Open Access Article
Open Access ArticleIQF characterization of a cathepsin B-responsive nanoprobe for report of differentiation of HL60 cells into macrophages†
Yanhui Zhang‡
 ab,
Dehua Huang‡b,
Chengxing Zhang
ab,
Dehua Huang‡b,
Chengxing Zhang b,
Jingjing Mengb,
Bo Tanb and
Zongwu Deng
b,
Jingjing Mengb,
Bo Tanb and
Zongwu Deng *ab
*ab
aSchool of Nano-Tech and Nano-Bionics, University of Science and Technology of China, Hefei, 230026, P. R. China. E-mail: zwdeng2007@sinano.ac.cn; Tel: +86 512-62872559
bCAS Key Laboratory of Nano-Bio Interface, Suzhou Institute of Nano-tech and Nano-bionics, Chinese Academy of Sciences, Suzhou, 215123, P. R. China
First published on 6th May 2021
Abstract
Tracking of in vivo fates of exogenous cell transplants in terms of viability, migration, directional differentiation and function delivery by a suitable method of medical imaging is of great significance in the development and application of various cell therapies. In this contribution directional differentiation of HL60 cells into macrophages and granulocytes, and a difference in the associated expression level of cathepsin B (Cat B) among the parent and daughter cells is used as a model to guide and evaluate the development of a Cat B-responsive Abz-FRFK-Dnp@PLGA nanoprobe for an optical report of the differentiation process. A well-documented internally quenched fluorescence (IQF) pair coupled with a peptide substrate FRFK of Cat B was synthesized and imbedded in PLGA to form the nanoprobe. The nanoprobe is resistant to leakage when dispersed in water for 10 days. Degradation of the nanoprobe is dominated by Cat B. HL60 cells were then labelled with the Abz-FRFK-Dnp@PLGA nanoprobe to track the differentiation process. Differentiation of labelled HL60 cells into macrophages exhibited a significantly higher fluorescence relative to the granulocytes or the labelled parent cells. The fluorescence difference allows the differentiation process to be followed. The established characterization and assessment procedure is to be used for the development and evaluation of nanoprobes for other imaging modalities.
Introduction
Tracking of in vivo fates of exogenous cell transplants in terms of viability, migration, directional differentiation and function delivery by a suitable method of medical imaging is of great significance in the development and application of various cell therapies. Numerous labelling and imaging strategies have been proposed for report of cell migration and homing, apoptosis, and directional differentiation.1–10 In this respect, magnetic resonance imaging (MRI) is thought to be one of the most promising imaging tools due to its inherent soft-tissue contrast, high spatial resolution and lack of ionizing radiation. MRI tracking of cell transplants requires labelling of the cells with contrast agents (CAs) to allow them to be distinguished from in vivo tissues and to report in vivo fates of cell transplants. Most labelling strategies have been devoted to report cell migration and homing although sometimes debatable.8–16 A few reports are devoted to report of cell apoptosis following cell transplantation. For example, Nejadnik et al. developed a caspase-3 activatable Gd agent for report of stem cell apoptosis in arthritic joints.17 Ngen et al. used an MRI dual-contrast (SPIONs + Gd-DTPA) method to report cell apoptosis.18 MRI report of in vivo cell differentiation remains to be a challenge.Recently, we have reported that labelling human mesenchymal stem cells (hMSCs) with (Gd-DOTA)i-TPP nanoclusters induces a significant signal reduction and allows long term tracking of hMSCs transplants under T2-weighted instead of T1-weighted MRI. The labelled hMSCs exhibit a persistent dark contrast that allows report of in vivo migration and cell viability of the hMSCs transplants.19–21 We learned from these work that depending on its structural and binding status in cells, Gd-chelate as a T1 contrast agent may exhibit both hypointensive (in a suitable nanostructural status) and hyperintensive (in small molecular status) effect. The findings prompt us to pursue a strategy for MRI report of in vivo cell differentiation that might deliver a contrast switch from dark to bright upon cell differentiation. This can be achieved if a change in the structural and binding status of the Gd agent in cells can be associated with the cell differentiation process.
To achieve this goal, a potential strategy is to develop a Gd-chelate nanoprobe that induce a hypointensive effect to labelled cells and that upon cell differentiation can be degraded into small molecular probes to deliver a hyperintensive effect. To demonstrate the feasibility of the strategy, an appropriate cell differentiation system with an appropriate enzymatic activity has to be selected. In this respect, given a low sensitivity of MRI, both the rate of the cell differentiation and the rate of the degradation have to be in a reasonable range that yet remains to be determined.
As a proof of concept study, we decide to use directional differentiation of HL60 cells into macrophages and granulocytes as a model system to verify the feasibility of the strategy. This cell differentiation system is selected because (1) it exhibits a fairly fast differentiation rate: 80% into macrophages or granulocytes within 3 days;22 (2) a fairly significant difference in the expression level of lysosomal cathepsin B (Cat B) between the parent and daughter cells is associated with the cell differentiation process: 5–10 times in macrophages and 1.5–2 times in granulocytes relative to HL60 cells;22 (3) peptide substrates of Cat B have been well documented in literature as both an digestive carboxydipeptidase in the lysosome and an endopeptidase in the extracellular matrix;23–25 (4) lysosomal Cat B delivers a moderately high proteolytic efficacy as a digestive carboxydipeptidase in the lysosome.26–28 We anticipate that this cell differentiation system provides excellent conditions for a proof of concept study toward the development and assessment of Gd-chelate nanoprobes for in vivo report of cell differentiation.
Various nanostructures can be used and need to be assessed for the development of Gd-chelate nanoprobes. The basic requirements for such a nanoprobe include a relatively long intracellular retention time, chemically stable, degradable by Cat B, and resistant to degradation by other enzymes than Cat B. In this respect, we yet need to establish some in vitro characterization and assessment methods on the cellular level for optimization of the probe structure and properties. To this goal we first try to use an optical instead of MRI nanoprobe to establish the in vitro characterization and assessment methods because it is unrealistic to use MRI itself given its inherent low sensitivity and complexity of data interpretation. In this work we use an optical polymer nanoprobe to explore the feasibility of fluorescence (FL) spectrometry as a characterization and assessment method based on an Internally Quenched Fluorescent (IQF) pair Abz-Dnp (Abz: ortho-aminobenzoic acid; Dnp: 2,4-dinitro-phenylene). This IQF pair is selected because the peptide substrates of Cat B have been documented by this method.24–27 The optical polymer nanoprobe contains poly(lactic-co-glycolic acid) (PLGA) and a peptide IQF probe Abz-FRFK-Dnp (Fig. 1A). This work serves as the first step of our march to the dedicated goal.
Results and discussion
Cat B in the lysosome functions as a digestive carboxy-dipeptidase. It cleaves the second amino bond from the C-terminal with relatively high proteolytic efficacy for many peptide substrates. The kinetic constants and specificity of the substrates have been well documented in literature.24–27 In this work we select the peptide sequence of FRFK because it presents the maximum kcat of ∼59 among the reported substrates with a Km of 3.28 × 10−5 and a kcat/Km of ∼1.8 × 106 when cleaved at the FR–FK site to give peptide fragments FR and FK. It delivers a moderately high proteolytic efficacy.28 The synthesized IQF molecular probe Abz-FRFK-Dnp is purified by HPLC (Fig. S1†) with its molecular weight confirmed by LC-MS (Fig. S2†), and structure confirmed by 1H and 13C NMR (Fig. S3†). The UV absorption spectrum of Abz-FRFK-Dnp is presented in Fig. 1B with two absorption bands peaked at 350 nm and 415 nm for Dnp, consistent with the UV absorption spectra of Dnp (Fig. S4†).We first tried to confirm the proteolytic efficacy of the FRFK sequence by Cat B as well as the enzymatic activity of Cat B by using the IQF molecular probe Abz-FRFK-Dnp. Fig. 1C presents IQF spectra of Abz-FRFK-Dnp as a function of incubation time with Cat B in solution. The FL spectra peak at ∼415 nm. The FL intensity exhibits a sharp increase within 8 hours incubation of Abz-FRFK-Dnp with Cat B and reaches its maximum at ∼8 hours. The maximum FL intensity is an indication of complete depletion of the substrate by Cat B. The results confirmed the enzymatic activity of Cat B and a high proteolytic efficacy of its peptide substrate FRFK. The proteolytic products were further confirmed to be Abz-FR-OH and FK(Dnp)-OH by LC-MS (Fig. S5†).
Second, we tried to confirm the expression level of Cat B in HL60 cells and its differentiated daughter cells by using the IQF molecular probe Abz-FRFK-Dnp and relevant cell lysates. Fig. 1D and E present IQF spectra of the molecular probe Abz-FRFK-Dnp after incubation with HL60, macrophage and granulocyte lysates for 1 and 24 hours, respectively. The FL spectra peak at ∼415 nm. The FL intensity resulting from incubation with the macrophage lysate for 1 hour is ∼5 times of that resulting from incubation with the HL60 lysate. Incubation with the granulocyte lysate for 1 hour induces FL intensity ∼2.5 times of that resulting from incubation with the HL60 cell lysate. The difference in FL intensity manifests the difference in expression level of Cat B in the parent HL60 cells and its differentiated daughter cells, which is consistent with literature report.22 The difference is also confirmed by Western blot assay (Fig. S6†). The difference between the daughter cells and the parent HL60 cells is narrowed after incubation with the relevant cell lysates for 24 hours as a result of a complete depletion of Abz-FRFK-Dnp by the daughter cell lysates and cumulated proteolysis of Abz-FRFK-Dnp by the HL60 cell lysate in the period, as manifested by the FL intensity measured as a function of incubation time in Fig. 1F.
The Abz-FRFK-Dnp@PLGA nanoprobe exhibits a yellow color (retentate of Fig. 2A). UV absorption spectrum of the Abz-FRFK-Dnp@PLGA nanoprobe exhibits two absorption bands peaked at 350 nm and 415 nm for Dnp as well (Fig. 2B), indicating successful imbedding of Abz-FRFK-Dnp in PLGA. The loading efficiency of Abz-FRFK-Dnp was determined to be about 87%. TEM indicates a structure of nanoneedles for the nanoprobe and the size of the nanoprobe is about 100–500 nm (Fig. 2C), as also confirmed by DLS (Fig. S7†). The as-prepared Abz-FRFK-Dnp@PLGA nanoprobe was subjected to ultrafiltration. IQF spectrometric measurements were taken on the retentate and the filtrate before and after incubation with Cat B (Fig. 2D). The FL intensity of the retentate after incubation with Cat B for 24 hours is ∼104 times of that before the incubation, suggesting that the Abz-FRFK-Dnp@PLGA nanoprobe is degradable by Cat B. That of the filtrate after incubation with Cat B for 24 hours is equivalent to that before the incubation, suggesting that free Abz-FRFK-Dnp in the filtrate is insignificant. The Abz-FRFK-Dnp@PLGA nanoprobe was then dispersed in water for 10 days followed by ultrafiltration. IQF spectrometric measurements were repeated on the retentate and the filtrate before and after incubation with Cat B (Fig. 2E). The FL intensity of the filtrate after incubation with Cat B for 24 hours is again equivalent to that before the incubation, suggesting that leakage of Abz-FRFK-Dnp from the Abz-FRFK-Dnp@PLGA nanoprobe is insignificant during the period. Degradation of the Abz-FRFK-Dnp@PLGA nanoprobe by Cat B is also confirmed by TEM result indicating the absence of nanoneedle (Fig. 2F).
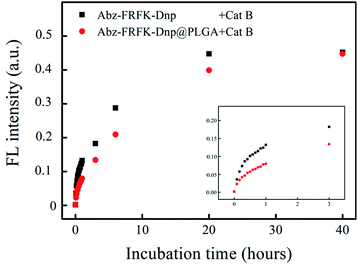
Fig. 3 presents results of a comparative measurement on the degradation rate of the molecular probe Abz-FRFK-Dnp and the Abz-FRFK-Dnp@PLGA nanoprobe by Cat B in solution. The degradation rate is manifested by the change in FL intensity as a function of incubation time. The results indicate that the degradation rate of the Abz-FRFK-Dnp@PLGA nanoprobe by Cat B was slowed down by a factor of 3 relative to the molecular probe Abz-FRFK-Dnp. The difference is significantly narrowed after incubation with Cat B for 40 hours as a result of a complete depletion of Abz-FRFK-Dnp and cumulated proteolysis of the Abz-FRFK-Dnp@PLGA nanoprobes in the period.
In order for a probe to be used for report of cell differentiation, an obvious difference in image contrast has to exhibit between the parent cells and the differentiated daughter cells, which is associated with a significant difference in the degradation rate of the probes. This difference is further required to persist for a certain period to cover the cell differentiation process. Fig. 4 presents the degradation rate of Abz-FRFK-Dnp@PLGA nanoprobe by cell lysates of HL60, macrophage and granulocyte as manifested by the change in FL intensity as a function of incubation time. The FL intensity resulting from the macrophage lysate reaches its maximum in 6 hours. That from the granulocyte lysate reaches ∼70% and 100% of the maximum in 6 hours and 2 days, and that from the HL60 cell lysate reaches ∼35% and ∼52% of the maximum in 6 hours and 2 days. As a result, a difference of 2–3 times in FL intensity between the daughter cell lysates and the parent HL60 cell lysate can persist over a period of 2 days, but the difference between the macrophage and granulocyte lysates is narrowed to the same level in this period.
PLGA used for preparation of the nanoprobe may also be degradable by enzymes. As a result, cellular enzymatic degradation of the Abz-FRFK-Dnp@PLGA nanoprobe may include contribution from several processes: cleavage of FRFK or degradation of PLGA by Cat B or other enzymes in cells. In order to assess the relative contribution from these relevant enzymatic processes, the degradation rate of Abz-FRFK-Dnp@ PLGA nanoprobe by cell lysates of macrophage or HL60 with and without addition of Cat B inhibitor was compared.
Fig. 5A presents the FL intensity as a function of incubation time for macrophage lysates in the presence and absence of the Cat B inhibitor. The FL intensity resulting from a macrophage lysate in the presence of the Cat B inhibitor indicates the contribution from other enzymes than Cat B, whereas that in the absence of the Cat B inhibitor indicates contribution from Cat B and other enzymes. The results in Fig. 5A indicate that the degradation rate by the macrophage lysate without the Cat B inhibitor is ∼6.4 times of that with the Cat B inhibitor, suggesting that the degradation process is dominated by Cat B.
To assess the possible contribution from degradation of PLGA by other enzymes than Cat B, an Abz-FRFK-Dnp@PLGA dispersion was incubated for 24 hours with a HL60 lysate saturated by the Cat B inhibitor followed by ultrafiltration. Both the retentate and the filtrate were collected and incubated with Cat B for 12 hours. IQF spectrometric measurements were taken before and after the incubation and the results are presented in Fig. 5B. The increase in FL intensity is significant for the retentate and insignificant for the filtrate after the incubation with Cat B for 12 hours, suggesting that release of Abz-FRFK-Dnp from degradation of PLGA by other enzymes than Cat B is insignificant in this period. These results also confirm that the degradation of Abz-FRFK-Dnp@PLGA nanoprobe was dominated by Cat B.
HL60 cells were then labelled by the Abz-FRFK-Dnp@PLGA nanoprobe, and were further induced to differentiate into macrophages or granulocytes. The labelled cells were subjected to microscopic fluorescent imaging at predetermined time intervals. Fig. 6A presents the microscopic fluorescent images of HL60 labelled with Abz-FRFK-Dnp@PLGA and its differentiated daughter cells as a function of incubation time. The microscopic FL images of the control HL60 cells labelled with Abz-FRFK-Dnp@PLGA are presented in the first row. Addition of dimethylsulfoxide (DMSO) into the cell culture medium induces HL60 cells to differentiate into granulocytes,22 the microscopic FL images of which are presented in the second row. Addition of 12-O-tetradecanoylphorbol 13-acetate (TPA) induces HL60 cells to differentiate into macrophages,22 the microscopic FL images of which are presented in the third row. The cellular FL intensity of the relevant cells is presented in Fig. 6B as a function of incubation time along with the a statistic analysis in Fig. 6C. The results are interpreted on the basis of that: (1) the FL intensity is correlated with the expression level of Cat B in the cells; (2) the cells with strong FL is correlated with macrophages because it expresses Cat B at a level much higher than HL60 cells and granulocytes; (3) HL60 cells and granulocytes also exhibit a weak FL as a result of their low expression level of Cat B.
Then, we note that HL60 cells incubated in the presence of TPA exhibits much stronger FL and delivers more number of cells with strong FL (third row) than the control HL60 cells (first row) and the HL60 cells incubated in the presence of DMSO (second row), which can serve as a clear indication of its differentiation into macrophages. Such a significant difference in FL intensity and the number of fluorescent cells between the macrophages and the parent or other daughter cells can persist over 3 days and allows a time window for observation of the differentiation process of HL60 cells when labelled with the Abz-FRFK-Dnp@PLGA nanoprobe. We also note that differentiation of HL60 cells into granulocytes is not distinguishable by this strategy, which is correlated with the expression level of Cat B as indicated in Fig. S6A and B.† In this respect, the results also suggest a minimum difference in the expression level of a relevant enzyme required for report of a cell differentiation process.
The findings presented here provide answer to several key questions that are important to the development of enzyme-responsive MRI nanoprobes for report of cell differentiation. First, a facile preparation method of IQF and MRI nanoprobes is provided. Second, the prepared nanoprobe is resistant to leakage prior to proteolysis. Third, although the rate of proteolysis of the nanoprobe is slowed down relative to the molecular probe, it is still fairly fast so that combined with a fast rate of cell differentiation, it may balance the low sensitivity of MRI. This work also established a series of experimental procedure for quantitative characterization and optimization of the probe structure and its performance when interacting with cellular enzymes. These findings and methods allow us to proceed to verify the proof of concept with an MRI nanoprobe of similar structure for report of cell differentiation.
Experimental
Synthesis of IQF molecular probe Abz-FRFK-Dnp
The IQF molecular probe Abz-FRFK-Dnp contains a peptide sequence of FRFK with its N-terminal connected to aminobenzoic (Abz) and the animo group of its lysine connected to 2,4-dinitrophenylene (Dnp). The peptide sequence was selected because it delivers a moderately high proteolytic efficacy with Cat B. Abz-FRFK-Dnp were prepared by a standard N-α-fluorenylmethoxy-carbonyl (Fmoc) peptide synthesis strategy according to the peptide sequence from C to N terminal. The purity of the monomers is confirmed by High Pressure Liquid Chromatography (HPLC, Waters, MA) with a C18 column (XBridge C18 5 μm 4.6 × 250 mm) under 220 nm UV absorbance. Molecular weight (MW) was determined by Electron Stimulated Ionization-Mass Spectrometry (ESI-MS, Agilent 6120 Single-Quadrupole LC/MS System). 1H NMR (d6-DMSO,TMS) and 13C NMR spectra was obtained on a Varian 400 MHZ spectrometer. UV adsorption spectra of Abz-FRFK-Dnp, ABz and Dnp was collected with UV-VIS Spectrophotometer (UV-2550, Shimadzu, Japan).Synthesis of Abz-FRFK-Dnp@PLGA nanoprobe
4 mg of Abz-FRFK-Dnp was dissolved in 40 μL of DMSO (100 mg mL−1). 10 μL of the solution was added to 200 μL 1% PVA water solution. The mixture turned muddy. 10 μL of PLGA solution dissolved in CH2Cl2 (20 mg mL−1) was added into the mixture followed by a supersonic stirring for 20 min. The mixture turned clear which is associated with the formation of Abz-FRFK-Dnp@PLGA nanoprobe. Then the mixture was subjected to ultrafiltration with a filter of 3000 Dalton at 4500 g for 20 min. The retentate with the Abz-FRFK-Dnp@PLGA nanoprobe was collected and washed three times with pure water. The product was dispersed in water for further use.The size and shape of the nanoprobe was characterized by Transmission Electron Microscopy (TEM, HT7700, Hitachi, Japan). The hydrodynamic diameter was measured by dynamic light scattering (DLS, ZEN3600 Nano ZS, Malvern). The entrapment and loading efficiency of Abz-FRFK-Dnp was evaluated by UV spectrometry. The stability of the Abz-FRFK-Dnp@PLGA nanoprobe and its degradability by Cat B were verified by FL spectrophotometry (F-4600, Hitachi, Japan) with an excitation wavelength of 320 nm and emission wavelength of 415 nm. The nanoprobe was dispersed in water and stored for 10 days. Then the sample was subjected to ultrafiltration with a filter of 3000 Dalton at 4500 g for 20 min. Both the retentate and the filtrate were collected followed by incubation with Cat B for 24 hours. TEM and fluorescent measurements were taken on both the retentate and the filtrate before and after incubation with Cat B.
Cell culture
HL60 cells were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). The cells were cultured at 37 °C in a 5% CO2 incubator (Thermo 3111, Waltham, MA) in IMDM medium with 20% FBS and 1% penicillin–streptomycin. All cell culture related reagents were purchased from Gibco.Proteolysis of the probes by cathepsin B in solution
A solution of Abz-FRFK-Dnp or dispersion of Abz-FRFK-Dnp@ PLGA in water with an equivalent concentration of Abz-FRFK-Dnp (10 μg mL−1) was incubated with Cat B (0.2 μg mL−1). Fluorescent measurements were taken on the samples at predetermined time intervals. LC-MS measurements were taken on Abz-FRFK-Dnp after incubation with Cat B to confirm the proteolytic products.Proteolysis of Abz-FRFK-Dnp@PLGA nanoprobe by cell lysates
Lysates of HL60 cells, macrophages or granulocytes was added into a dispersion of Abz-FRFK-Dnp@PLGA in water at an Abz-FRFK-Dnp concentration of 10 μg mL−1. Fluorescent measure-ments were taken on the samples at predetermined time intervals.To evaluate the contribution of enzymes other than Cat B to the proteolysis of Abz-FRFK-Dnp@PLGA nanoprobe, a Cat B inhibitor (methyl(2S)-1-[(2S)-3-methyl-2-[[(2S, 3S)-3-(propyl-carbamoyl)oxirane-2-carbonyl]amino]pentanoyl]pyrrolidine-2-carboxylate(CA-074 methyl easter, MedChem Express, USA)) was added into a cell lysate of macrophage differentiated from HL60, which was then added into a dispersion of Abz-FRFK-Dnp @PLGA in water with an Abz-FRFK-Dnp concentration of 10 μg mL−1. Fluorescent measurements were taken on the samples at predetermined time intervals. A parallel experiment was conducted on a cell lysate without addition of the Cat B inhibitor for comparison. In another experiment, a HL60 cell lysate saturated with the Cat B inhibitor was added to an Abz-FRFK-Dnp@PLGA dispersion (10 μg mL−1) followed by an incubation of 24 hours. Then the mixture was subjected to ultrafiltration with a filter of 3000 Dalton at 4500 g for 20 min. Both the retentate and the filtrate were collected and incubated with Cat B for 24 hours. FL measurements were taken on both the retentate and the filtrate before and after incubation with Cat B.
Intracellular proteolysis of Abz-FRFK-Dnp@PLGA nanoprobe
HL60 cells were seeded into 100 mm × 20 mm style cell culture dishes at a density of about 1 × 106 cells per dish and maintained for 2 days. Abz-FRFK-Dnp@PLGA nanoprobe was added into the cell dish followed by an incubation of 24 hours. After the completion of cell labelling, the cells were rinsed three times with 2–3 mL PBS to remove the residual materials. The cells were then divided into three groups and transferred to 6-well plates. The labelled cells were cultured at 37 °C in a 5% CO2 incubator in IMDM medium with 20% FBS and 1% penicillin–streptomycin in the presence of TPA (48 nM) or DMSO (1.25%) for a given period to allow cell differentiation and intracellular proteolysis to proceed. A parallel experiment was also conducted as a control without addition of TPA or DMSO. After a given period, the cells were subjected to microscopic fluorescent imaging using broadband FL microscopy (Suzhou NIR-Optical Technology Co., Ltd., China). The mean FL intensity of cells with different treatment was measured using ImageJ software, and about 200 cells (12 images in total) were selected to obtain a mean value. Cat B expression levels of HL60, granulocyte and macrophage were quantified by Western blot assay.Statistical analysis
Statistical analysis was performed using the origin Software 8.5 and data were expressed as mean values ± standard deviation (SD). The differences between the median values were evaluated using one-sample Student's t-test. Levels of significance were determined as follows: **p < 0.001, #p > 0.01.Conclusions
A Cat B-responsive IQF nanoprobe Abz-FRFK-Dnp@PLGA has been synthesized for report of directional differentiation of HL60 cells into macrophages. Combination of IQF spectro-metry with cell lysate experiments demonstrates that proteolysis of Abz-FRFK-Dnp@PLGA by Cat B is significantly slowed down relative to proteolysis of Abz-FRFK-Dnp. It creates a significant difference in IQF FL between the parent cells and differentiated macrophage cells that can persist to cover the cell differentiation process and serve as an indicator of the differentiation process. The nanoprobe is resistant to other enzymes than Cat B in HL60 cells. Cell experiments further confirms that directional differentiation of HL60 cells labelled with Abz-FRFK-Dnp@PLGA into macrophages can be monitored by microscopic FL images of the cells. The experimental and characterization methods established in this work pave a way toward the development and assessment of an MRI nanoprobe for report of in vivo fates of cell transplants.Author contributions
Y. H. Z., Z. W. D. and D. H. H. designed and synthesized the IQF molecular and nanoprobe, performed experiments and analysed data; Y. H. Z., C. X. Z. and J. J. M. performed fluorescent measurements; D. H. H. performed FL microscopy experiments, Y. H. Z. and D. H. H. analysed FL microscopy data; Z. W. D., Y. H. Z. and B. T. wrote the original manuscript; all authors contributed to the final manuscript; Z. W. D. supervised the project.Conflicts of interest
There is no conflict to declare.Acknowledgements
This work was funded by general projects from the Natural Science Foundation of China (21673281, 31371010), and the National Key R&D Program from MOST of China (2017YFA0104301).Notes and references
- R. J. C. Bose and R. F. Mattrey, Drug Discovery Today, 2019, 24, 492–504 CrossRef PubMed.
- P. K. Nguyen, J. Riegler and J. C. Wu, Cell Stem Cell, 2014, 14, 431–444 CrossRef CAS PubMed.
- A. Gera, G. K. Steinberg and R. Guzman, Regener. Med., 2010, 5, 73–86 CrossRef PubMed.
- Y. Q. Wang, C. J. Xu and H. Ow, Theranostics, 2013, 3, 544–560 CrossRef PubMed.
- W. Cui, S. Tavri, M. J. Benchimol, M. Itani, E. S. Olson, H. Zhang, M. Decyk, R. G. Ramirez, C. V. Barback, Y. Kono and R. F. Mattre, Biomaterials, 2013, 34, 4926–4935 CrossRef CAS PubMed.
- L. Faivre, M. Chaussard, L. Vercellino, V. Vanneaux, B. Hosten, K. Teixera, V. Parietti, P. Merlet, L. Sarda-Mantel, N. Rizzo-Padoin and J. Largheroabh, Curr. Res. Transl. Med., 2016, 64, 141–148 CrossRef CAS PubMed.
- G. C. Chen, S. Y. Lin, D. H. Huang, Y. J. Zhang, C. Y. Li, M. Wang and Q. B. Wang, Small, 2018, 14, 1702679 CrossRef PubMed.
- D. L. Kraitchman and J. W. M. Bulte, Basic Res. Cardiol., 2008, 103, 105–113 CrossRef CAS PubMed.
- G. Genove, U. DeMarco, H. Y. Xu, W. F. Goins and E. T. Ahrens, Nat. Med., 2005, 11, 450–454 CrossRef CAS PubMed.
- B. Cohen, K. Ziv, V. Plaks, T. Israely, V. Kalchenko, A. Harmelin, L. E. Benjamin and M. Neeman, Nat. Med., 2007, 13, 498–503 CrossRef CAS PubMed.
- W. J. Rogers, C. H. Meyer and C. M. Kramer, Nat. Clin. Pract. Cardiovasc. Med., 2006, 3, 554–562 CrossRef CAS PubMed.
- L. S. Politi, Neuroradiolog, 2007, 49, 523–534 CrossRef PubMed.
- O. Lunov, T. Syrovets, C. Rocker, K. Tron, G. U. Nienhaus, V. Rasche, V. Mailander, K. Landfester and T. Simmet, Biomaterial, 2010, 31, 9015–9022 CrossRef CAS PubMed.
- M. Mahmoudi, H. Hosseinkhani, M. Hosseinkhani, S. Boutry, A. Simchi, W. Shane Journeay, K. Subramani and S. Laurent, Chem. Rev., 2011, 111, 253–280 CrossRef CAS PubMed.
- H. Li, G. Parigi, C. Luchinat and T. J. Meade, J. Am. Chem. Soc., 2019, 141, 6224–6233 CrossRef CAS PubMed.
- A. Khurana, H. Nejadnik, R. Gawande, G. Lin, S. Lee, S. Messing, R. Castaneda, N. Drugin, L. Pisani, T. F. Lue and H. E. Daldrup-Link, Radiology, 2012, 264, 803–811 CrossRef PubMed.
- H. Nejadnik, D. Ye, O. D. Lenkov, J. S. Doing, J. E. Martin, R. Castillo, N. Derugin, B. Sennino, J. Rao and H. Daldrup-Link, ACS Nano, 2015, 9, 1150–1160 CrossRef CAS PubMed.
- E. J. Ngen, L. Wang, Y. Kato, B. Krishnamachary, W. L. Zhu, N. Gandhi, B. Smith, M. Armour, J. Wong, K. Gabrielson and D. Artemov, Sci. Rep., 2015, 5, 13628 CrossRef PubMed.
- Y. H. Zhang, H. Y. Zhang, B. B. Li, H. L. Zhang, B. Tan and Z. W. Deng, Nano Res., 2018, 11(3), 1625–1641 CrossRef CAS.
- Y. H. Zhang, H. Y. Zhang, L. J. Ding, H. L. Zhang, B. Tan and Z. W. Deng, Mol. Med. Rep., 2017, 16, 4068–4074 CrossRef CAS PubMed.
- P. L. Zhang, Y. H. Zhang, B. B. Li, H. L. Zhang, H. X. Lin, Z. W. Deng and B. Tan, J. Pept. Sci., 2018, 24, e3077 CrossRef PubMed.
- D. Burnett, J. Crocker and A. T. M. Vaughan, J. Cell. Physiol., 1983, 115, 249–254 CrossRef CAS PubMed.
- J. Schmitza, E. Gilberga, R. Loserd, J. Bajorathc, U. Bartzb and M. Gutschow, Bioorg. Med. Chem., 2019, 27, 1–15 CrossRef PubMed.
- M. Fonović and B. Turk, Biochim. Biophys. Acta, 2014, 1804, 2560–2570 CrossRef PubMed.
- F. C. Portaro, A. B. Santos, M. H. Cezari, M. A. Juliano, L. Juliano and E. Carmona, Biochem. J., 2000, 347, 123–129 CrossRef CAS.
- M. H. Cezari, L. Puzer, M. A. Juliano, A. K. Carmona and L. Juliano, Biochem. J., 2002, 368, 365–369 CrossRef CAS PubMed.
- S. S. Cotrin, L. Puzer, W. A. de Souza Judice, L. Juliano, A. K. Carmona and M. A. Juliano, Anal. Biochem., 2004, 335, 244–252 CrossRef CAS PubMed.
- A. Bar-Even, E. Noor, Y. Savir, W. Liebermeister, D. Davidi, D. S. Tawfik and R. Milo, Biochem, 2011, 50, 4402–4410 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI:10.1039/d1ra01549d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |