and Mg2+ chemical species are not present as pure residual salts coming from sea water (i.e., NaCl and MgCl2), but are structurally present in colorless and red cells surfaces, respectively. Particularly the Mg2+ ions can be coordinated to negative active sites present on the cells surfaces. Indeed, Mg2+ ions were not removed from RSC surfaces by chelation processes, after addition of the anticoagulant solution (ISO-EDTA) to coelomic fluid. On the contrary, the removal of calcium, if present, was effective on both red and colorless cell surfaces as evidenced by the almost a total absence of Ca2+ ions (see Table 1). The essential role of magnesium in the sea urchin was already highlighted during larval development. In A. lixula embryos, Mg2+ deficiency has a negative effect on mesodermal derivatives, including the RSC, inducing developmental delay and severe skeletal malformations.40 The remarkable presence of magnesium chemically available found in RSC underlines the importance of these ions and the possible role of this cell type. It is known that the skeleton of adult as well as the larval spicules of sea urchins are composed of magnesium calcite41 and Mg2+ incorporation can increase skeletal hardness.42 Taking also into account the role of Mg in biogenic mineral formation,43 we may suppose that red cells, occurring in cases of skeletal damage, could also represent a reserve of chemically available magnesium to be used during skeletal damage repair.
2. Based on Cl 2p3/2 binding energies (198.8–199.0 eV), these signals have been attributed to Cl− ions.19 However, it is important to point out that the peak fit shapes differ in FWHM between red and colorless cells. In detail, the best fit of colorless cells Cl 2p peak was obtained with FWHM 1.2 eV, otherwise a FWHM of 1.5 eV, was necessary to fit the Cl 2p peak shape for red cells samples. This behavior can suggest that Cl− ions chlorine species present on both samples are not equivalent because a chemically different environment. In fact, the colorless cells are rich in Na+ counter-ions, (Fig. 5, panel C and Table 1)14,21, with Cl−/Na+ ratio of 0.7 that is quite dissimilar to 1.2 reported value for Cl− and Na+ content in seawater.39 Instead, red cells are enriched in Mg2+ ions (Fig. 5, panel D, Table 1) with Cl−/Mg2+atomic ratio of 1.4. In both cases, an excess in positive ions, respectively Na+ and Mg2+, content has been observed, considering NaCl and MgCl2 pure salts stoichiometry. The excess in positive charge, probably is offset by carboxylate, phosphate and sulphone/sulphate non protonate functional groups that can be present on the external membrane.14 This data is also confirmed by the total absence of MgCO3 and Na2CO3. No C 1s peaks are observed at 290.1 and 290.3 eV (see Table 1) as assigned to carbonate groups C 1s experimentally acquired respectively on MgCO3 and Na2CO3 standard powder samples (Fig. S6†). These data confirm that Na+ and Mg2+ chemical species are not present as pure residual salts coming from sea water (i.e., NaCl and MgCl2), but are structurally present in colorless and red cells surfaces, respectively. Particularly the Mg2+ ions can be coordinated to negative active sites present on the cells surfaces. Indeed, Mg2+ ions were not removed from RSC surfaces by chelation processes, after addition of the anticoagulant solution (ISO-EDTA) to coelomic fluid. On the contrary, the removal of calcium, if present, was effective on both red and colorless cell surfaces as evidenced by the almost a total absence of Ca2+ ions (see Table 1). The essential role of magnesium in the sea urchin was already highlighted during larval development. In A. lixula embryos, Mg2+ deficiency has a negative effect on mesodermal derivatives, including the RSC, inducing developmental delay and severe skeletal malformations.40 The remarkable presence of magnesium chemically available found in RSC underlines the importance of these ions and the possible role of this cell type. It is known that the skeleton of adult as well as the larval spicules of sea urchins are composed of magnesium calcite41 and Mg2+ incorporation can increase skeletal hardness.42 Taking also into account the role of Mg in biogenic mineral formation,43 we may suppose that red cells, occurring in cases of skeletal damage, could also represent a reserve of chemically available magnesium to be used during skeletal damage repair.
 | ||
| Fig. 5 XPS high-resolution region of Cl 2p (panels A and B), Na 1s (panel C, no fit) and Mg1s (panel D, no fit) for colorless and red cells, respectively. | ||
3.8. Sulfur and phosphate functional groups
As expected, all samples presented S 2p and P 2p photo-peak signals as evident in the HR region (Fig. 6). However, the S 2p and P 2p signals were quite different when compared between colorless and red samples. In particular, there are two asymmetric S 2p peak signals on red cells, respectively at 163.9 eV and 169.8 eV. The most intense peak was assigned to sulphate species (C–O–![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) O3−, 169.8 eV),44,45 according to a recent work where the presence of sulphate groups was attributed to sulphated derivatives of spinochrome (polyhydroxylated naphthoquinone compounds) in isolated red cells.13 Therefore, this result confirms that these substances, can be also segregated on cell surface. The less intense S 2p peak was attributed to organic sulfides species,23 as reported in incorporating peptide cysteine (C–S–, 163.6–164 eV).21,34,46 In colorless cells, there was only the S 2p peak at 168.2 eV attributed to traces of sulphonate species (C–SO3−, 168.8–168.6 eV). The phosphorus peaks, presenting traces on both cells (Table 1), can be attributed to phosphate organic components from phospholipid which chemically differs between red and colorless cells (see related chemical shift in Table 1). However, unlike sulfur, the differences in phosphate chemical shift are difficult to be attributed at specific phosphorous functional groups when involved in bio-organic compounds.14–16 The importance of sulphate species has been reported in some studies highlighting that variations in the number of sulphate esters are responsible for various changes in biological activities of natural products. These changes can result in opposite (increase or decrease) biological activity or even in reversal effects.13
O3−, 169.8 eV),44,45 according to a recent work where the presence of sulphate groups was attributed to sulphated derivatives of spinochrome (polyhydroxylated naphthoquinone compounds) in isolated red cells.13 Therefore, this result confirms that these substances, can be also segregated on cell surface. The less intense S 2p peak was attributed to organic sulfides species,23 as reported in incorporating peptide cysteine (C–S–, 163.6–164 eV).21,34,46 In colorless cells, there was only the S 2p peak at 168.2 eV attributed to traces of sulphonate species (C–SO3−, 168.8–168.6 eV). The phosphorus peaks, presenting traces on both cells (Table 1), can be attributed to phosphate organic components from phospholipid which chemically differs between red and colorless cells (see related chemical shift in Table 1). However, unlike sulfur, the differences in phosphate chemical shift are difficult to be attributed at specific phosphorous functional groups when involved in bio-organic compounds.14–16 The importance of sulphate species has been reported in some studies highlighting that variations in the number of sulphate esters are responsible for various changes in biological activities of natural products. These changes can result in opposite (increase or decrease) biological activity or even in reversal effects.13
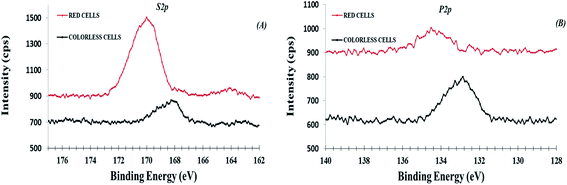 | ||
| Fig. 6 XPS high-resolution region of S 2p (panel A, no-fit) and P 2p (panel B, no fit) for colorless and red cells. | ||
4. Conclusions
A complete XPS analysis of averaged chemical composition of two different cell populations from the coelomic fluid of A. lixula, has been reported. The XPS technique could provide speciation of characteristic carbon, oxygen, nitrogen, sulphur, phosphorus as well as other characteristic elements like chlorine, sodium, magnesium and calcium present on these cells types.Results showed that the composition and the chemical state of several species were strongly depending on cellular population. Total carbon and nitrogen content were larger on the uncolored cells with respect to the red cells, while the oxygen content was reduced. Differences in terms of speciation of these elements with related content are also evidenced.
Colorless cells, characterized by some functional groups (i.e. alkyl, amide, alcohol etc.) compatible with protein, phospholipide and (lipo)-polysaccharides molecules, are particularly enriched in chlorine and sodium ions.
RSC, besides the expected carbon alkyl groups, showed chemical differences particularly in terms of nitrogen- and oxygen-carbon functional groups, that are more compatible with pyrimidin and quinone based molecules presence. Consistently the presence of sulphated groups was associated to the presence of sulphated derivatives of polyhydroxylated naphthoquinone (i.e. spinochrome) recently reported as responsible for various changes in biological activities of natural products. The presence on the RSC surface suggests deepening investigation about them to clarify their biological role. In addition on RSC the typical high content in chlorine is associated to an high magnesium cations content, that seems to replace sodium. These high content in Cl, Na and Mg, not found in previous studied cells surfaces by XPS, could be related to the marine origin of the analysed cells. A role of the magnesium chemically coordinated with red cell membrane in the cells was to be considered. Future investigations will be directed to better understand the role of Mg and its contribution in performing immune processes involving RSC. Our observations suggest a deep investigation of the behavior of these cells to understand if they use this ion in safeguarding the integrity of the sea urchin skeleton. This could also allow us to better define the role of these cells and probably expand the number of functions they perform or processes in which they are involved. Finally, no traces of toxic elements are detectable on both cells analyzed populations.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.References
- G. L. Russo, M. Russo, I. Castellano, A. Napolitano and A. Palumbo, Mar. Drugs, 2014, 12, 4069–4085 CrossRef CAS.
- F. Ramírez-Gómez and J. E. García-Arrarás, Invertebr. Surviv. J., 2010, 7, 211–220 Search PubMed.
- L. C. Smith, J. P. Rast, V. Brockton, D. P. Terwilliger, S. V. Nair, K. M. Buckley and A. J. Majeske, Invertebr. Surviv. J., 2006, 3(1), 25–39 Search PubMed.
- C. W. Chang, J. Chem. Educ., 1978, 55, 684 CrossRef CAS.
- C. J. Coates, C. McCulloch, J. Betts and T. Whalley, J. Innate Immun., 2018, 10, 119–130 CrossRef CAS PubMed.
- B. M. Heatfield and D. F. Travis, J. Morphol., 1975, 145, 51–71 CrossRef CAS.
- M. Service and A. C. Wardlaw, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1984, 79, 161–165 CrossRef.
- P. T. Johnson, J. Invertebr. Pathol., 1969, 13, 42–62 CrossRef CAS.
- P. Gerardi, M. Lassegues and C. Canicatti, Biol. Cell., 1990, 70, 153–157 CrossRef.
- T. Haug, A. K. Kjuul, O. B. Styrvold, E. Sandsdalen, Ø. M. Olsen and K. Stensvag, J. Invertebr. Pathol., 2002, 81, 94–102 CrossRef CAS.
- P. T. Johnson and F. A. Chapman, J. Invertebr. Pathol., 1970, 16, 268–276 CrossRef.
- K. A. Coffaro and R. T. Hinegardner, Science, 1977, 197, 1389–1390 CrossRef CAS.
- J. Hira, D. Wolfson, A. J. C. Andersen, T. Haug and K. Stensvag, Sci. Rep., 2020, 10, 1–9 CrossRef.
- G. F. Lorusso, G. De Stasio, P. Casalbore, D. Mercanti, M. T. Ciotti, A. Cricenti and G. Margaritondo, J. Phys. D: Appl. Phys., 1997, 30, 1794 CrossRef CAS.
- P. G. Rouxhet and M. J. Genet, Surf. Interface Anal., 2011, 43, 1453–1470 CrossRef CAS.
- A. Skallberg, C. Brommesson and K. Uvdal, Biointerphases, 2017, 12, 02C408 CrossRef PubMed.
- G. De Stasio, D. Mercanti, M. T. Ciotti, T. C. Droubay, P. Perfetti, G. Margaritonda and B. P. Tonner, J. Phys. D: Appl. Phys., 1996, 29, 259 CrossRef CAS.
- V. J. Smith, The echinoderms, Invertebrate Blood Cells, ed. Ratcliffe, N. and Rowley, A., 1981, pp. 513–562 Search PubMed.
- D. Baer, K. Artyushkova, H. Cohen, C. D. Easton, M. Engelhard, T. R. Gengenbach, G. Greczynski, P. Mack, D. J. Morgan and A. Roberts, J. Vac. Sci. Technol., A, 2020, 38, 031204 CrossRef CAS.
- ISO 19318:2004, Surface chemical analysis - X-ray photoelectron spectroscopy - Reporting of methods used for charge control and charge correction, International Organization for Standardization, Geneva, Switzerland, 2004 Search PubMed.
- NIST, X-Ray Photoelectron Spectroscopy Database, https://srdata.nist.gov/xps/main_search_menu.aspx Search PubMed.
- C. D. Wagner and A. Josh, J. Electron Spectrosc. Relat. Phenom., 1988, 47, 283–313 CrossRef CAS.
- A. M. Salvi and J. E. Castle, J. Electron Spectrosc. Relat. Phenom., 1998, 95, 45–56 CrossRef.
- D. A. Shirey, Phys. Rev. B: Solid State, 1972, 5, 4709–4714 CrossRef.
- G. H. Major, N. Fairley, P. M. A. Sherwood, M. R. Linford, J. Terry, V. Fernandez and K. Artyushkova, J. Vac. Sci. Technol., A, 2020, 38, 061203 CrossRef CAS.
- D. Briggs and M. P. Seah, Practical Surface Analysis: Auger and X-Ray Photoelectron Spectroscopy, J. Wiley, Chichester, 1990, 1, pp. 151–152 Search PubMed.
- D. Briggs and J. T. Grant, Surface Analysis by Auger and X-Ray Photoelectron Spectroscopy, Surface Spectra, IM Publications, Chichester, UK, 2003 Search PubMed.
- A. Pinsino, C. Della Torre, V. Sammarini, R. Bonaventura, E. Amato and V. Matranga, Cell Biol. Toxicol., 2008, 24, 541–552 CrossRef CAS.
- M. T. de Faria and J. R. M. C. da Silva, J. Invertebr. Pathol., 2008, 98, 58–62 CrossRef PubMed.
- E. Hobaus, Proceedings of the European Colloquium on Echinoderms, AA Balkema Rotterdam, The Netherlands, 1979, pp. 247–249 Search PubMed.
- A. J. Barlow, S. Popescu, K. Artyushkova, O. Scott, N. Sano, J. Hedley and P. J. Cumpson, Carbon, 2016, 107, 190–197 CrossRef CAS.
- A. M. Salvi, R. Pucciariello, M. R. Guascito, V. Villani and L. Intermite, Surf. Interface Anal., 2002, 33, 850–861 CrossRef CAS.
- L. Ploux, K. Anselme, A. Dirani, A. Ponche, O. Soppera and V. Roucoules, Langmuir, 2009, 25, 8161–8169 CrossRef CAS.
- J. S. Stevens, S. J. Byard and S. L. Schroeder, Cryst. Growth Des., 2010, 10, 1435–1442 CrossRef CAS.
- Y. Zubavichus, A. Shaporenko, V. Korolkov, M. Grunze and M. Zharnikov, J. Phys. Chem. B, 2008, 112, 13711–13716 CrossRef CAS PubMed.
- S. Fleutot, J. C. Dupin, G. Renaudin and H. Martinez, Phys. Chem. Chem. Phys., 2011, 13, 17564–17578 RSC.
- M. Kjærvik, K. Schwibbert, P. Dietrich, A. Thissen and W. E. Unger, Surf. Interface Anal., 2018, 50, 996–1000 CrossRef.
- SPECS, Application Note #000399, XPS surface chemical analysis of bacterial samples with EnviroESCA, SPECS Surface Nano Analysis GmbH, Voltastrasse 5, 13355 Berlin/Germany|www.EnviroESCA.com.
- F. Prodi, F. Belosi, D. Contini, G. Santachiara, L. Di Matteo, A. Gambaro, A. Donateo and D. Cesari, Atmos. Res., 2009, 92, 141–150 CrossRef CAS.
- C. Martino, R. Chiarelli, M. C. Roccheri, V. Matranga and M. Byrne, Invertebr. Reprod. Dev., 2019, 63, 165–176 CrossRef CAS.
- P. Dubois and C. P. Chen, Calcification in echinoderms, Echinoderm Studies, 3, M. Jangoux and J.M. Lawrence, AA Balkema Rotterdam/Brookfield, 1989, pp. 109–178 Search PubMed.
- M. Byrne, A. M. Smith, S. West, M. Collard, P. Dubois, A. Graba-Landry and S. A. Dworjanyn, Environ. Sci. Technol., 2014, 48, 12620–12627 CrossRef CAS.
- Y. Politi, D. R. Batchelor, P. Zaslansky, B. F. Chmelka, J. C. Weaver, I. Sagi, S. Weiner and L. Addadi, Chem. Mater., 2010, 22, 161–166 CrossRef CAS.
- J. Song and P. A. Peng, Aerosol Sci. Technol., 2009, 43, 1230–1242 CrossRef CAS.
- M. R. Guascito, D. Cesari, D. Chirizzi, A. Genga and D. Contini, Atmos. Environ., 2015, 116, 146–154 CrossRef CAS.
- C. D. Wagner, A. V. Naumkin, A. Kraut-Vass, J. W. Allison, C. J. Powell and J. R. Rumble Jr, NIST Standard Reference Database 20, Version 3.4, National Institute of Standards and Technology, Gaithersburg, 2003 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra03156b |
| This journal is © The Royal Society of Chemistry 2021 |


