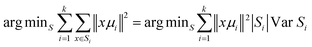Open Access Article
Open Access ArticleOxidative stress induced by tBHP in human normal colon cells by label free Raman spectroscopy and imaging. The protective role of natural antioxidants in the form of β-carotene
B. Brozek-Pluska * and
K. Beton
* and
K. Beton
Lodz University of Technology, Institute of Applied Radiation Chemistry, Laboratory of Laser Molecular Spectroscopy, Wroblewskiego 15, 93-590 Lodz, Poland. E-mail: beata.brozek-pluska@p.lodz.pl
First published on 4th May 2021
Abstract
The present study aimed to investigate the protective effect of β-carotene on the oxidative stress injury of human normal colon cell line CCD-18Co triggered by tert-butyl hydroperoxide (tBHP). XTT examination was used to determine cell viability after β-carotene supplementation and to determine the optimal concentration of antioxidant in spectroscopic studies. Cell biochemistry for the CCD-18Co control group, after tBHP addition and for cells in the β-carotene–tBHP model was studied using label-free Raman microspectroscopy. Results for stress treated CCD-18Co human colon normal cells and human colon cancer cells Caco-2 based on vibration features were also compared. Pretreatment with β-carotene alleviated damage in CCD-18Co human normal colon cells induced by tBHP and showed the preventative effect on cell apoptosis. Treatment with β-carotene altered the level of ROS investigated based on intensities of Raman peaks typical for lipids, proteins and nucleic acids. The present study confirmed the antioxidant, protective role of β-carotene against ROS by using spectroscopic label-free Raman techniques.
Introduction
Carotenoids, also called tetraterpenoids are yellow, red and orange pigments produced by plants and fungi as well as bacteria. They are responsible for specific and well recognizable color of e.g. pumpkins, carrots, corn, tomatoes, flamingos, salmon and lobsters. There are about 600 plant pigments in the world, but only six of them have a significant impact on the human body – α- and β-carotene, β-cryptoxanthin, lutein, lycopene and zeaxanthin. To date, around thousand known carotenoids and their derivatives have been categorized into two major classes, xanthophylls (including oxygen in the molecular structure) and carotenes (which are purely hydrocarbons). In plants carotenoids are accumulated in the plastids and are inactive in photosynthesis but play an auxiliary role as light-harvesting pigments, absorbing light energy for use in photosynthesis; simultaneously they provide photoprotection via non-photochemical quenching.1–3Mammals obtain carotenoids predominantly through plant foods, carnivorous animals obtain them also from animal fat.4 In organism carotenoids are absorbed by the intestines into the blood, which transports them to various tissues in the body using lipoproteins. Approximately twenty of fat-soluble carotenoids are found in human blood and tissues.5 In mammals β-carotene, vitamin E and other carotenoids are commonly perceived as antioxidants, but their biological activities are different and distinguish them from each other.6,7 The unique biological properties of carotenoids, which influence the immune system,6,8 intercellular communication control, differentiation cells growth regulation9 and apoptosis10 have activated the interest in this group of compounds for many years and are crucial for the understanding of their metabolism and beneficial role for human health.11
Plant foods were shown to be inversely associated with cancer risk in epidemiologic studies.12–14 The significant association with cancer risk has been reported for α-carotene, β-carotene and lycopene with breast cancer,15–22 and with ovarian cancer,23,24 for lycopene with prostate cancer,25,26 for lycopene, lutein, vitamin E and β-carotene with colorectal cancer.27–34 However, not all reported results are consistent and more studies are needed to find clear relation between cancer risk and supplementation using carotenoids.35–37
The protective role of β-carotene against reactive oxygen species (ROS) has been shown also by e.g. by Zhou et al.38 Based on animal model their proved that β-carotene can serve as a neuro-protective compound in the spinal cord injury (SCI) model and could improve functional recovery as well as inhibit oxidative stress and inflammation via inhibiting NF-κB in the spinal cord of SCI rats. Palozza et al.39 has shown that at low concentrations, the carotenoids may do duty as an antioxidant, inhibiting free radical production based on human colon adenocarcinoma as well as human leukemia studies. Antioxidative properties of β-carotene evinced by neutralization of reactive oxygen species and free radicals that can damage DNA in development of stomach cancer have been discussed by McCullough et al.40 Extraordinary antioxidative properties of carotenoids against ROS-mediated diseases, i.e. cancer, cardiovascular or photosensitivity disorders has been discussed by Fiedor et al.41 The special role of β-carotene, which can act as light filter and prevent oxidative stress by diminishing light exposure and can serve as a part of the antioxidant defense system in animals and humans has been discussed also by Stahl et al.42
The purpose of this study was to show that the protective role of β-carotene against reactive oxygen species (ROS) can be investigated based on analysis of human colon single cells by using Raman spectroscopy and imaging. The influence of β-carotene supplementation time was also taken into cognizance.
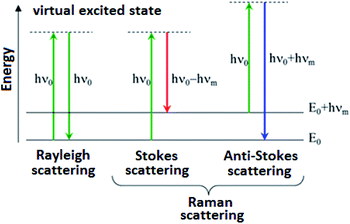
Raman spectroscopy is an analytical technique where inelastic scattered light is used to obtain the information about the vibrational energy of analysed molecules.
In the vast majority of scattering events, the energy of the molecule is unchanged after its interaction with the photon; and the energy, and therefore the wavelength, of the scattered photon is equal to that of the incident photon. This is called elastic (energy of scattering particle is preserved) or Rayleigh scattering and is the dominant process during interaction of photon with the molecule.
In a much rarer event (approximately 1 in 10 million photons) Raman scattering occurs, which is an inelastic scattering process with a transfer of energy between the molecule and scattered photon. If the molecule gains energy from the photon during the scattering (excited to a higher vibrational level) then the scattered photon loses energy and its wavelength increases which is called Stokes Raman scattering. Inversely, if the molecule loses energy by relaxing to a lower vibrational level the scattered photon gains the corresponding energy and its wavelength decreases; which is called Anti-Stokes Raman scattering. Quantum-mechanically, Stokes and Anti-Stokes are equally probable processes. However, with an ensemble of molecules, the majority of molecules will be in the ground vibrational level (Boltzmann distribution) and Stokes scattering is statistically more probable process. In consequence, the Stokes Raman scattering is always more intense than the Anti-Stokes component and for this reason, it is nearly always the Stokes Raman scattering that is measured wherewithal Raman spectroscopy (Scheme 1).
The results for normal human colon cells, normal human colon cells exposed to oxidative stress conditions by using tBHP, normal human colon cells upon of β-carotene supplementation and tBHP adding and human colon cancer cells were compared. The influence of β-carotene supplementation time was also taken into cognizance.
Materials and methods
Cell culture
CCD-18Co (CRL-1459) cell line was purchased from ATCC and cultured using ATCC-formulated Eagle's Minimum Essential Medium, Catalog no. 30-2003. To make the complete growth medium, the fetal bovine serum to a final concentration of 10% was added. Every 2 to 3 days the new medium was used. CaCo-2 cell line was also purchased from ATCC and cultured according to the ATCC protocols. The base medium for this cell line was ATCC-formulated Eagle's Minimum Essential Medium, Catalog no. 30-2003. To make the complete growth medium, we have added fetal bovine serum to a final concentration of 20%. Medium was renewed 1 to 2 times a week. Cells used in the experiments were stored in an incubator providing environmental conditions at 37 °C, 5% CO2, 95% air. For all results presented in this manuscript we have recorded the Raman spectra and imaging for paraformaldehyde fixed cells. The procedure for fixed cells was as follows: cells were seeded onto CaF2 windows (25 × 1 mm) at a low density of 103 cells per cm3. After 24 h or 48 h incubation on the CaF2 slides with β-carotene (c = 10 μM) the tBHP for a final concentration of 200 μM was added for 30 min, after a half hour the cells were rinsed with phosphate-buffered saline (PBS, SIGMA P-5368, pH 7.4 at 25 °C, c = 0.01 M) to remove any residual medium or/and any residual β-carotene, fixed in paraformaldehyde (4% buffered formaldehyde) for 10 minutes and washed twice with distilled water. The Raman confocal measurements were performed immediately after the preparation of the samples.Raman spectra and imaging acquisition and analysis
All Raman images and spectra reported in this manuscript were recorded using the alpha 300 RSA+ confocal microscope (WITec, Ulm, Germany) using a 50 μm core diameter fiber, 532 nm excitation line, an imaging spectrograph/monochromator (Acton-SP- 2300i), a CCD camera (Andor Newton DU970-UVB-353) and an Ultra High Throughput Spectrometer (UHTS 300). Excitation laser line was focused on the samples through a 40× water dipping objective (NA: 1.0). The average laser excitation power was 10 mW, with integration time of 1.0 s for low-frequency region and 0.5 s for high-frequency region. Edge filters were used to remove the Rayleigh scattered light. A piezoelectric table was used to record Raman images. Spectra were collected at one acquisition per pixel. The cosmic rays were removed from each Raman spectrum (model: filter size: 2, dynamic factor: 10) and the Savitzky–Golay smoothing procedure was also implemented (model: order: 4, derivative: 0). Data acquisition and processing were performed using WITec Project Plus software.All imaging data were analyzed using the Cluster Analysis method (CA) implemented in WITec Project Plus software.
Cluster analysis method
Briefly, Cluster Analysis is a form of exploratory data analysis in which observations are divided into different groups that have some common characteristics – vibrational features in our case. Cluster Analysis constructs groups (or classes or clusters) based on the principle within a group the observations must be as similar as possible, while observations belonging to different groups must be as different.The partition of n observations (x) into k (k ≤ n) clusters S should be done to minimize the variance (Var) according to the formula:
The normalization was performed using Origin software (model: divided by the norm).
Chemical compounds
β-Carotene catalogue number C9750, Luperox® TBH70X, tert-butyl hydroperoxide solution catalogue number 458139, bisbenzimide H 33342 trihydrochloride catalogue number B2261, Red Oil-O catalogue number O0625 were purchased from Sigma-Aldrich, and used without additional purification. XTT proliferation kit with catalogue number 20-300-1000 was purchased from Biological Industries.XTT cells viability tests
Tetrazolium salts have been widely used for many years as detection reagents in histochemical and cell biology tests.43,44 The second generation tetrazolium dye, XTT, can be effectively used in tests to assess viability and proliferation, metabolic cytotoxicity, respiratory chain activity and apoptic cells.44–46The use of the XTT reagent is based on the statistical calculation of the metabolic activity of cells using a colorimetric technique which is a reaction to the changed environmental conditions in which they are located. The XTT test requires the use of a wavelength of 450 nm, from which the specific signal of the sample is obtained, and 650 nm constituting the reference sample during the test. The XTT test was used to calculate the metabolic activity of a living cell.
Tetrazolium salts are converted in cells with the help of special enzymes called diformazan, however, this reaction occurs properly only in cells with undamaged metabolism. The XTT test counts the final amount of formazan and converts it to the number of live cells. The advantage of the XTT test is the fact that this reagent is soluble in water, thanks to which the first representative readings can be obtained after about 3 hours of incubation. The sensitivity of the XTT test can be significantly increased thanks to the use of an intermediate electron carrier – PMS (n-methyldibenzopyrazine methyl sulfate) – which catalyzes the reduction of XTT and formation of a formazan derivative. The PMS activation reagent is included with the XTT in the ATCC XTT Cell Proliferation Assay Kit (ATCC®30-1011K™) that was used for the cell culture studies in this work.
XTT tests on cell lines were carried out on a multi-sensing BioTek Synergy HT model reader designed for microplate testing. The test protocol was created especially for the presented experiments. After the microplate was applied to the reader, the inside of which was at a room temperature of 20–22 °C, the sample underwent a pre-mixing process. Then, the actual measurement was carried out, consisting in examining the signal coming from the sample with excitation at 450 nm. After measuring the samples, the instrument carried out a reference measurement with excitation at 650 nm wavelength. After this sequence the microplate was removed from the reader.
The stress compound solutions were prepared by pre-diluting it in the PBS solvent. After obtaining the initial concentration of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μM, it was diluted in a medium intended for a given type of cells, so as to obtain the final concentration of 200 μM.
000 μM, it was diluted in a medium intended for a given type of cells, so as to obtain the final concentration of 200 μM.
Scheme 2 shows the results of XTT test obtained for CCD-18Co human, normal, colon cells incubated with β-carotene in various concentrations and in various time.
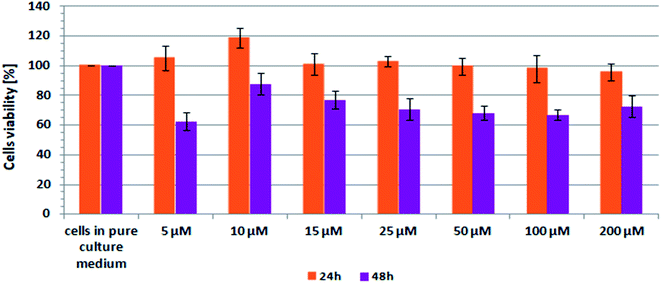 | ||
| Scheme 2 Results of XTT analysis obtained for CCD-18Co human, normal, colon cells supplemented with different concentrations of β-carotene in different time intervals. | ||
Results
In this section, the data obtained by using Raman spectroscopy and Raman imaging for human normal colon cells before and after ROS generation, including cells supplemented with β-carotene before ROS injuring are presented. We will show also the comparison on results obtained for normal human colon cells CCD-18Co in oxidative stress conditions and cancer human colon cell line CaCo-2.Generally, in the Raman vibrational spectra there are two regions of interest: the Raman fingerprint region: 500–1800 cm−1 and the high frequency region: 2700–3100 cm−1 (the region 1800–2700 cm−1 is not considered for analysis due to the lack of Raman bands).
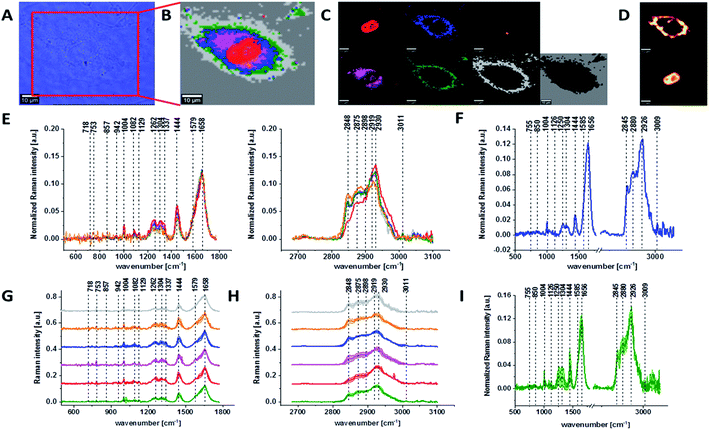
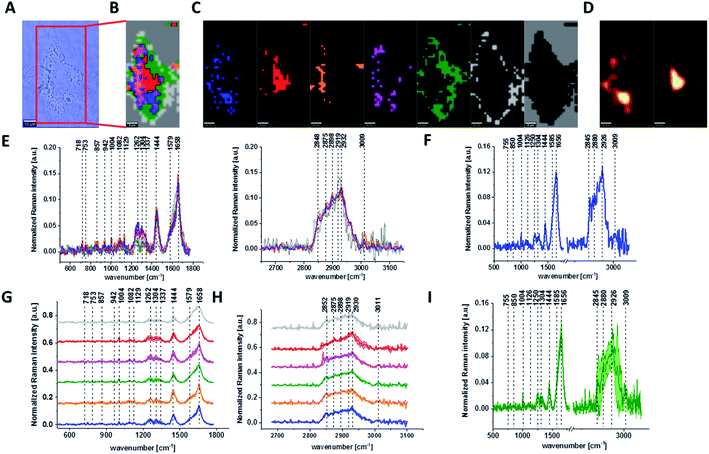
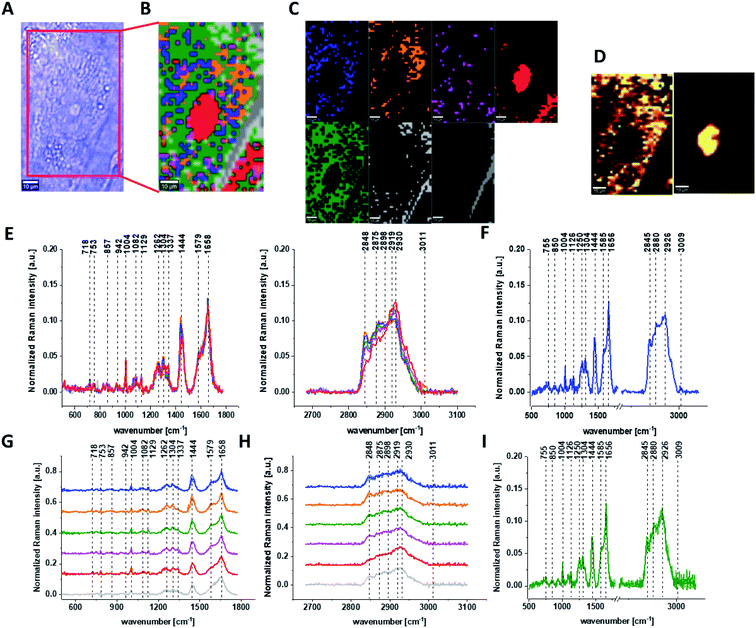
Fig. 1 presents the microscopy image, Raman image of single human normal colon cell CCD-18Co constructed based on Cluster Analysis (CA) method, Raman images of all clusters identified by CA assigned to: nucleus, mitochondria, lipid-rich regions, membrane, cytoplasm, and cell environment, fluorescence images of lipid-rich regions and nucleus, the average Raman spectrum typical for single human normal colon cell CCD-18Co presented on microscopy image, the average Raman spectra typical for CCD-18Co human normal colon cells mean ± SD for all identified clusters for low frequency and high frequency region, and the average Raman spectrum for human normal colon cells mean ± SD for cells as a whole, all data for experiments performed without any supplementation of β-carotene, cells measured in PBS, colors of the spectra correspond to the colors of clusters.
Because β-carotene is soluble only in organic compounds, we decided to compare the Raman spectra typical for CCD-18-Co human normal colon cells analyzed in PBS (shown in Fig. 1) with results obtained for CCD-18-Co human normal colon cells after adding the same volume of organic solvent (THF/EtOH) as for 10 μM β-carotene supplementation (10 μM it was the final concentration obtained in culturing medium). The results of Raman imaging analysis of CCD-18Co human colon normal cells after adding the mixture of organic solvents to PBS including the comparison on the average Raman spectra typical for cells as whole analyzed in PBS and in PBS/THF/EtOH are shown on Fig. 2.
One can see from Fig. 2 that the differences between the average Raman spectra (for cells as a whole) recorded in PBS and in PBS/THF/EtOH solution are subtle and fluctuate around 2%. Even though the difference is very slight, as the control group in all further comparisons and calculations we used results obtained for CCD-18-Co cells in PBS/THF/EtOH solution.
Results presented in Fig. 1 and 2 confirm that Raman spectroscopy and imaging can be used to characterize the biochemical composition of human normal colon cells. Table 1 shows the main chemical components which can be identified based on their vibrational features in analyzed cells and the tendency in Raman peaks intensities observed for CCD-18Co human, normal colon cells in oxidative stress conditions generated by tBHP adding (discussed later in the manuscript).
| Wavenumber [cm−1] | Tentative assignments of Raman bands | Tendency in Raman peak intensity observed for CCD-18Co human, normal colon cells in oxidative stress conditions generated by adding tBHP |
|---|---|---|
| 716 | C–N (membrane phospholipids head)/adenine, CN2(CH3)3 (lipids), choline group | ↘ |
| 812 | Tyrosine | ↘ |
| 820 | Structural protein modes of tumors, proteins, including collagen I | ↘ |
| 832 | Tyrosine (Fermi resonance of ring fundamental and overtone), asymmetric O–P–O stretching, tyrosine | ↘ |
| 869 | C–C stretching modes of protein | ↘ |
| 891 | Protein bands, structural protein modes of tumors | ↘ |
| 932 | Skeletal C–C, α-helix, proline, hydroxyproline, ν(C–C) skeletal of collagen backbone | ↘ |
| 1004 | Symmetric ring breathing of phenylalanine | ↘ |
| 1078 | Symmetric PO2− of DNA (represents more DNA in cell) | ↘ |
| 1254 | Amide III, adenine, cytosine | ↘ |
| 1304 | CH2 deformation (lipid), adenine, cytosine | ↘ |
| 1444 | δ(CH2), lipids, fatty acids | ↘ |
| 1602 | δ(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), phenylalanine (protein assignment) C), phenylalanine (protein assignment) |
↘ |
| 1626 | Amide C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching absorption for the b-form polypeptide films O stretching absorption for the b-form polypeptide films |
↘ |
| 1654 | Amide I | ↘ |
| 1720 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
↘ |
| 1754 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (lipid) O (lipid) |
↘ |
| 2854 | CH2 symmetric stretch of lipids & CH2 asymmetric stretch of lipids and proteins | ↘ |
| 2880 | CH2 asymmetric stretch of lipids and proteins | ↘ |
| 2926 | Symmetric CH3 stretch, due primarily to protein | ↘ |
| 3009 | νas(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), lipids, fatty acids C–H), lipids, fatty acids |
↘ |
The various signals seen on Fig. 1 and 2 have been assigned to nucleic acids, proteins or lipids and provide adequate information to assess variations in spectral characteristics.
As we mentioned above the main goal of our experiments was to prove the protective role of natural antioxidants in oxidative stress conditions by using Raman spectroscopy and imaging, therefore the next step was the analysis of human colon normal cells CCD-18Co after ROS generation. The main advantage of Raman imaging method that should be highlighted in this point is that vibrational imaging allows to investigate biochemical composition of cells without any cells destruction. Moreover, the high spatial resolution allows to track any changes on subcellular level.
Before we start the analysis of spectroscopic data we have to underline a few regularities.
ROS are produced by living organisms as a result of natural cellular metabolism, but in such a case the concentrations of them is low to moderate their functions in physiological cell processes, but at high ROS concentrations adverse modifications to cell components, such as: lipids, proteins, and DNA can be noticed.48–53 The shift in balance between oxidant/antioxidant in favor of oxidants seems to be crucial for understanding many dysfunctions of human cells.
It has been shown in literature that oxidative stress contributes to many pathological conditions, including cancer,14–36 neurological disorders,54–57 atherosclerosis, hypertension, ischemia/perfusion,58–61 acute respiratory distress syndrome, idiopathic pulmonary fibrosis, chronic obstructive pulmonary disease,62 and asthma.63–65
Generally, ROS can be divided into two groups: free radicals and nonradicals. Free radicals contain one or more unpaired electrons and are characterize by high reactivity to molecules. Free radicals can recombine sharing electrons and create nonradical forms. The three ROS of physiological significance are: superoxide anion (O2−˙), hydroxyl radical (OH˙), and hydrogen peroxide (H2O2). Superoxide anion is formed by the addition of one electron to the molecular oxygen. This process is mediated by nicotine adenine dinucleotide phosphate [NAD(P)H] oxidase, xanthine oxidase or by mitochondrial electron transport system. The main place where the superoxide anion is produced are mitochondria because 1–3% of all electrons “leak” from respiration chain system and produce superoxide. Superoxide is then converted into hydrogen peroxide, which diffuses across the plasma membrane by the action of superoxide dismutases.
The most reactive of ROS is hydroxyl radical, which can damage proteins, lipids, carbohydrates and DNA. Hydroxyl radical can start lipid peroxidation by taking an electron from polyunsaturated fatty acids. Other oxygen-derived free radicals are the peroxylradicals (ROO˙). Simplest form of these radicals is hydroperoxyl radical (HOO˙), which plays a crucial role in fatty acid peroxidation. Free radicals can trigger lipid peroxidation chain reactions by abstracting a hydrogen atom from a sidechain methylene carbon. The lipid radical then reacts with oxygen to produce peroxyl radical. Peroxyl radical initiates a chain reaction and transforms polyunsaturated fatty acids into lipid hydroperoxides.66 Lipid hydroperoxides are very unstable and easily decompose to secondary products, such as aldehydes. Isoprostanes are another group of lipid peroxidation products that are generated via the peroxidation of unsaturated fatty acids e.g. arachidonic acid.
tert-Butyl hydroperoxide (tBHP) used in our experiments to produce ROS is well-known as a model substance for oxidative stress generation and subsequent analysis of cellular alterations in cells and tissues. Generally, in living organisms two pathways by which tBHP is metabolized can be distinguish; both of them induce oxidative stress. The first one provided by cytochrome P450, leads to production of peroxyl and alkoxyl radicals.67 The second pathway employs glutathione peroxidase. tert-Butyl hydroperoxide is detoxified to tert-butanol and reduced glutathione is depleted by oxidation to its disulphide form.21
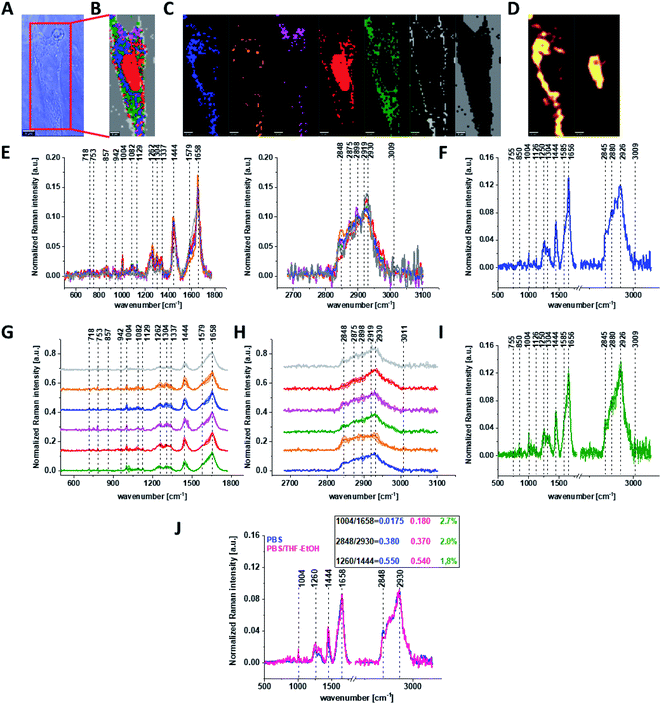
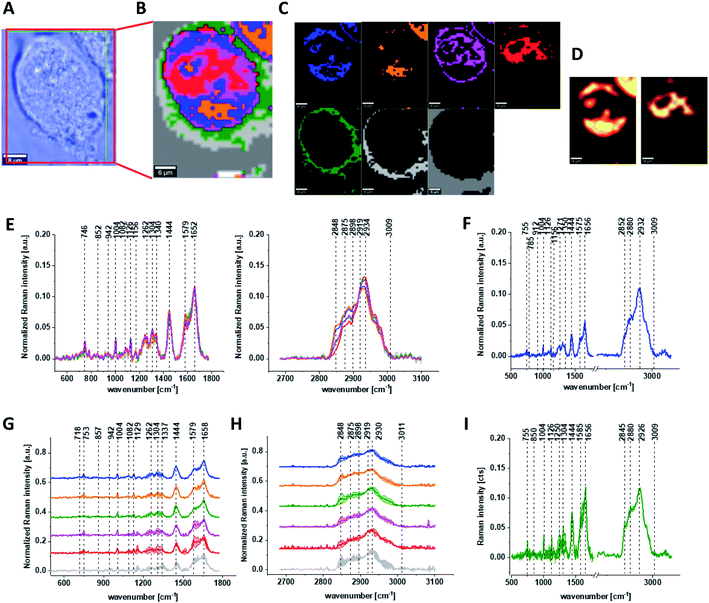
Fig. 3 shows the microscopic image, Raman image constructed based on Cluster Analysis (CA) method, Raman images of all clusters identified by CA assigned to: nucleus, mitochondria, lipid-rich regions, membrane, cytoplasm, and cell environment, fluorescence images of lipid-rich regions and nucleus, the average Raman spectra typical of all clusters identified by CA for single human normal colon cell for low frequency and high frequency region, the average Raman spectrum typical of single CCD-18Co cell as a whole, and the average Raman spectra for human normal colon CCD-18Co cells mean ± SD after adding tBHP for 30 minutes, colors of the spectra correspond to the colors of clusters.
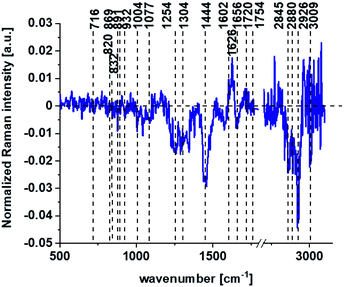
Based on high resolved Raman spectra recorded for normal human colon cells CCD-18Co and for individual organelles without and after adding tBHP for ROS generation shown in Fig. 2 and 3 one can obtained the complex information about the biochemistry of single cell as a whole or we can track changes in cell's nucleus, mitochondria, and lipid structures. Fig. 4 and Table 1 show the main spectroscopic differences detected based on vibrational Raman spectra before and after oxidative stress generation.
Fig. 4 shows the difference Raman spectrum of human normal colon cells from control sample (CCD-18Co in PBS/THF/EtOH solution) and human normal colon cells in oxidative stress conditions generated by tBHP adding (data based on the Raman vibrational spectra for cells as a whole).
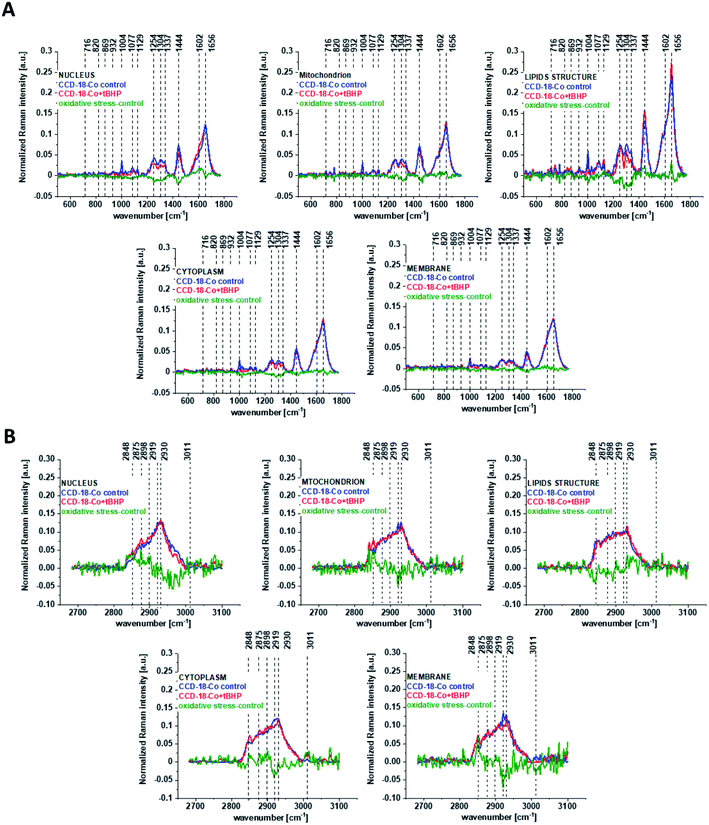
One can see from Fig. 4 that ROS generation has changed the biochemical composition of normal human colon cells CCD-18Co. In detail changes noticed in Fig. 4 will be discussed in the manuscript later. The tendency in Raman peaks intensities observed for CCD-18Co human, normal colon cells after ROS generation by tBHP adding for cells as a whole is also shown in Table 1. As we have stressed above Raman microspectroscopy and Cluster Analysis of Raman data allow to observed the influence of ROS generation on individual organelles of human, normal, CCD18-Co cells. Fig. 5 shows the difference spectra (green) calculated for nucleus, mitochondria, lipids structures, cell membrane and cytoplasm in high frequency region (A) and in low frequency region (B) based on the average Raman spectra typical for all clusters assigned to the subcellular structures for cells in control group (blue) and after tBHP adding (red).
Fig. 5 shows that the generation of ROS affects the intensity of the Raman signal assigned to all selected organelles of human normal CCD18-Co colon cells. This observation confirms that ROS generation can affect the function of all cell substructures.
The next step of the analysis was the interpretation of Raman data obtained for CCD-18Co human normal colon cells at first supplemented by β-carotene for 24 or 48 hours and then treated using tBHP for 30 min.
Fig. 6 shows all results obtained for human normal colon cells CCD-18Co after 24 h of β-carotene supplementation and then adding of tBHP for 30 min.
Fig. 7 shows all results obtained for human normal colon cells CCD-18Co after 48 h of β-carotene supplementation and then adding of tBHP for 30 min.
One can see form Fig. 6 and 7 that also for conditions including β-carotene supplementation and ROS generation we can obtain high resolved Raman spectra based on which the influence on biochemistry of single cells of antioxidant supplementation and oxidative stress can be analyzed.
Discussion
Having reached this point when the analysis based on Raman vibrational spectra for different human normal colon CCD-18Co cells groups has been performed we can compare and discuss results shown on Fig. 1–7.Considering that CCD-18Co human normal colon cells are basically composed of three types of macromolecules: proteins, nucleic acids and lipids, in order to explore characteristic changes under ROS conditions we will focused the attention to these class of compounds. The qualitative and quantitative comparison between paired bands assigned to proteins, lipids and nucleic acids will continue to be discussed according to the biological attribution of them.
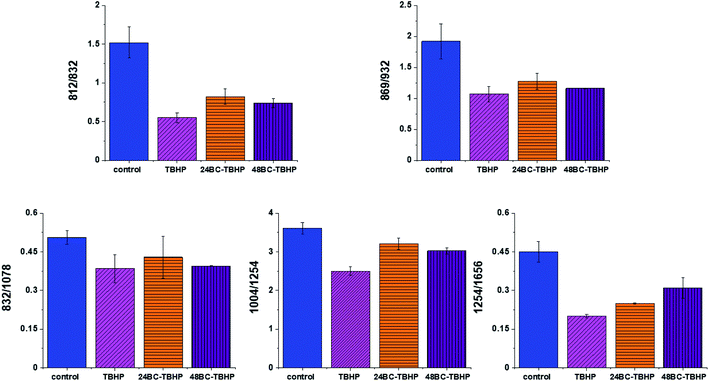
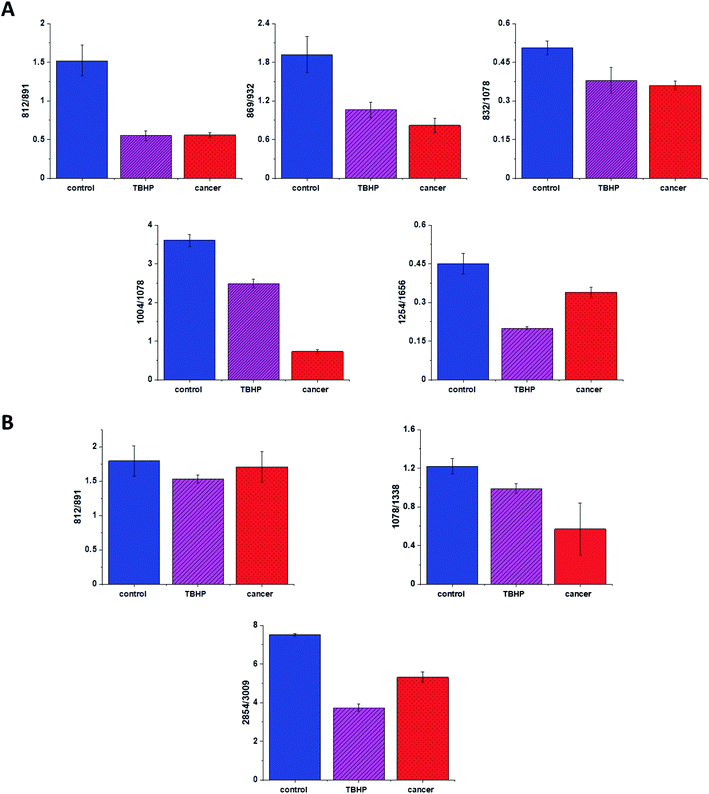
Based on the results shown in Table 1 and frequencies highlighted in Fig. 1–7 we have chosen the following Raman bands to compare: 812, 832, 869, 932, 1004, 1254, 1654 cm−1.
Fig. 8 shows the comparison of Raman band intensity ratios for four different human normal colon cells groups: control group, group treated by using tBHP for 30 min, group at first supplemented with β-carotene for 24 h and after that incubated with tBHP for 30 min, and group at first supplemented with β-carotene for 48 h and after that incubated with tBHP for 30 min.
Compared with the oxidation group (magenta), the relative intensities of several protein bands 812/832, 869/932, 1004/1254 and 832/1078 (the ratio for Raman bands typical for protein (tyrosine) and DNA) in the control (blue) and protective groups (at first supplemented by β-carotene, orange and violet) show the same variation trend.
In detail for Raman bands at 812 and 832 cm−1 for tBHP group (after ROS generation) compare to control one the decreasing for Raman bands ratio I812/I832 was observed. Decreasing observed in the oxidation group, indicates that tyrosine and its internal ‘out of plane ring breathing mode’ was affected by oxidation. Such interaction suggests that the molecular vibration of tyrosine is sensitive to oxidation, resulting in structural changes. The adding of β-carotene before ROS generation results in increasing of analyzed ratio compared to the oxidation group.
The bands at 869 and 932 cm−1 also correspond to proteins. The band 869 cm−1 came from C–C stretching modes of proteins, while the band at 932 cm−1 correspond to C–C stretching vibrational mode of proline, valine and protein backbone (α-helix conformation). One can see form Fig. 8 that the ratio of Rama band intensities I869/I932 is significantly decreased in the oxidation group, and once again, increased after adding β-carotene. Although the observed increasing confirms the protective role of β-carotene, still the relative intensity is lower than observed for the control group. Summarizing, all observations made for the ratio I869/I932 indicate that tBHP changes the content and structure of some amino acids by reaction of ROS with proteins, resulting in concentration modulation and functional damage of them. This observation is consistent with data published in the literature.68 The same trend was observed for the ratio calculated based on the Raman bands corresponding to phenylalanine (1004 cm−1) and Amide III. Once again one can observe the decreasing of the ratio for oxidation group and protective role of the antioxidant even if the values for 24BC + tBHP and 48BC + tBHP groups are lower than typical for control cells. The same variation for all analyzed cells groups was noticed for Raman bands 1245 and 1656 cm−1. The decreasing of the ratio I1254/I1656 for ROS injured cells and systematic increase of this ratio as a function of incubation time with β-carotene was observed. Moreover, the change of band ratio of amides I1254/I1656 may suggest also that a desamidization was triggered and resulted in modification of the spatial structure of proteins, in loss of protein activity or modification of their biological function.69
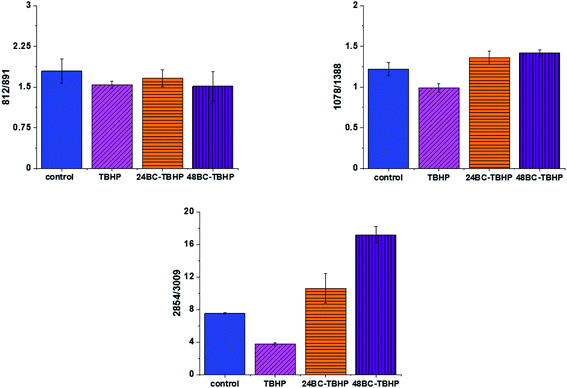
The analysis of Raman bands ratios has been performed also for bands typical for nucleic acids and lipids. Fig. 9 shows the results obtained for these types of compounds.
For bands typical for nucleic acids (812, 891, 1078, 1388 cm−1) and lipids (2854, 3009 cm−1) once again as for bands typical for proteins we have noticed changes induced by ROS and modulations triggered by β-carotene supplementation. Lipids are the main component of subcellular structures like lipid droplets and the cell membrane, which is susceptible to peroxidation induced by ROS, products from ROS reaction can cause damage to either the cell membrane or the organelle membrane.70,71 The main protective role of β-carotene in this case was seen for lipids fraction, especially unsaturated one observed in high-frequency region. We have to remember that lipids peroxidation is a process that consists of three phases: initiation, propagation and termination. In the initiation phase, the hydrogen atom is separated from the molecule of polyunsaturated fatty acid or the rest of such acid that is part of the phospholipid under the influence of e.g. the hydroxyl radical (OH˙). In the propagation phase reactions alkyl free radicals react with oxygen to form peroxide free radicals, which detach hydrogen atoms from subsequent, undamaged molecules of unsaturated fatty acids. This reaction produces fatty acid peroxide and another alkyl radical which can be oxidized another molecule of fatty acid. The above reactions can repeat many times, which leads to the transformation into peroxides – several, several dozen or even several hundred fatty acid molecules. Lipid peroxidation products change the physical properties of all cell membranes. For phospholipids located inside the lipid bilayer, introduced polar peroxide, ketone and aldehyde groups remain or hydroxy. This lowers the hydrophobicity of the lipid interior of cell membranes, as well as changes the organization of the lipid bilayer, which to disturbance of lipid asymmetry of membranes.72
We also noticed also that the β-carotene may affect the ROS damages induced in nucleic acids (812/893, 1078/1386). DNA ROS damage effect may involve two aspects, nucleic acid bases and DNA phosphoric acid skeleton.73 ROS are well known as mediators of double strand breaks (DSBs), which can be mutagenic due to chromosomal rearrangements or loss of genetic information due to erroneous DNA repair. ROS have also been reported to directly induce other forms of DNA damage through oxidizing nucleoside bases e.g. formation of 8-oxo guanine,74 which can lead to G–T or G–A transversions if unrepaired. Oxidized bases are typically recognized and repaired by the Base Excision Repair (BER) pathway, but when they occur simultaneously on opposing strands, attempted BER can lead to the generation of DSBs.75 It has been shown that ROS accumulation also induces mitochondrial DNA lesions, strand breaks and degradation of mitochondrial DNA.76 Data presented on Fig. 9 confirm that based on Raman spectra the changes in DNA after ROS generation can be tracked.
It has been shown that permanent changes in the genome of cells e.g. as a result of oxidative stress, it is the first stage characteristic of the process of mutagenesis, carcinogenesis and cell aging. The appearance of a mutation in DNA is a critical step in the process of cancer development and more than 100 DNA modifications have been identified in cancer cells products.77
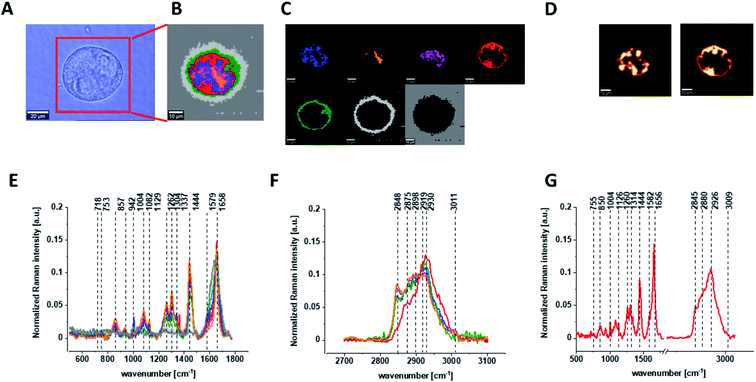
Finally, we have decided to compare the results obtained for CCD18-Co human normal colon cells in oxidative stress conditions with data obtained for CaCo-2 human cancer cell line. Fig. 10 shows the data obtained for human cancer colon cells CaCo-2. Fig. 11 the comparison for selected Raman bands typical for proteins, nucleic acids and lipids.
Fig. 11 shows the comparison of ratios for selected Raman bands typical for proteins, nucleic acids and lipids for CCD-18Co human normal colon cells, CCD-18Co human normal colon cells after ROS generation by adding of tBHP for 30 min and CaCo-2 human cancer colon cells.
One can see form Fig. 11 that intensities of Raman band typical for proteins, nucleic acids and lipids are comparable for CCD-18Co human normal colon cells in oxidative stress conditions and for CaCo-2 human colon cancer cell and simultaneously differ significantly from control group. This observation confirm that Raman band intensities analysis allow to track biochemical changes induced by ROS and during cancerogenesis. Moreover, comparable ratios for ROS injured cells and cancer cells may confirm that cells altered by ROS shows many common characteristic with pathological cells.
Conclusions
In the presented work, we performed label-free detection of tBHP-induced oxidative stress by using Raman imaging and spectroscopy. We have shown that Raman imaging and spectroscopy are capable to characterize and differentiate human normal colon cells CCD-18Co, human normal colon cells after ROS generation, human normal colon cells at first incubated with β-carotene and after that treated by using tBHP, and finally human, cancer colon cells CaCo-2.Moreover, we have confirmed that substructures of human colon single cells such as: nucleus, mitochondria lipid-rich regions, membrane, and cytoplasm can be precisely visualized based on Raman spectra. Moreover, biochemical changes typical for selected cells substructures can be tracked based on vibrational features.
We have shown also that fluorescence based images accurately correspond to the Raman images confirming localization of cells substructures such as: nucleus and lipids-rich structures.
Based on Raman band intensities attributed to proteins, nucleus acids and lipids as well as ratios 812/832, 869/932, 832/1078, 1004/1254, 1254/1656, 812/891, 1078/1368 2854/3009 calculated based on them, we have confirmed the protective role of β-carotene for cells in oxidative stress conditions for label-free and nondestructive spectroscopic method.
Author contributions
Conceptualization: BB-P; funding acquisition: BB-P; investigation: BB-P, KB; methodology: BB-P, KB, writing – original draft: BB-P; manuscript editing: BB-P, KB. All authors reviewed and provide feedback on the manuscripts.Conflicts of interest
There are no conflicts to declare. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.Funding
This work was supported by the National Science Centre of Poland (Narodowe Centrum Nauki) UMO-2017/25/B/ST4/01788.References
- W. Stahl and H. Sies, Am. J. Clin. Nutr., 2012, 96, 1179S–1184S CrossRef CAS PubMed.
- W. Stahl and H. Sies, Biochim. Biophys. Acta, Mol. Basis Dis., 2005, 1740, 101–107 CrossRef CAS PubMed.
- G. E. Bartley and P. A. Scolnik, Plant Cell, 1995, 7, 1027–1038 CAS.
- J. A. Olson, Arch. Latinoam. Nutr., 1999, 49, 7S–11S CAS.
- E. J. Johnson, Nutr. Clin. Care, 2002, 5, 56–65 CrossRef.
- D. A. Hughes, A. J. A. Wright, P. M. Finglas, A. G. J. Polley, A. L. Bailey, S. B. Astley and S. Southon, J. Infect. Dis., 2000, 182(suppl. 1), S11–S15 CrossRef CAS.
- D. A. Hughes, A. J. A. Wright, P. M. Finglas, A. C. J. Peerless, A. L. Bailey, S. B. Astley, A. C. Pinder and S. Southon, J. Lab. Clin. Med., 1997, 129, 309–317 CrossRef CAS.
- S. Moriguchi and M. Muraga, Vitam. Horm., 2000, 59, 305–336 CrossRef CAS.
- P. Palozza, G. Calviello, S. Serini, N. Maggiano, P. Lanza, F. O. Ranelletti and G. M. Bartoli, Free Radicals Biol. Med., 2001, 30, 1000–1007 CrossRef CAS.
- J. M. Turley, S. Funakoshi, F. W. Ruscetti, J. Kasper, W. J. Murphy, D. L. Longo and M. C. Birchenall-Roberts, Cell Growth Differ., 1995, 6, 655–663 CAS.
- A. Kaczor and M. Baranska, Carotenoids: Nutrition, Analysis and Technology, John Wiley & Sons, Ltd, Chichester, UK, 2016 Search PubMed.
- K. Hultén, A. L. Van Kappel, A. Winkvist, R. Kaaks, G. Hallmans, P. Lenner and E. Riboli, Cancer Causes Control, 2001, 12, 529–537 CrossRef PubMed.
- K. A. Steinmetz and J. D. Potter, Cancer Causes Control, 1991, 2, 325–357 CrossRef CAS PubMed.
- J. D. Potter, M. L. Slattery, R. M. Bostick and S. M. Gapstur, Colon Cancer A Review of the Epidemiology, 1993, vol. 15 Search PubMed.
- L. Slattery Martha, J. D. Potter, A. Coates, K.-N. Ma, B. T. Dennis, M. Duncan Debra and J. Caan Bette, Cancer Causes Control, 1997, 8, 575–590 CrossRef PubMed.
- B. Brozek-Pluska, A. Jarota, J. Jablonska-Gajewicz, R. Kordek, W. Czajkowski and H. Abramczyk, Technol. Cancer Res. Treat., 2012, 11, 317–331 CrossRef CAS PubMed.
- H. Abramczyk, B. Brozek-Pluska, J. Surmacki, J. Musial, R. Kordek and R. Microspectroscopy, Analyst, 2014, 139, 5547–5559 RSC.
- H. Abramczyk, A. Imiela, B. Brozek-Pluska and M. Kopec, Nanomedicine – Futur. Med., 2019, 14, 1873–1888 CrossRef CAS PubMed.
- B. Brozek-Pluska, M. Kopeć and H. Abramczyk, Anal. Methods, 2016, 8, 8542–8553 RSC.
- H. Abramczyk and B. Brozek-Pluska, Chem. Rev., 2013, 113, 5766–5781 CrossRef CAS PubMed.
- H. Abramczyk, B. Brozek-Pluska, A. Jarota, J. Surmacki, A. Imiela and M. Kopec, Expert Rev. Mol. Diagn., 2020, 20, 99–115 CrossRef CAS PubMed.
- H. Abramczyk, B. Brozek-Pluska, J. Surmacki, J. Jablonska-Gajewicz and R. Kordek, Prog. Biophys. Mol. Biol., 2012, 108, 74–81 CrossRef CAS PubMed.
- D. W. Cramer, H. Kuper, B. L. Harlow and L. Titus-Ernstoff, Int. J. Cancer, 2001, 94, 128–134 CrossRef CAS PubMed.
- C. La Vecchia, Exp. Biol. Med., 2002, 227, 860–863 CrossRef CAS PubMed.
- T. M. Vogt, S. T. Mayne, B. I. Graubard, C. A. Swanson, A. L. Sowell, J. B. Schoenberg, G. M. Swanson, R. S. Greenberg, R. N. Hoover, R. B. Hayes and R. G. Ziegler, Am. J. Epidemiol., 2002, 155, 1023–1032 CrossRef CAS PubMed.
- E. Giovannucci, E. B. Rimm, Y. Liu, M. J. Stampfer and W. C. Willett, J. Natl. Cancer Inst., 2002, 94, 391–398 CrossRef CAS PubMed.
- M. L. Slattery, J. Benson, K. Curtin, K. N. Ma, D. Schaeffer and J. D. Potter, Am. J. Clin. Nutr., 2000, 71, 575–582 CrossRef CAS PubMed.
- M. L. Slattery, S. L. Edwards, K. Anderson and B. Caan, Nutr. Cancer, 1998, 30, 201–206 CrossRef CAS PubMed.
- K. Wu, W. C. Willett, J. M. Chan, C. S. Fuchs, G. A. Colditz, E. B. Rimm and E. L. Giovannucci, Cancer Epidemiol., Biomarkers Prev., 2002, 11, 1298–1304 CAS.
- M. Ferraroni, C. La Vecchia, B. D'Avanzo, E. Negri, S. Franceschi and A. Decarli, Br. J. Cancer, 1994, 70, 1150–1155 CrossRef CAS PubMed.
- R. M. Bostick, J. D. Potter, D. R. McKenzie, T. A. Sellers, L. H. Kushi, K. A. Steinmetz and A. R. Folsom, Cancer Res., 1993, 53, 4230–4237 CAS.
- P. Ghadirian, A. Lacroix, P. Maisonneuve, C. Perret, C. Potvin, D. Gravel, D. Bernard and P. Boyle, Cancer, 1997, 80, 858–864 CrossRef CAS PubMed.
- B. Brozek-Pluska, K. Miazek, J. Musiał and R. Kordek, RSC Adv., 2019, 9, 40445–40454 RSC.
- B. Brozek-Pluska, J. Musiał, R. Kordek and H. Abramczyk, Int. J. Mol. Sci., 2019, 20, 3398 CrossRef CAS PubMed.
- P. Terry, M. Jain, A. B. Miller, G. R. Howe and T. E. Rohan, Nutr. Cancer, 2002, 42, 167–172 CrossRef CAS PubMed.
- A. Shin, H. Li, X. O. Shu, G. Yang, Y. T. Gao and W. Zheng, Int. J. Cancer, 2006, 119, 2938–2942 CrossRef CAS PubMed.
- N. Malila, J. Virtamo, M. Virtanen, P. Pietinen, D. Albanes and L. Teppo, Eur. J. Clin. Nutr., 2002, 56, 615–621 CrossRef CAS PubMed.
- L. Zhou, L. Ouyang, S. Lin, S. Chen, Y. J. Liu, W. Zhou and X. Wang, Int. Immunopharmacol., 2018, 61, 92–99 CrossRef CAS PubMed.
- P. Palozza, Biochim. Biophys. Acta, Mol. Basis Dis., 2005, 1740, 215–221 CrossRef CAS PubMed.
- M. L. McCullough and E. L. Giovannucci, Oncogene, 2004, 23, 6349–6364 CrossRef CAS PubMed.
- J. Fiedor and K. Burda, Nutrients, 2014, 6, 466–488 CrossRef PubMed.
- W. Stahl and H. Sies, Mol. Aspects Med., 2003, 24, 345–351 CrossRef CAS PubMed.
- F. P. Altman, Prog. Histochem. Cytochem., 1976, 9, 3–6 CrossRef.
- M. V. Berridge, P. M. Herst and A. S. Tan, Biotechnol. Annu. Rev., 2005, 11, 127–152 CAS.
- N. J. Marshall, C. J. Goodwin and S. J. Holt, Growth Regul., 1995, 5, 69–84 CAS.
- D. A. Scudiere, R. H. Shoemaker, K. D. Paul, A. Monks, S. Tierney, T. H. Nofziger, M. J. Currens, D. Seniff and M. R. Boyd, Cancer Res., 1988, 48, 4827–4833 Search PubMed.
- Z. Movasaghi, S. Rehman and I. U. Rehman, Appl. Spectrosc. Rev., 2007, 42, 493–541 CrossRef CAS.
- M. Valko, C. J. Rhodes, J. Moncol, M. Izakovic and M. Mazur, Chem.-Biol. Interact., 2006, 160, 1–40 CrossRef CAS PubMed.
- B. Halliwell and J. M. C. Gutteridge, Free Radicals in Biology and Medicine, Oxford University Press, New York, 3rd edn, 1999 Search PubMed.
- L. J. Marnett, Mutat. Res., Fundam. Mol. Mech. Mutagen., 1999, 424, 83–95 CrossRef CAS PubMed.
- W. G. Siems, T. Grune and H. Esterbauer, Life Sci., 1995, 57, 785–789 CrossRef CAS.
- E. R. Stadtman, Curr. Med. Chem., 2004, 11, 1105–1112 CrossRef CAS PubMed.
- M. Wang, K. Dhingra, W. N. Hittelman, J. G. Liehr, M. De Andrade and D. Li, Cancer Epidemiol., Biomarkers Prev., 1996, 5, 710 Search PubMed.
- P. Jenner, Ann. Neurol., 2003, 53(suppl. 3), S26–S38 CrossRef CAS PubMed.
- L. Lyras, N. J. Cairns, A. Jenner, P. Jenner and B. Halliwell, J. Neurochem., 1997, 68, 2061–2069 CrossRef CAS PubMed.
- L. Sayre, M. Smith and G. Perry, Curr. Med. Chem., 2012, 8, 721–738 CrossRef PubMed.
- P. K. Toshniwal and E. J. Zarling, Neurochem. Res., 1992, 17, 205–207 CrossRef CAS PubMed.
- N. S. Dhalla, R. M. Temsah and T. Netticadan, J. Hypertens., 2000, 18, 655–673 CrossRef CAS PubMed.
- S. Kašparová, V. Brezová, M. Valko, J. Horecký, V. Mlynárik, T. Liptaj, O. Vančová, O. Uličná and D. Dobrota, Neurochem. Int., 2005, 46, 601–611 CrossRef PubMed.
- S. Kerr, M. J. Brosnan, M. McIntyre, J. L. Reid, A. F. Dominiczak and C. A. Hamilton, Hypertension, 1999, 33, 1353–1358 CrossRef CAS PubMed.
- R. C. Kukreja and M. L. Hess, Cardiovasc. Res., 1992, 26, 641–655 CrossRef CAS PubMed.
- S. Asami, H. Manabe, J. Miyake, Y. Tsurudome, T. Hirano, R. Yamaguchi, H. Itoh and H. Kasai, Carcinogenesis, 1997, 18, 1763–1766 CrossRef CAS PubMed.
- A. A. Andreadis, S. L. Hazen, S. A. A. Comhair and S. C. Erzurum, Free Radicals Biol. Med., 2003, 35, 213–225 CrossRef CAS PubMed.
- S. A. A. Comhair, K. S. Ricci, M. Arroliga, A. R. Lara, R. A. Dweik, W. Song, S. L. Hazen, E. R. Bleecker, W. W. Busse, K. F. Chung, B. Gaston, A. Hastie, M. Hew, N. Jarjour, W. Moore, S. Peters, W. G. Teague, S. E. Wenzel and S. C. Erzurum, Am. J. Respir. Crit. Care Med., 2005, 172, 306–313 CrossRef PubMed.
- S. A. A. Comhair, W. Xu, S. Ghosh, F. B. J. M. Thunnissen, A. Almasan, W. J. Calhoun, A. J. Janocha, L. Zheng, S. L. Hazen and S. C. Erzurum, Am. J. Pathol., 2005, 166, 663–674 CrossRef CAS.
- D. Crane, D. Häussinger, P. Graf and H. Sies, Hoppe-Seyler's Z. Physiol. Chem., 1983, 364, 977–988 CrossRef CAS PubMed.
- M. J. Davies, Biochem. J., 1989, 257, 603–606 CrossRef CAS PubMed.
- E. R. Stadtman, Ann. N. Y. Acad. Sci., 2001, 928, 22–38 CrossRef CAS PubMed.
- P. Pacher, J. S. Beckman and L. Liaudet, Physiol. Rev., 2007, 87, 315–424 CrossRef CAS PubMed.
- S. X. Zhang, J. J. Wang, A. Dashti, K. Wilson, M. H. Zou, L. Szweda, J. X. Ma and T. J. Lyons, J. Mol. Endocrinol., 2008, 41, 135–143 CAS.
- R. Kohen and A. Nyska, Toxicol. Pathol., 2002, 30, 620–650 CrossRef CAS PubMed.
- E. J. McConnell, A. M. Bittelmeyer and B. U. Raess, Arch. Biochem. Biophys., 1999, 361, 252–256 CrossRef CAS PubMed.
- U. S. Srinivas, B. W. Q. Tan, B. A. Vellayappan and A. D. Jeyasekharan, Redox Biol., 2019, 25, 101084 CrossRef CAS PubMed.
- F. Salehi, H. Behboudi, G. Kavoosi and S. K. Ardestani, Sci. Rep., 2018, 8, 13902 CrossRef PubMed.
- W. J. Cannan, B. P. Tsang, S. S. Wallace and D. S. Pederson, J. Biol. Chem., 2014, 289, 19881–19893 CrossRef CAS PubMed.
- I. Shokolenko, N. Venediktova, A. Bochkareva, G. I. Wilson and M. F. Alexeyev, Nucleic Acids Res., 2009, 37, 2539–2548 CrossRef CAS PubMed.
- H. Pelicano, D. Carney and P. Huang, Drug Resist. Updates, 2004, 7, 97–110 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |