Open Access Article
Open Access ArticleA PCR-free screen-printed magnetic electrode for the detection of circular RNA from hepatocellular cancer based on a back-splice junction†
Bin Zhang‡
 ab,
Yitao Liang‡
ab,
Yitao Liang‡ a,
Liang Bo
a,
Liang Bo a,
Mingyu Chenb,
Bobo Huanga,
Qingpeng Cao
a,
Mingyu Chenb,
Bobo Huanga,
Qingpeng Cao a,
Jinwei Weia,
Tianyu Liab,
Xiujun Cai*bc and
Xuesong Ye
a,
Jinwei Weia,
Tianyu Liab,
Xiujun Cai*bc and
Xuesong Ye *ac
*ac
aBiosensor National Special Laboratory, Key Laboratory of Biomedical Engineering of Ministry of Education, College of Biomedical Engineering and Instrument Science, Innovation Centre for Minimally Invasive Technique and Device, Zhejiang University, Hangzhou 310027, PR China. E-mail: yexuesong@zju.edu.cn; Tel: +86 571 87952756
bDepartment of General Surgery, Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University, 3 East Qingchun Road, Hangzhou 310016, PR China. E-mail: srrsh_cxj@zju.edu.cn; Tel: +86 571 86006617
cZhejiang Research and Development Engineering Laboratory of Minimally Invasive Technology and Equipment, School of Medicine, Zhejiang University, 3 East Qingchun Road, Hangzhou 310016, PR China
First published on 17th May 2021
Abstract
Circular RNA (circRNA) has the potential to be applied to disease diagnosis and therapy. However, the currently available circRNA detection techniques are limited. This work proposes a sensitive and selective approach for circRNA detection based on gold nanoparticle-modified screen-printed magnetic electrodes (AuNPs-SPME). Magnetic beads (MBs) with capture probes based on specific back-splice junction (BSJ) sites were employed to identify and selectively isolate the target circRNA, which could be directly adsorbed onto the AuNPs-SPME. Then, the circRNA attached to the surface was detected by changes in the methylene blue redox signal. The simple and time-saving AuNPs-SPME is highly sensitive (LOD = 1.0 pM) to circCDYL, one of the biomarkers of hepatocellular cancer (HCC). The analytical performance of the method presented has also been verified in human serum samples, holding great promise for clinical diagnosis.
Introduction
Hepatocellular carcinoma (HCC) is the most common cancer and the fourth-leading cause of cancer-associated mortality worldwide.1 Patients diagnosed with advanced or unresectable HCC have very poor outcomes, due to limited therapeutic options, while early detection of HCC using imaging and tumor markers could dramatically improve patient survival rates.2 CircCDYL (chromodomain Y like) plays an important role in several pathological processes, such as hepatocellular carcinoma (HCC),3 mantle cell lymphoma (MCL)4 and bladder cancer.5 Besides, it is specifically up regulated and is an independent marker for detecting early stages of HCC which makes it a potential biomarker.6 Though circular RNA (circRNA) was discovered nearly 40 years ago,7 its function in physical and pathological conditions were only characterized recently.8,9 Considered as a result of proactive back-splicing events, in which the 3′ tail of one exon is joined to the 5′ head of an upstream exon,9 due to this unique structure, circRNAs are highly stable and highly enriched in whole blood, plasma, and platelets,10 when compared with linear mRNAs, miRNAs, and other RNA molecules.Northern blotting (NB) and real-time quantitative PCR (RT-PCR) amplification are traditional circRNA detection methods. RNase R were used to enrich and identify circRNA in total RNA to decrease linear RNA in traditional detection methods. Afterward, northern blotting (NB) is applied to separate circRNAs in polyacrylamide gel electrophoresis (PAGE). However, low abundance of circRNA cannot be monitored when the samples are insufficient. RT-PCR has shown high sensitivity and specificity. But it is not negligible that detection errors caused by the special cyclic structure of circRNA and reverse transcription (RT) steps are not compatible with detection in the complete blood. What's more, RNA sequencing (RNA-seq) is the most commonly used method for genome-wide profiling of circRNAs,11 but its application is limited by detection efficiency, high-cost, complicated processes and device-dependence.
Recently, electrochemical biosensors have shown potential for addressing challenges related to the detection of nucleic acids,12,13 because of the high sensitivity, simplicity, cost-effectiveness and compatibility with miniaturization. Noncoding RNA detected by electrochemical gene sensors have gained increasing attention, such as microRNA14–16 and lncRNA.17,18 Unlike linear RNAs, the back-splice junction (BSJ) sites of circRNA are considered to be a conserved motif (Fig. S1†). Most detection methods employ alignment-based strategies to recognize specific BSJ reads for circRNAs. Jiao et al.19 first proposed an electrochemical method for the detection of circRNAs based on the combination of BSJ sites recognition and duplex-specific nuclease (DSN)-assisted target recycling signal amplification without reverse transcription. However, nuclease was also needed in the analysis, which may hinder the application of point-of-care detection.
Screen-printed electrodes (SPE) have been used extensively for biosensors because of their low cost, reproducibility, portability and ability to be mass produced.20,21 On the other hand, functionalized magnetic nanoparticles (magnetic beads, MBs) have been widely used for the preconcentration of analytes to achieve a small sample volume for ultra-sensitive detection,22 which offered very attractive characteristics in terms of sensitivity and fast assay kinetics, due to the possibility of achieving high loads of immobilized biomolecules since the beads are under continuous stirring in suspension and the minimization of the matrix effect in the development of electrochemical biosensors.23 Hence, an electrochemical biosensor, combined with SPE and MBs, meets the requirements of low cost and easy operation for the circRNA detection.
Herein, we developed a PCR-free detection assay for the analysis of circCDYL based on SPE and functional MBs. Total RNA sequences were incubated with streptavidin-labeled magnetic beads modified with a biotinylated synthetic capture probe from BSJ sites. Then MBs were adsorbed directly onto the gold nanoparticles (AuNPs) modified screen-printed magnetic electrode (AuNPs-SPME), after which the level of circCDYL was then analyzed by differential pulse voltammetry (DPV) in the presence of the methylene blue redox system. We have proposed an electrochemical biosensor with joint application of SPME and MBs targeting the BSJ sites of circRNA so as to analyze the circCDYL expression levels with highly sensitive and specificity for diagnosis HCC patients. The advantages of this sensing platform are as follows. Firstly, we applied the magnetic rubber sheet as substrate for SPE, which provided uniform magnetism to achieve homogeneous electrode performance for different SPME, which will improve the consistency of SPME if printed industrially. Secondly, total RNA could incubate directly with MBs in our analysis, which is functionalized captured probes targeted to BSJ of circRNA and that makes it suitable to select other probes with different sequences for distinguishable circRNA in a convenient way. Thirdly, no RNase R treatments are needed in our system to enrich circRNA. The detection process is more convenient and manageable, without additional enzyme treatment.
Experimental
Chemicals and materials
DNase/RNase-free distilled water (Dalian Meilun Biotechnology Co., Ltd, China) was used throughout the experiments. Methylene blue and gold chloride (HAuCl4·4H2O) were supplied by Sigma (St. Louis, MO, USA). Screen printing inks were purchased from Jujo Chemical Co., Ltd (Tokyo, Japan) and a 0.3 mm rubber magnet sheet were purchased from Guangzhou Magnetic Material Co., Ltd (Guangzhou, China). Capture probes, RNA sequences and 20× SSC buffers were purchased from Sangon Biotechnology Inc. (Shanghai, China). The base sequences are in Table S1.† The normal human serum was obtained from Sir Run Run Shaw Hospital. Streptavidin-labeled magnetic beads (XFNANO, Materials Tech Co. Ltd. China) were 1 μm in diameter. All other chemicals not mentioned here were of analytical reagent grade. AuNPs modification and electrochemical measurements were performed using an electrochemical workstation (IVIUM, CompactStat.h). For evaluation of the morphology and structures of MBs, a field emission scanning electron microscope (Gemini 300, Zeiss) was used which was equipped with X-ray energy-dispersive spectrometry (EDS) for elemental composition analysis.Preparation of functional magnetic beads and isolation of circRNA
10 μL of streptavidin-labeled magnetic beads (10 mg mL−1) were washed twice with 5× SSC buffer (pH 7) and then 10 μL of 10 μM biotinylated captured probes were added. The incubation was conducted at 4 °C to facilitate the capture of probes with magnetic beads. Afterwards, the magnetic beads were washed three times and resuspended in 100 μL of 5× SSC buffer (pH 7). For circRNA capture, 5 μL of previously prepared different concentrations of synthetic circCDYL (spanning over 1.0 pM to 100 nM) were mixed with 7.5 μL of functional magnetic beads. The mixture was incubated at 4 °C to allow the hybridization of capture probe and target circRNA. After being washed twice and separated using a magnet, 7.5 μL of RNase-free water was used to resuspend the magneto-bioconjugates. The released solution was then directly put onto the working surface of an AuNPs modified SPME.Fabrication of AuNPs-SPME
The AuNPs-SPME consisted of three printed electrodes: an AuNPs modified working electrode (WE) and two electrodes acting as reference electrodes (RE) and counter electrodes (CE), which were previously reported in our group's paper.21 The diameter of working electrode was 2.5 mm. In short, silver, carbon and isolated oil were printed onto the rubber magnet sheet in the sequence of several templates. After each printing step, the film was heated for 15 min at 90 °C to eliminate the residual solvent. The pretreated SPME was then immersed into a 3 mM HAuCl4 solution containing 0.1 M KNO3. A constant potential of −0.48 V was applied for 200 s to deposit AuNPs onto the working electrode. The obtained AuNPs-SPME was washed with double distilled deionized water and dried under nitrogen.Electrochemical measurements
All the methods used for electrochemical measurements were conducted at room temperature. The cyclic voltammetry (CV) measurement was conducted in 10 mM PBS (containing 2 mM [Fe(CN)6]3−/4− and 0.5 M KCl) with a scan rate of 10 mV s−1 from 0 to 0.6 V. The differential pulse voltammetric (DPV) experiments were recorded at 0 to −0.5 V with a pulse amplitude of 50 mV and a pulse width of 50 ms in 0.1 M methylene blue (containing 1 M KCl, 1 M Tris·HCl). For synthetic RNA samples, a 5.0 μL solution was incubated with 7.5 μL functional magnetic beads and washed twice. Then the mixture was applied to a AuNPs modified SPME surface. The Ipeak is regarded as the background signal without MBs and the peak current change of methylene blue signal (ΔIpeak) has a corresponding relationship with the circCDYL concentration after DNA/RNA hybridization.Results and discussion
The schematic illustration of the circRNA biosensor is shown in Scheme 1. It has been previously shown that the methylene blue redox system alone can be used for the quantification of surface-bound nucleotide on unmodified gold electrodes.24,25 Methylene blue could intercalate into the duplex DNA/RNA and interact with DNA/RNA in three ways: electrostatic interaction, intercalation and specific interaction with exposed guanine residues. Although the high affinity between methylene blue and guanine residues was greatly obstructed because of guanine residues enwrapped in hybrid duplexes, DNA–RNA hybrid duplexes still improved the electrostatic interaction between methylene blue and the deoxyribose–phosphate backbone. Moreover, the residual sites of circRNA could interact with methylene blue. Therefore, less methylene blue was adsorbed on single-stranded DNA (ssDNA) than on hybrid duplexes, resulting in a smaller peak current.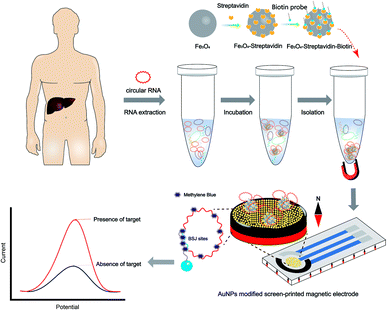 | ||
| Scheme 1 Gold nanoparticles (AuNPs) modified screen-printed magnetic electrode (AuNPs-SPME) for the detection of circular RNA from hepatocellular cancer. | ||
Characterization of AuNPs-SPME
Fig. 1A shows the real picture of the SPME. WE, RE, CE and the contact part of the SPME were illustrated in Fig. 1B. The beads can be directly adsorbed on the working electrode surface of the SPME, which simplified and standardized the operation for further analysis. To verify the magnetism of the SPME, magnetic powders were dropped onto the WE surface. As shown in Fig. 1C, even in the vertical state, powders were still fixed onto the surface of the SPME, and the magnetic field of SPME was sufficient to adsorb the MBs. In contrast with other magnetic SPCEs,26,27 which capture MBs onto SPEs by using an external magnet, we first applied magnetic substrates in our SPME to obtain a simple and stable magnetic field suitable for industrial production. In addition, different magnetic fields could be achieved by controlling the thicknesses of magnetic substrates.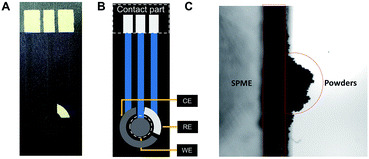 | ||
| Fig. 1 (A) A photo of a screen-printed magnetic electrode (SPME); (B) illustration of electrode; (C) powders were absorbed on the surface of SPMEs while vertical. | ||
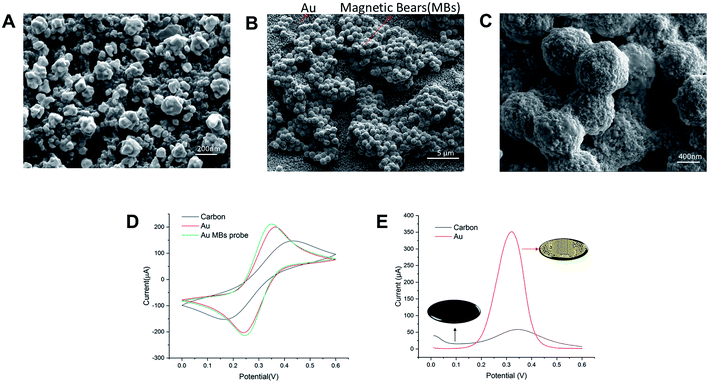
AuNPs were electrodeposited onto the working electrode in a HAuCl4 solution using the amperometry technique, which can increase the effective area and facilitate electron transfer. We have calculated the effective areas of AuNPs-SPME by the measurement of the peak current obtained as a function of scan rate. Surface areas of most AuNPs-SPMEs were in the range of 1.5 to 1.6 × 10−2 cm2. The surface morphology of modified AuNPs and MBs loaded onto SPME were examined with a scanning electron microscope (SEM). In Fig. 2A, the SEM image showed that the distribution of AuNPs on WE of SPME was uniform and dense, and the diameters of AuNPs were about 100 to 200 nm. During the gold-plating process, gold nanoparticles are randomly anchored on the surface of the electrode. Different conditions may cause different effective areas of electrodes, which should be taken into account. MBs were dropped closely upon the AuNPs modified electrodes, owing to the magnetic substrate. In a magnified view of MBs (Fig. 2B), the diameter of streptavidin-labeled magnetic beads is about 1 μm. This shows that the quality of the modified AuNPs and MBs loaded onto the SPME are good. Besides, the EDS of streptavidin-labeled magnetic beads and functional magnetic beads are also shown in Fig. S2.† Biotinylated probes can be immobilized on magnetic beads labeled with streptavidin through the interaction between biotin and streptavidin with high affinity. It can be seen that the P element is increased distributed on the magnetic beads, verifying the presence of biotin-capture probes.
Modification of the electrode surface will cause a change of the electrochemical signal. CV technique was employed and five consecutive cycles (0 to 0.6 V; 50 mV s−1) were recorded in carbon-SPME, AuNPs-SPME and MBs loaded AuNPs-SPME. As shown in Fig. 2D, an anodic and a cathodic peak were observed at 0.18 and 0.42 V (vs. Ag|AgCl), respectively. The carbon-SPME exhibited a pair of redox peaks and the peak difference (ΔEp) was 260 mV. After AuNPs were electrodeposited onto the SPME, the peak current (Ip) increased and ΔEp was lowered to 110 mV. Compared with bare carbon electrode, the peak current of the AuNPs modified electrode increased noticeably and the peak-to-peak potential difference decreased (Fig. 2D). These results showed that the AuNPs enlarged the surface of the electrode, leading to more [Fe(CN)6]3− ions on it. After MBs adsorbed on the SPME, the ΔEp decreased and the Ip increased. This may be due to the oligonucleotides presenting coulomb repulsion to the bulk [Fe(CN)6]3− ions. Furthermore, the DPV signal increased dramatically in Fig. 2E. Such results illustrated that AuNPs-SPME could facilitate the electron-transfer of signals and improve the electrochemical performances for low concentration detection.
Optimization of experimental conditions
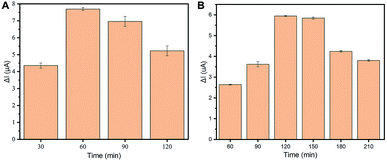
Firstly, the relationship between the magnetic bead amounts and the DPV signal needed to be investigated. To explore the optimal electrochemical performance of the electrodes, various amounts of suspension MBs ranging from 0 to 20 μg were explored in AuNPs-SPME surfaces (Fig. S3†). An increased DPV signal was observed with MBs on AuNPs-SPME, compared with blank AuNPs-SPME. The highest DPV signal was observed with 10 μg MBs and the lowest was 5 and 7.5 μg MBs. This may be explained by uneven distribution of MBs possibly as barriers for tuning current. Under the microscope view, 7.5 μg functional MBs display uniform distribution on the SPME. In order to maximize DPV signal detection, 7.5 μg MBs were selected as the optimal amounts for the assay with the lowest DPV and uniform distribution for the background signals.Incubation time was correlated with the number of probes on the beads, which was related to the signal response. In order to optimize the incubation time of streptavidin labeled magnetic beads and biotinylated probes, various periods of time ranging from 30–120 minutes were measured for the largest DPV signal at 4 °C. As shown in Fig. 3A, the signal increased at first, then decreased with incubation time. A strong signal was achieved within 60 minutes and decrease from 90 minutes to 120 minutes. This is mainly because the adsorption sites become occupied towards 60 minutes and adsorption efficiency decreases with an increase in adsorption time. Therefore, 60 minutes was the most suitable time for this procedure.
To achieve the best performance for circRNA detection, the incubation time required for RNA and functional MBs was investigated next. The incubation time was related to the amount of target RNA captured by the functional MBs. The RNA solution and functional MBs were incubated over various periods ranging from 60–210 min at 4 °C. As shown in Fig. 3B, an increased signal was observed after 120 minutes of incubation time. 120 minutes adsorption time led to the highest level, indicating a good discrimination DPV signal. However, extended incubation times resulted in a reduced signal. The reduction of the signal after 120 minutes could be explained by the decomposition of oligonucleotides, which means decreased RNA/DNA in the analysis. Accordingly, 120 min was selected as the optimal incubation time between RNA and functional MBs for our following experiments.
Electrochemical performance of circRNA biosensor
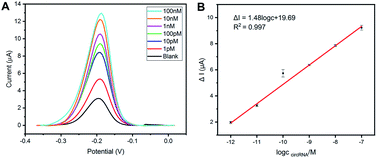
After experimental optimization, linear range and limit of detection (LOD) of this method have been investigated by analyzing peak currents in different concentrations of the circCDYL. As displayed in Fig. 4A, the peak current of methylene blue increases with the increased target circCDYL concentration (from 1 pM to 100 nM). Fig. 4B shows the linearity between peak current and the logarithm of target circCDYL concentrations and the dynamic range is from 1 pM to 100 nM, with a low limit of detection (LOD) of 1 pM. The linear equation with a correlation coefficient (R2) of 0.997 is obtained: ΔI = 1.48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (circCDYL) + 19.69, in which ΔI represents the increased peak current of the different concentrations of circCDYL.
C (circCDYL) + 19.69, in which ΔI represents the increased peak current of the different concentrations of circCDYL.
The AuNPs-SPME biosensor for circRNA detection showed high sensitivity and wide linear range. Also, the performance of this biosensor was compared with duplex-specific nuclease (DSN) assisted methods for circRNA,19 which was acceptable. It offers several practical advantages, such as a considerable time without enzyme incubation and a much simpler working protocol. Our analysis time required as short as less than three hours, which is significantly shorter than the several hours required for nucleic acid extraction, cDNA synthesis and PCR reaction (Table S2†). A shorter detection time offers the possibility of POC diagnosis. Notably, this biosensor does not require nucleic acid amplification and reverse transcription. Samples do not need to be transferred to laboratories. The electrical output provided by this sensor is compatible with automation and a low-cost device for near-POC test.
Specificity, reproducibility, and real application of the electrochemical method
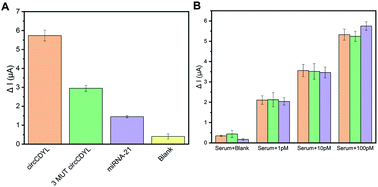
Another important feature of a good biosensor is high specificity. The specificity of the biosensor was examined by comparing the current response of the target circCDYL with that of 3-base mutation sequences (3 MUT circCDYL), a non-complementary RNA (miRNA-21) and blank RNA under the same conditions. As can be seen in Fig. 5A, the peak current variation (ΔI) of the complementary circCDYL was much higher than that of the 3 mutation circCDYL sequence and noncomplementary RNA. It is believed that the developed biosensor is capable of fulfilling excellent specificity. Furthermore, the reproducibility of the biosensor was also investigated. Five different electrodes were used to measure synthesized circCDYL of the same concentration. Relative standard deviations (RSD) of our biosensors was 7.33%, which showed good reproducibility of the method (Fig. S4†). The performance of the electrodes will be kept consistent in the future by using industrial-produced SPME and calculating the effective areas.To evaluate the practical applications and analytical reliability of the designed SPME biosensor, different concentrations of synthesized circCDYL in serum were detected. Repeated data is shown in Fig. 5B and demonstrates that the signal was relevant to ΔI in human serum in the presence of circRNA. Compared with blank RNA, an increased response was shown in serum solutions from 1 pM to 100 pM concentration in serum solution was shown. The recovery values ranged from 119.7% to 115.1%, and the corresponding RSDs ranged from 8.34% to 8.94% for circCDYL (Table S3†), which is in good comparable with results reported by other researchers.19 It is important to remark that this electrochemical platform for circRNA detection is highly satisfying and stable, even in the presence of serum. The performance of the electrodes will be kept consistent in clinical application. These features are consistent with the possibility of this biosensor being adopted as a useful tool for screening HCC at POCT.
Conclusions
In summary, we have developed a screen-printed electrode with magnetic substrates, showing the advantages of electrochemical detection. Since the biosensor is magnetic, magnetic beads with probes can be absorbed onto the working electrode to achieve enhanced analytical performance in terms of simplicity, selectivity and stability. In addition, the BSJ sequence of circRNA-targeted magnetic probe beads with assistance from methylene blue can avoid the disadvantages of enzyme-assisted translation. In less than 3 hours, the electrochemical SPME was able to detect circCDYL down to 1 pM with high sensitivity and was applied to serum samples with satisfactory results. The features of this biosensor implicated the suitability in terms of time production, sensitivity and capability in complex matrix such as serum. In fact, there is a possibility of translating this strategy to detect different types of biomolecules in real samples with specific probes for future clinical analysis.Author contributions
Bin Zhang: methodology, investigation, data curation, writing – original draft. Yitao Liang: investigation, data curation, writing – review & editing. Bo Liang: conceptualization, funding acquisition. Mingyu Chen: supervision, software, resources. Bobo Huang: investigation, writing – review & editing. Qingpeng Cao: methodology, supervision. Jinwei Wei: data curation, writing – review & editing. Tianyu Li: data curation. Xiujun Cai: conceptualization, resources. Xuesong Ye: project administration, resources, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank the Hendi Maher for his assistance in English edition. This work was supported by the Natural Science Foundation of Zhejiang Province (LQ20F010011, LY18H180006); Key Research and Development Program of Zhejiang Province (2019C03066); National Natural Science Foundation of China (61501400, 81501555); Fundamental Research Funds for the Central Universities.References
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Ca-Cancer J. Clin., 2018, 68, 394–424 CrossRef PubMed
.
- S. Colagrande, A. L. Inghilesi, S. Aburas, G. G. Taliani, C. Nardi and F. Marra, World J. Gastroenterol., 2016, 22, 7645–7659 CrossRef PubMed
.
- Y. Wei, X. Chen, C. Liang, Y. Ling, X. Yang, X. Ye, H. Zhang, P. Yang, X. Cui, Y. Ren, X. Xin, H. Li, R. Wang, W. Wang, F. Jiang, S. Liu, J. Ding, B. Zhang, L. Li and H. Wang, Hepatology, 2020, 71, 130–147 CrossRef CAS PubMed
.
- M. Mei, Y. Wang, Q. Wang, Y. Liu, W. Song and M. Zhang, Cancer Manage. Res., 2019, 11, 10215–10221 CrossRef CAS PubMed
.
- J. Sun, H. Zhang, D. Tao, F. Xie, F. Liu, C. Gu, M. Wang, L. Wang, G. Jiang, Z. Wang and X. Xiao, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 1349–1356 CrossRef CAS PubMed
.
- Y. Wei, X. Chen, C. Liang, Y. Ling, X. Yang, X. Ye, H. Zhang, P. Yang, X. Cui, Y. Ren, X. Xin, H. Li, R. Wang, W. Wang, F. Jiang, S. Liu, J. Ding, B. Zhang, L. Li and H. Wang, Hepatology, 2020, 71, 130–147 CrossRef CAS PubMed
.
- M. T. Hsu and M. Coca-Prados, Nature, 1979, 280, 339–340 CrossRef CAS PubMed
.
- S. Memczak, M. Jens, A. Elefsinioti, F. Torti, J. Krueger, A. Rybak, L. Maier, S. D. Mackowiak, L. H. Gregersen, M. Munschauer, A. Loewer, U. Ziebold, M. Landthaler, C. Kocks, F. le Noble and N. Rajewsky, Nature, 2013, 495, 333–338 CrossRef CAS PubMed
.
- X. Li, L. Yang and L. L. Chen, Mol. Cell, 2018, 71, 428–442 CrossRef CAS PubMed
.
- I. L. Patop and S. Kadener, Curr. Opin. Genet. Dev., 2018, 48, 121–127 CrossRef CAS PubMed
.
- Z. Liu, Y. Ran, C. Tao, S. Li, J. Chen and E. Yang, Genome Biol., 2019, 20, 99 CrossRef PubMed
.
- R. Tavallaie, S. R. De Almeida and J. J. Gooding, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2015, 7, 580–592 CAS
.
- R. Tavallaie, N. Darwish, M. Gebala, D. B. Hibbert and J. J. Gooding, ChemElectroChem, 2014, 1, 165–171 CrossRef
.
- H. Tang, J. Zhu, D. Wang and Y. Li, Biosens. Bioelectron., 2019, 131, 88–94 CrossRef CAS PubMed
.
- L. Tian, K. Qian, J. Qi, Q. Liu, C. Yao, W. Song and Y. Wang, Biosens. Bioelectron., 2018, 99, 564–570 CrossRef CAS PubMed
.
- R. Salahandish, A. Ghaffarinejad, E. Omidinia, H. Zargartalebi, A. K. Majidzadeh, S. M. Naghib and A. Sanati-Nezhad, Biosens. Bioelectron., 2018, 120, 129–136 CrossRef CAS PubMed
.
- X. Li, G. Peng, F. Cui, Q. Qiu, X. Chen and H. Huang, Biosens. Bioelectron., 2018, 113, 116–123 CrossRef CAS PubMed
.
- M. N. Islam, S. Moriam, M. Umer, H. P. Phan, C. Salomon, R. Kline, N. T. Nguyen and M. J. A. Shiddiky, Analyst, 2018, 143, 3021–3028 RSC
.
- J. Jiao, C. Li, L. Ning, L. Shi, L. Wang, Y. Xiang and G. Li, Sens. Actuators, B, 2020, 302, 127166 CrossRef CAS
.
- Q. Zhu, B. Liang, Y. Cai, Q. Cao, T. Tu, B. Huang, L. Fang and X. Ye, Talanta, 2018, 190, 70–77 CrossRef CAS PubMed
.
- B. Huang, L. Ji, B. Liang, Q. Cao, T. Tu and X. Ye, Analyst, 2019, 144, 3282–3288 RSC
.
- J. B. Haun, T. J. Yoon, H. Lee and R. Weissleder, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2010, 2, 291–304 CAS
.
- L. Jirakova, R. Hrstka, S. Campuzano, J. M. Pingarrón and M. Bartosik, Electroanalysis, 2019, 31, 293–302 CrossRef CAS
.
- M. Chen, C. Hou, D. Huo, H. Fa, Y. Zhao and C. Shen, Sens. Actuators, B, 2017, 239, 421–429 CrossRef CAS
.
- L. Zhu, R. Zhao, K. Wang, H. Xiang, Z. Shang and W. Sun, Sensors, 2008, 8, 5649–5660 CrossRef CAS PubMed
.
- M. A. Pereira-Barros, M. F. Barroso, L. Martin-Pedraza, E. Vargas, S. Benede, M. Villalba, J. M. Rocha, S. Campuzano and J. M. Pingarron, Biosens. Bioelectron., 2019, 137, 171–177 CrossRef CAS PubMed
.
- E. Povedano, V. Ruiz-Valdepenas Montiel, M. Gamella, M. Pedrero, R. Barderas, A. Pelaez-Garcia, M. Mendiola, D. Hardisson, J. Feliu, P. Yanez-Sedeno, S. Campuzano and J. M. Pingarron, Anal. Chem., 2020, 92, 5604–5612 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available: Schematic illustration of BSJ sites of circRNA. EDS map of magnetic beads. Various amounts of suspension volume ranging from 0 to 20 μg MBs in electrochemical performance and surface of the electrode. The electrochemical DPV response of independently fabricated 5 biosensors in 10 pM concentrations. Oligonucleotides used in this work. Comparison of different methods for circRNA analysis. Determination of circRNA added in human serum. See DOI: 10.1039/d1ra01033f |
| ‡ Contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |