DOI:
10.1039/D1RA02869C
(Paper)
RSC Adv., 2021,
11, 17603-17610
Rare bioparticle detection via deep metric learning
Received
13th April 2021
, Accepted 7th May 2021
First published on 13th May 2021
Abstract
Recent deep neural networks have shown superb performance in analyzing bioimages for disease diagnosis and bioparticle classification. Conventional deep neural networks use simple classifiers such as SoftMax to obtain highly accurate results. However, they have limitations in many practical applications that require both low false alarm rate and high recovery rate, e.g., rare bioparticle detection, in which the representative image data is hard to collect, the training data is imbalanced, and the input images in inference time could be different from the training images. Deep metric learning offers a better generatability by using distance information to model the similarity of the images and learning function maps from image pixels to a latent space, playing a vital role in rare object detection. In this paper, we propose a robust model based on a deep metric neural network for rare bioparticle (Cryptosporidium or Giardia) detection in drinking water. Experimental results showed that the deep metric neural network achieved a high accuracy of 99.86% in classification, 98.89% in precision rate, 99.16% in recall rate and zero false alarm rate. The reported model empowers imaging flow cytometry with capabilities of biomedical diagnosis, environmental monitoring, and other biosensing applications.
1. Introduction
Rare bioparticle detection is essential to various applications such as cancer diagnosis and prognosis, viral infections, and implementing early warning systems in water monitoring.1–6 In these applications, the target bioparticles in the sample are extremely rare with a huge abundance of background particles. For example, the ratio of the target bioparticle and background bioparticles could be 1 in 1000 (0.1%) or even less.7 Currently, bioimage analysis has made a huge progress, benefitting from rich-dataset supervised learning using deep neural networks.4,5,8 However, conventional deep neural networks only use simple classifiers such as SoftMax to obtain highly accurate results with the confidence that the deep neural network learns more distinct features than traditional machine learning in classification. Thus, they sometimes get unexpected results in many practical applications, e.g., rare bioparticle detection7,9–11 and bioparticle sorting,8,12,13 because it is hard to collect representative image data in those applications and the input images in inference time may be distinct from those during training. These applications also require the model to have a performance of low false alarm as well as high recovery rate in practical environments. For example, a large amount of false alarms will introduce high-cost consequential actions.14 Up to now, it remains a great challenge in the detection of rare bioparticles in practical applications.
Conventional deep neural networks use simple classifier to make the decision of seen/unseen classes. Therefore, they often make wrong predictions, and do so confidently.15–18 For example, the conventional deep neural network model predicts wrongly (it predicts the pollutants as Cryptosporidium or Giardia) with a high confidence level (>99.99%) as shown in Fig. 1. These inaccuracies arise from the conventional classification approaches such as convolutional neural networks (CNNs) with a linear Softmax classifier11 (Fig. 2(a)) that limit their ability to detect novel examples.9,15,17,19,20 As a result, conventional Softmax-based approaches are not suitable for open-set rare bioparticle detection. For example, a highly accurate algorithm based on a sophisticated densely connected neural network for bioparticle classification was developed for rare bioparticle detection,8 but it only achieved a sensitivity and specificity of 77.3% and 99.5%, respectively.
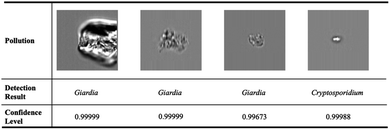 |
| | Fig. 1 Wrong prediction in conventional deep neural networks. Conventional deep neural networks often make wrong predictions and do so confidently when the images are not seen in the training process. The pollutions are predicted as targets, Giardia or Cryptosporidium, with a confidence level >99.99%. | |
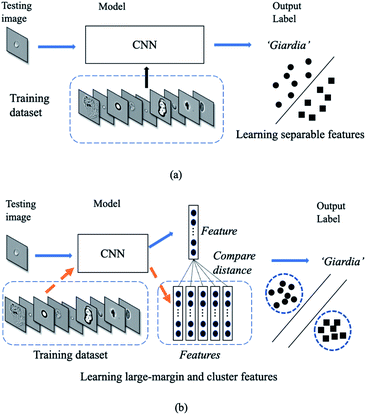 |
| | Fig. 2 Deep classification vs. deep metric learning. (a) In deep classification, the model only studies a boundary. (b) In deep metric learning, the model studies a more generative representation with similar classes are close and the unsimilar classes are far away. | |
Deep metric learning10 in Fig. 2(b) provides a possible direction to improve open-set detection by learning a map from the input image space to an output embedding features in the latent space. Instead of using the SoftMax classifier, this approach uses semantic similarity such as the Euclidean distance to constrain the models. It does not rely on the cross-entropy loss but proposes another class of network loss, i.e., the contrastive loss. Thus, the sum of the output class probabilities is not doom to be one and this provides it a generatability.9 Generative model is essentially a metric learning problem whereby the key is to learn a large margin distance metric within the latent space when the testing data are usually disjoint from the training dataset.
Unsupervised deep metric learning is used to learn a low-dimensional subspace and preserve useful geometrical information of the samples. On the other hand, supervised deep metric learning is used to learn a projection from the sample space to the feature space and measure the Euclidean metric in this feature space to discriminate the results. The metric learning is defined to study a map function f with a dataset χ = {x, y, z,…}, whereby 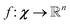 is well defined mapping and
is well defined mapping and 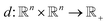 is the Euclidean distance over
is the Euclidean distance over 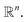 df(x, y) = d(f(x), f(y)) = ‖f(x) − f(y)‖2 is close to zero when x and y are similar.
df(x, y) = d(f(x), f(y)) = ‖f(x) − f(y)‖2 is close to zero when x and y are similar.
The mathematical definition of Euclidean distance d(x, y) between x and y is expressed as10
| |
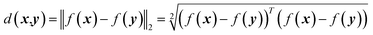 | (1) |
where
x,
y ∈
χ, and it is assumed that metric
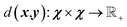
satisfies the following properties as
| | |
d(x, z) ≤ d(x, y) + d(y, z)
| (2c) |
Deep metric learning is widely applied in signature verification,21 face verification and recognition,22 and person re-identification.23
In this paper, a rare bioparticle detection system is demonstrated (Fig. 3), which consists of an imaging flow cytometry system to capture the images of all pollutants and create an image database. A deep neural network based on deep metric learning and a decision algorithm are designed to detect rare bioparticles of Cryptosporidium and Giardia. The model leverages convolutional neural network to study the rich features in the dataset and learning distinct metric by using Siamese network21 and contrastive loss, which maximizes the distance of different classes and minimizes the distance of similar classes. Experimental results showed that the deep metric learning studies good features and performs better than conventional deep learning, which was manifested to be a solid network model for rare bioparticle detection problems.
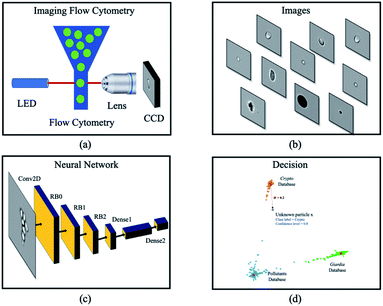 |
| | Fig. 3 Overview of a deep metric neural network for rare bioparticle detection using an imaging flow cytometry. Water sample is processed using (a) the imaging flow cytometry system (Amnis® ImageStream®X Mk II), capturing the images of all pollutants and creating (b) an image database. (c) Deep metric neural network, and (d) decision algorithms are used for classification and detection. | |
2. Methods and materials
2.1 Bioparticle profiling
First, the samples were imaged using the imaging flow cytometry (Amnis® ImageStream®X Mk II). Bioparticles such as Cryptosporidium, Giardia, microplastics and other pollutants such as dirt and cell debris with size ranging from 3 to 14 μm that naturally exist in drinking water were included in the study. The naturally existing pollutants were obtained by concentrating 10 litres of drinking water using a water filtration system. Formalin-treated Cryptosporidium oocysts, Giardia cysts (Waterborne™ Inc.) and synthetic microplastic beads (Thermo Fisher Scientific, Duke Scientific and Polysciences Inc.) of different sizes (1.54 μm, 3 μm, 4 μm, 4.6 μm, 5 μm, 5.64 μm, 8 μm, 10 μm, 12 μm and 15 μm) were spiked separately in 200 μL water. Bioparticles were hydrodynamically focused by a sheath flow and flowed through the detection region with phosphate buffered saline solution (PBS) used as the sheath medium. Single bioparticles were illuminated with an LED light source, and brightfield images (Fig. 4) were acquired with a charge-coupled device (CCD) camera24 using a 60× objective in Fig. 3(a).
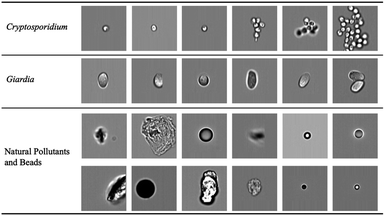 |
| | Fig. 4 Bioparticle image dataset. Each row represents one type of bioparticle. From the top to bottom are Cryptosporidium, Giardia, natural pollutants and beads. All subfigures share the same scale bar. | |
2.2 Bioparticle image dataset
The raw image sequence files (.RIF) of different samples were captured. The raw brightfield images were extracted from the image sequence files by IDEAS software (accompanying with the ImageStream) and patched to 120 × 120 pixels as in Fig. 3(b). From millions of acquired raw images, 89![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 663 images were selected to construct the dataset by experts. The image dataset consists of three classes: Cryptosporidium (2078 images), Giardia (3438 images), and natural pollutants and beads (84
663 images were selected to construct the dataset by experts. The image dataset consists of three classes: Cryptosporidium (2078 images), Giardia (3438 images), and natural pollutants and beads (84![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 147 images). The brightfield images of protozoa had complex patterns, such as distinct sizes, degree of aggregation and different internal structures, which complicated the learning task.
147 images). The brightfield images of protozoa had complex patterns, such as distinct sizes, degree of aggregation and different internal structures, which complicated the learning task.
2.3 Deep metric learning for rare bioparticle detection
Siamese network21 is the most popular deep metric learning network structure which is used to train the deep learning model shown in Fig. 5(a). The base network structure of deep metric learning model is illustrated in Fig. 5(b). The input of embedding network is a grayscale image with 120 × 120 pixels and a convolutional layer with a filter size of 7 × 7 is used in the first stage. Then, three residual network blocks (RB0 to RB2)25 are attached to the first convolution neural network layer. The output of the last residual network block RB2 is flattened, and then followed by a fully-connected layer26 together with a parametric ReLU (PReLU) layer.27 Finally, a fully-connected layer with 2 output units is attached to the PReLU layer to generate the latent feature vector of bioparticles. The detail parameters of the embedding network are listed in Table 1.
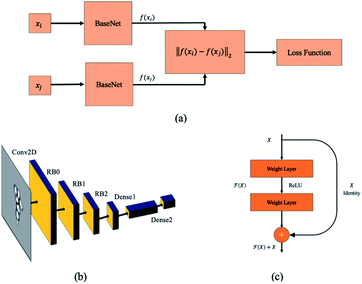 |
| | Fig. 5 Siamese network. (a) The structure of Siamese network for training deep metric learning. The twin networks share the same network parameters. A loss function is attached to this twin network to regularize the network. (b) Embedding network. (c) Residual block. | |
Table 1 Network parameters of the embedding network
| Layer/block |
Type |
Output dimension |
Params |
| Conv2d |
Convolution |
60 × 60 × 64 |
9536 |
| RB0 |
Residual block |
30 × 30 × 64 |
147![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 968 968 |
| RB1 |
Residual block |
15 × 15 × 128 |
525![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 568 568 |
| RB2 |
Residual block |
8 × 8 × 256 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 099 099![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 712 712 |
| Pool |
Average pool |
2 × 2 × 256 |
0 |
| Dense1 |
Fully connected |
256 |
262![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
| PReLU |
Parametric ReLU |
256 |
1 |
| Dense2 |
Fully connected |
2 |
514 |
The first convolutional 2D layer (Conv2D in Fig. 5(b))28 takes an h × w × n input feature map Xi, where h is the spatial height, w is the spatial width and n is the output channels of the feature map (120 × 120 × 1). The input Xi is transformed into a 60 × 60 × 64 output feature maps Xo and expressed as29
| |
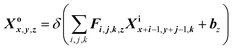 | (3) |
where
z = 1, 2, …,
m and
k = 1, 2, …,
n. The input feature map
Xi is convolved with a number of feature detectors, each of which has a three-dimensional filter
F in the present layer (7 × 7 × 1), and a bias
b. A ReLU function
δ(
x) is attached to this convolution operator.
Three cascaded residual network blocks (RB0 to RB2 in Fig. 5(b))25 with down sampling (stride = (2, 2)) are attached to the first convolution layer. The RB has two 3 × 3 convolutional layers and the same number of output channels as shown in Fig. 5(c). In the end, a batch normalization layer and a ReLU activation function follow each convolutional layer. In addition, an identify path is added to connect the input to the output directly.
The classifier is implemented by two fully connected layers (dense layer in Fig. 5(b)). It takes the last output of RB2 as the input and applies cascaded matrix multiplications and non-linear function to the weight matrix F and bias b to produce a vector with two dimensions in the latent space. The equation of fully connected layer can be expressed as
The PReLU layer is used after the fully-connected layer, which is expressed as
| |
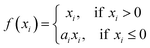 | (5) |
where
xi is the input value and
ai is the parameter of the PReLU layer.
2.4 Model training
The model is trained with Siamese network-based structure. Siamese network is proposed for the signature's verification in 1994 and used for training the neural network. The network consists of two base embedding networks and a joint output neuron. Residual network blocks are used as the embedding networks to extract the features. The two embedding networks share the same weights, and the identical sub-networks extract feature vectors from two images simultaneously and the joined neuron measures the distance between the two feature vectors in the latent space by using a metric. In the training process, the similar and dissimilar pairs (xi and xj) are passed through the network and generate features vector in the latent space named f(xi) and f(xj). In the loss function, the distance metric d(x, y) = ‖f(x) − f(y)‖2 is regressed to minimize the distance between the similar pairs and keep the distance of the dissimilar pairs. The contrastive loss is used to train the Siamese network. For the pair of input (xi, xj), it is a positive pair if xi and xj are semantically similar and negative pair if they are dissimilar. The training process of Siamese network deals with minimizing the contrastive loss, which is expressed as| |
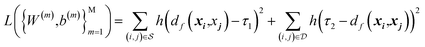 | (6) |
where h(x) = max(0, x) is the hinge loss function, and τ1 = 0.9 and τ2 = 1.0 are two positive thresholds with τ1 < τ2, respectively. 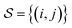 is the similar pair and
is the similar pair and 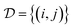 is the dissimilar pair.
is the dissimilar pair.
The deep metric learning model is implemented with deep learning framework-PyTorch30 and trained over an Ubuntu GPU server31 with four Nvidia GeForce RTX 2080 cards and the Intel Xeon CPU E5-2650. To train and evaluate the performance of the model, the selected image dataset is randomly split into a training, validation and testing dataset with 48%, 12% and 40% of the total number of images, respectively. Later, the images in the training dataset are augmented to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 images, and each image is randomly sampled from the dataset and processed by position transformation, horizontal and vertical flipping, rotation or zooming. The weight of the deep neural networks is initialized with the Glorot uniform initializer32 at a mean value of zero and a standard deviation at 10−2, and the network is trained in an end-to-end fashion using the Adam stochastic optimizing algorithm.33 The parameters for Adam are β1 = 0.9, β2 = 0.999, and a learning rate decay is used for training. The positive or negative pair is generated on the fly. First, it enumerates current image anchor from the image list. Then, the positive image is randomly selected from rest images of the same class and the negative image is randomly selected from the images in rest classes. Early stop is also used to prevent overfitting by stopping the training when the model's performance on validation dataset start to degrade.34 A maximum of 300 epochs is used to train the model.
000 images, and each image is randomly sampled from the dataset and processed by position transformation, horizontal and vertical flipping, rotation or zooming. The weight of the deep neural networks is initialized with the Glorot uniform initializer32 at a mean value of zero and a standard deviation at 10−2, and the network is trained in an end-to-end fashion using the Adam stochastic optimizing algorithm.33 The parameters for Adam are β1 = 0.9, β2 = 0.999, and a learning rate decay is used for training. The positive or negative pair is generated on the fly. First, it enumerates current image anchor from the image list. Then, the positive image is randomly selected from rest images of the same class and the negative image is randomly selected from the images in rest classes. Early stop is also used to prevent overfitting by stopping the training when the model's performance on validation dataset start to degrade.34 A maximum of 300 epochs is used to train the model.
2.5 Deep metric learning based classifier
The deep metric network studies a map from the images into a latent space and cannot be used directly to classify the images. In order to classify the rare bioparticle images with deep metric learning model, further processing is needed to be added in the end of this neural network model. It converts the values in the latent vector into a target class label and a confidence score. As shown in Fig. 6, the class label is assigned by the closed cluster center, which can be calculated by either mean latent vectors (mean center) or Gaussian Mixture Models (GMM)35 of a known class, such as Cryptosporidium, in the training dataset. The confidence score is used to present the similarity between the target bioparticle to the certain classes, which are collected in the training phase. The confidence score can be calculated by the distance of the target bioparticle to the center of certain class on the distribution diagram of the latent space or a Gaussian estimator.
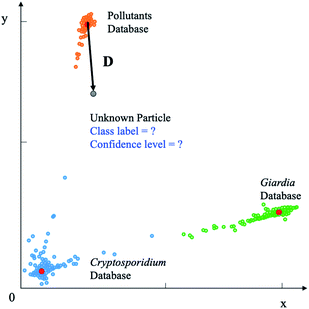 |
| | Fig. 6 Deep metric learning based classification. The unknown bioparticle is classified to correspond classes. Class label is assigned to classify the unknown particles by the closed cluster center (red). Confidence level is used to present the similarity of unknown particles to certain databased collected by Cryptosporidium and Giardia samples. | |
Gaussian distribution36 is a continuous probability distribution, which has a characteristic with symmetric “Bell curve” shape that quickly falls off toward 0. GMM is a probabilistic model, which assumes that the underlying data belong to a linear combination of several Gaussian distributions. A GMM model gives a posterior distribution over K Gaussian distributions, which shows better performance on optimizing model complexity.37 The GMM can be represented by38
| |
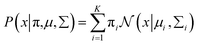 | (7) |
where
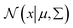
is a normal distribution,
x is a multidimension vector variable,
μ is the mean of this
x and ∑ is the covariance matrix. The
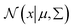
is given by
38| |
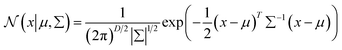 | (8) |
where
D is the number of dimensions of the feature vector. The π
i are mixing coefficients. It satisfied 0 ≤ π
i ≤ 1 and
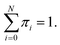
With the assumption that
xi come from independent
K mixture distributions insider
C, the equation can be expressed as
38| |
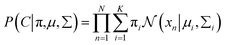 | (9) |
Expectation-maximization (EM) algorithm is used to find the local maximum likelihood and estimates of individual parameters in GMM (μ and ∑). EM is an iterative algorithm, which follows the rule that every iteration strictly increases the maximum likelihood. EM algorithm may not reach the global optimal point, but it can guarantee to local saddle point. The EM algorithm consists of two main steps: expectation and maximization. The expectation step calculates the expectation of the clusters when each xi ∈ X is assigned to the clusters with given μ, ∑, π. The maximization step maximizes the expectation in previous step by find suitable parameters.
First, the program randomly assigned samples X = {x1, x2, …, xn} to components estimated mean ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 1,
1, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 2, …,
2, …, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) k. For example,
k. For example, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 1 = x6,
1 = x6, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 2 = x20,
2 = x20, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 3 = x21,
3 = x21, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 4 = x33,
4 = x33, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 5 = x60 when N = 100 and K = 5. Then,
5 = x60 when N = 100 and K = 5. Then, 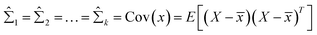 is assigned where
is assigned where ![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) = E(X), and all mixing coefficients are set to a uniform distribution with
= E(X), and all mixing coefficients are set to a uniform distribution with 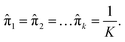 In the expectation step,
In the expectation step, 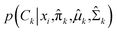 is given by38
is given by38
| |
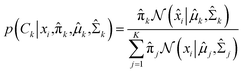 | (10) |
In the maximization step, 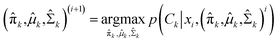 and can be calculated as38
and can be calculated as38
| |
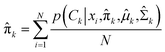 | (11a) |
| |
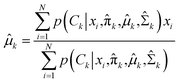 | (11b) |
| |
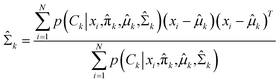 | (11c) |
The whole EM process repeats iteratively until the EM algorithm converges to a point and gives a maximum likelihood estimate for each 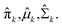
3. Results and discussions
3.1 Classification evaluation
The output latent vectors of the deep metric neural network are mapped to a 2D latent space by embedding network as shown in Fig. 7. Compared with the conventional deep classification method in Fig. 7(a), the deep metric learning model is trained using the Siamese network in Fig. 7(b) and the contrastive loss shows better performance. The dots of the similar images in deep metric learning are closer and the dissimilar images are kept far away from others, providing the ability of generatability. Moreover, the t-SNE graphs of RB0 to RB2 from low level to high level features in Fig. 8 also show that the data is well separated in the deep metric learning based model in Fig. 8(a)–(c) even in the shallow layers by comparing with the conventional classification-based method in Fig. 8(d)–(f).
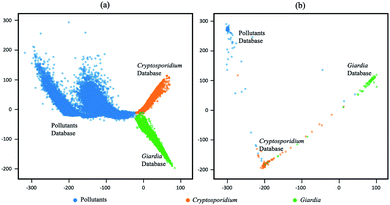 |
| | Fig. 7 Visualization on 2D latent space of conventional deep classification-based model and deep metric learning which mapped by embedding network. (a) Conventional deep classification-based model, (b) deep metric learning based model. | |
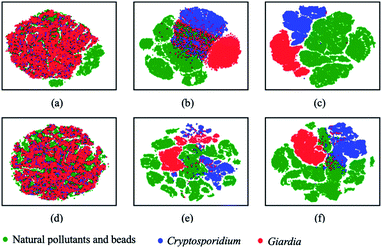 |
| | Fig. 8 Visual on intermediate layers with t-SNE on deep metric learning and conventional classification-based model. (a–c) The lower, middle, and high level of deep metric learning, (d–f) the lower, middle, and high level of conventional deep classification-based model. | |
For classification, the GMM model is selected because it can show how much confidence is associated to the target cluster, and it has the same accuracy of 99.86% with the mean center. The final results of the model comparison between deep metric learning and conventional deep classification are summarized in Table 2. The model based on deep metric learning is superior to the model based on conventional deep classification neural networks in terms of accuracy, precision, recall and F1 score. The model based on deep metric learning network achieves 99.86% in accuracy, 98.89% in precision rate, 99.16% in recall rate and 99.02% in F1 score. On the other hand, the model based on conventional deep classification gives 99.71% in accuracy, 97.84% in precision, 98.55% in recall and 98.19% in F1 score. The results in Tables 3 and 4 also show that the performance of the individual class in deep metric learn-based model is better when it has a large quantity of training data. For example, the classification result on contaminated particles is much better in deep metric learning.
Table 2 Precision, recall and F1 score on test dataset
| Methods |
Measurement (%) |
| Accuracy |
Precision |
Recall |
F1 score |
| Deep classification |
99.71 |
97.84 |
98.55 |
98.19 |
| Deep metric learning |
99.86 |
98.84 |
99.17 |
99.00 |
Table 3 Confusion matrix of conventional deep classification
| Class |
Prediction |
| Pollutants |
Cryptosporidium |
Giardia |
| Actual |
Pollutants |
29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 610 610 |
35 |
14 |
| Cryptosporidium |
20 |
807 |
4 |
| Giardia |
10 |
8 |
1357 |
Table 4 Confusion matrix of deep metric learning-based classification
| Class |
Prediction |
| Pollutants |
Cryptosporidium |
Giardia |
| Actual |
Pollutants |
29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 639 639 |
17 |
3 |
| Cryptosporidium |
9 |
820 |
2 |
| Giardia |
6 |
9 |
1360 |
3.2 Model verification using spiked samples
In order to evaluate the performance of the deep metric learning model on rare bioparticle detection in real situation, Cryptosporidium and Giardia were spiked into the concentrated water sample to simulate rare bioparticle in contaminated water. In total, ten testing were run and the captured images were detected by the software with a confidence level at 0.98 and verified by biological experts based on their morphologies. The final results are summarised in Table 5. The deep metric learning gives zero false warning signal, which is vital to implement the early warning system that needs the specificity of 100%. In comparison, the conventional deep classification gives false positive signal in test 1, 3, 5, 6, 7, 10, especially false warning signals in test 7 and 10 are not acceptable. Some confused images are listed in Fig. 9. The first row shows the false positive images detected from the background pollution. They are easy to be identified for human, but failure to be obtained in conventional deep neural network. The second row shows some examples that Cryptosporidium are classified to Giardia and vice versa. On the contrary, deep metric learning-based model serves as a paradigm to deal with the rare cell detection. For the recovery rate, the deep metric learning gives an average of 85.5%.
Table 5 Cryptosporidium and Giardia detection using deep metric learning
| No. |
Spike |
Image number |
Manual counting |
Sensitivity |
Specificity |
Alarm |
Recovery rate |
| 1 |
20C |
23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 483 483 |
7 |
85.7% |
100% |
Yes |
85.7% |
| 2 |
20C |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 422 422 |
8 |
75.0% |
100% |
Yes |
75.0% |
| 3 |
20C |
21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 834 834 |
10 |
80.0% |
100% |
Yes |
80.0% |
| 4 |
20G |
19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 383 383 |
7 |
100.0% |
100% |
Yes |
100.0% |
| 5 |
20G |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320 320 |
9 |
88.9% |
100% |
Yes |
88.9% |
| 6 |
20G |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 872 872 |
6 |
83.3% |
100% |
Yes |
88.3% |
| 7 |
0 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0 |
— |
100% |
No |
— |
| 8 |
0 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0 |
— |
100% |
No |
— |
| 9 |
0 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0 |
— |
100% |
No |
— |
| 10 |
0 |
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
0 |
— |
100% |
No |
— |
| Mean |
85.5% |
100% |
— |
85.5% |
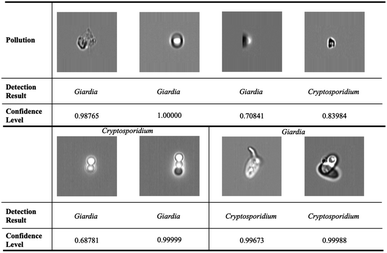 |
| | Fig. 9 Wrong prediction in conventional deep neural networks. The first row shows the false positive images detected from the background pollution. They are easy to be identified for human, but failure to be obtained in conventional deep neural network. The second row shows some examples that Cryptosporidium are classed to Giardia and vice versa. | |
4. Conclusions
Siamese-based deep metric learning provides a set of new tools for learning latent vectors by leveraging both convolutional neural network and deep metric learning. In this paper, we present a deep neural network based on deep metric learning for rare bioparticle detection by incorporating Siamese constraint in the learning process. The model can learn interpretable latent representation that preserves semantic structure of similar and dissimilar images. The experimental results demonstrate that Siamese-based deep metric learning can achieve classification-based accuracy while encoding more semantic structural information in the latent embedding. Thus, it is suitable for rare bioparticle detection, and achieves 99.86% in accuracy and zero false alarm. The model empowers intelligent imaging flow cytometry with the capability of rare bioparticle detection, benefiting the biomedical diagnosis, environmental monitoring, and other biosensing applications.
Author contributions
S. L., A. Q. L. and T. B. jointly conceived the idea. B. T. T. N. performed experiments. Y. Z., A. E., X. H. Z., B. H. W., G. C., H. T., Y. S., T. B., X. D. J. and A. Q. L. were involved in the discussion and data analysis. S. L., A. Q. L., Y. Z., X. D. J, L. K. C. and Y. Z. S. wrote the manuscript. T. B., X. D. J. and A. Q. L. supervised and coordinated all of the work.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by the Singapore National Research Foundation under the Competitive Research Program (NRF-CRP13-2014-01), Ministry of Education Tier 1 RG39/19, and the Singapore Ministry of Education (MOE) Tier 3 grant (MOE2017-T3-1-001).
References
- N. Meng, E. Lam, K. K. M. Tsia and H. K.-H. So, IEEE J. Biomed. Health Inform., 2018, 23, 2091–2098 Search PubMed.
- Z. Göröcs, M. Tamamitsu, V. Bianco, P. Wolf, S. Roy, K. Shindo, K. Yanny, Y. Wu, H. C. Koydemir and Y. Rivenson, Light: Sci. Appl., 2018, 7, 1–12 CrossRef PubMed.
- Y. Wu, A. Calis, Y. Luo, C. Chen, M. Lutton, Y. Rivenson, X. Lin, H. C. Koydemir, Y. Zhang and H. Wang, ACS Photonics, 2018, 5, 4617–4627 CrossRef CAS.
- G. Kim, Y. Jo, H. Cho, H.-s. Min and Y. Park, Biosens. Bioelectron., 2019, 123, 69–76 CrossRef CAS PubMed.
- A. Isozaki, H. Mikami, H. Tezuka, H. Matsumura, K. Huang, M. Akamine, K. Hiramatsu, T. Iino, T. Ito and H. Karakawa, Lab Chip, 2020, 20, 2263–2273 RSC.
- X. Mao and T. J. Huang, Lab Chip, 2012, 12, 1412–1416 RSC.
- Y. Chen, P. Li, P.-H. Huang, Y. Xie, J. D. Mai, L. Wang, N.-T. Nguyen and T. J. Huang, Lab Chip, 2014, 14, 626–645 RSC.
- Y. Zhang, M. Ouyang, A. Ray, T. Liu, J. Kong, B. Bai, D. Kim, A. Guziak, Y. Luo and A. Feizi, Light: Sci. Appl., 2019, 8, 1–15 CrossRef CAS PubMed.
- M. Masana, I. Ruiz, J. Serrat, J. van de Weijer and A. M. Lopez, 2018, arXiv preprint arXiv:1808.05492.
- J. Lu, J. Hu and J. Zhou, IEEE Signal Process. Mag., 2017, 34, 76–84 Search PubMed.
- I. Goodfellow, Y. Bengio, A. Courville and Y. Bengio, Deep learning, MIT press Cambridge, 2016 Search PubMed.
- Y. Z. Shi, S. Xiong, Y. Zhang, L. K. Chin, Y. Y. Chen, J. B. Zhang, T. Zhang, W. Ser, A. Larrson and S. Lim, Nat. Commun., 2018, 9, 1–11 CrossRef PubMed.
- Y. Shi, S. Xiong, L. K. Chin, J. Zhang, W. Ser, J. Wu, T. Chen, Z. Yang, Y. Hao and B. Liedberg, Sci. Adv., 2018, 4, eaao0773 CrossRef PubMed.
- T. A. Reichardt, S. E. Bisson, R. W. Crocker and T. J. Kulp, Presented in Proc. SPIE 6945, Optics and Photonics in Global Homeland Security IV, 69450R, April 2008 Search PubMed.
- A. Bendale and T. Boult, IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recogn., 2015, 1893–1902 Search PubMed.
- M. A. Pimentel, D. A. Clifton, L. Clifton and L. Tarassenko, Signal Process., 2014, 99, 215–249 CrossRef.
- A. Bendale and T. E. Boult, IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recogn., 2016, 1563–1572 Search PubMed.
- D. Hendrycks and K. Gimpel, 2016, arXiv preprint arXiv:1610.02136.
- B. J. Meyer, B. Harwood and T. Drummond, IEEE Int. Conf. Image Process., 2018, 151–155 Search PubMed.
- D. S. Trigueros, L. Meng and M. Hartnett, 2018, arXiv preprint arXiv:1811.00116.
- J. Bromley, I. Guyon, Y. LeCun, E. Säckinger and R. Shah, NIPS (News Physiol. Sci.), 1994, 6, 737–744 Search PubMed.
- Y. Taigman, M. Yang, M. A. Ranzato and L. Wolf, IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recogn., 2014, 1701–1708 Search PubMed.
- R. R. Varior, M. Haloi and G. Wang, ECCV, 2016 Search PubMed.
- E. R. Fossum and D. B. Hondongwa, IEEE J. Electron Devices Soc., 2014, 2(3), 33–43 CAS.
- K. He, X. Zhang, S. Ren and J. Sun, IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recogn., 2016, 770–778 Search PubMed.
- Y. LeCun, Y. Bengio and G. Hinton, Nature, 2015, 521, 436–444 CrossRef CAS PubMed.
- K. He, X. Zhang, S. Ren and J. Sun, IEEE International Conference on Computer Vision, 2015, 1026–1034 Search PubMed.
- Y. LeCun, B. E. Boser, J. S. Denker, D. Henderson, R. E. Howard, W. E. Hubbard and L. D. Jackel, NIPS (News Physiol. Sci.), 1990, 396–404 Search PubMed.
- A. G. Howard, M. Zhu, B. Chen, D. Kalenichenko, W. Wang, T. Weyand, M. Andreetto and H. Adam, 2017, arXiv preprint arXiv:1704.04861.
- A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury, G. Chanan, T. Killeen, Z. Lin, N. Gimelshein and L. Antiga, NIPS (News Physiol. Sci.), 2019, 32 Search PubMed.
- M. Hodak, M. Gorkovenko and A. Dholakia, Towards power efficiency in deep learning on data center hardware, in 2019 IEEE International Conference on Big Data (Big Data), IEEE, 2019, pp. 1814–1820 Search PubMed.
- X. Glorot and Y. Bengio, ICAIS, 2010 Search PubMed.
- D. P. Kingma and J. Ba, 2014, arXiv preprint arXiv:1412.6980.
- R. Caruana, S. Lawrence and C. L. Giles, NIPS (News Physiol. Sci.), 2001, 402–408 Search PubMed.
- D. A. Reynolds, Encyclopedia of Biometrics, 2009, p. 741 Search PubMed.
- W. M. Mendenhall and T. L. Sincich, Statistics for Engineering and the Sciences, CRC Press, 2016 Search PubMed.
- A. Corduneanu and C. M. Bishop, ICAIS, 2001 Search PubMed.
- D. A. Forsyth and J. Ponce, Computer vision: a modern approach, Pearson, 2012 Search PubMed.
|
| This journal is © The Royal Society of Chemistry 2021 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article bd,
Yi Zhangc,
Bihan Wenb,
Ying Sunf,
Binh T. T. Nguyenb,
Giovanni Chierchiaa,
Hugues Talbote,
Tarik Bourouina
bd,
Yi Zhangc,
Bihan Wenb,
Ying Sunf,
Binh T. T. Nguyenb,
Giovanni Chierchiaa,
Hugues Talbote,
Tarik Bourouina *a,
Xudong Jiang*b and
Ai-Qun Liu
*a,
Xudong Jiang*b and
Ai-Qun Liu *bg
*bg
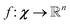 is well defined mapping and
is well defined mapping and 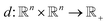 is the Euclidean distance over
is the Euclidean distance over 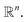 df(x, y) = d(f(x), f(y)) = ‖f(x) − f(y)‖2 is close to zero when x and y are similar.
df(x, y) = d(f(x), f(y)) = ‖f(x) − f(y)‖2 is close to zero when x and y are similar.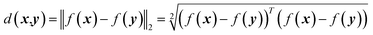
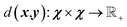 satisfies the following properties as
satisfies the following properties as![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 663 images were selected to construct the dataset by experts. The image dataset consists of three classes: Cryptosporidium (2078 images), Giardia (3438 images), and natural pollutants and beads (84
663 images were selected to construct the dataset by experts. The image dataset consists of three classes: Cryptosporidium (2078 images), Giardia (3438 images), and natural pollutants and beads (84![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 147 images). The brightfield images of protozoa had complex patterns, such as distinct sizes, degree of aggregation and different internal structures, which complicated the learning task.
147 images). The brightfield images of protozoa had complex patterns, such as distinct sizes, degree of aggregation and different internal structures, which complicated the learning task.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 968
968![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 568
568![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 099
099![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 712
712![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400
400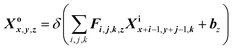
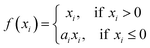
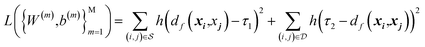
 is the similar pair and
is the similar pair and  is the dissimilar pair.
is the dissimilar pair.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 images, and each image is randomly sampled from the dataset and processed by position transformation, horizontal and vertical flipping, rotation or zooming. The weight of the deep neural networks is initialized with the Glorot uniform initializer32 at a mean value of zero and a standard deviation at 10−2, and the network is trained in an end-to-end fashion using the Adam stochastic optimizing algorithm.33 The parameters for Adam are β1 = 0.9, β2 = 0.999, and a learning rate decay is used for training. The positive or negative pair is generated on the fly. First, it enumerates current image anchor from the image list. Then, the positive image is randomly selected from rest images of the same class and the negative image is randomly selected from the images in rest classes. Early stop is also used to prevent overfitting by stopping the training when the model's performance on validation dataset start to degrade.34 A maximum of 300 epochs is used to train the model.
000 images, and each image is randomly sampled from the dataset and processed by position transformation, horizontal and vertical flipping, rotation or zooming. The weight of the deep neural networks is initialized with the Glorot uniform initializer32 at a mean value of zero and a standard deviation at 10−2, and the network is trained in an end-to-end fashion using the Adam stochastic optimizing algorithm.33 The parameters for Adam are β1 = 0.9, β2 = 0.999, and a learning rate decay is used for training. The positive or negative pair is generated on the fly. First, it enumerates current image anchor from the image list. Then, the positive image is randomly selected from rest images of the same class and the negative image is randomly selected from the images in rest classes. Early stop is also used to prevent overfitting by stopping the training when the model's performance on validation dataset start to degrade.34 A maximum of 300 epochs is used to train the model.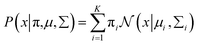
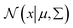 is a normal distribution, x is a multidimension vector variable, μ is the mean of this x and ∑ is the covariance matrix. The
is a normal distribution, x is a multidimension vector variable, μ is the mean of this x and ∑ is the covariance matrix. The 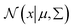 is given by38
is given by38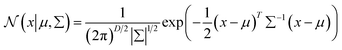
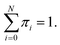 With the assumption that xi come from independent K mixture distributions insider C, the equation can be expressed as38
With the assumption that xi come from independent K mixture distributions insider C, the equation can be expressed as38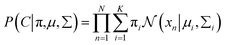
![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 1,
1, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 2, …,
2, …, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) k. For example,
k. For example, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 1 = x6,
1 = x6, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 2 = x20,
2 = x20, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 3 = x21,
3 = x21, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 4 = x33,
4 = x33, ![[small mu, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e0b3.gif) 5 = x60 when N = 100 and K = 5. Then,
5 = x60 when N = 100 and K = 5. Then, 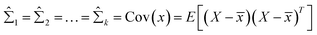 is assigned where
is assigned where ![[x with combining macron]](https://www.rsc.org/images/entities/i_char_0078_0304.gif) = E(X), and all mixing coefficients are set to a uniform distribution with
= E(X), and all mixing coefficients are set to a uniform distribution with 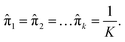 In the expectation step,
In the expectation step, 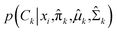 is given by38
is given by38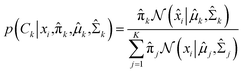
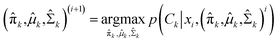 and can be calculated as38
and can be calculated as38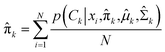
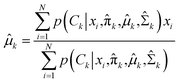
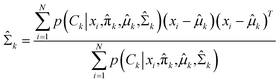
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 610
610![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 639
639![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 483
483![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 422
422![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 834
834![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 383
383![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320
320![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 872
872![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000








