Open Access Article
Open Access ArticlePEI-assisted boronate affinity magnetic nanoparticle-based SELEX for efficient in vitro evolution of saponin-binding aptamers†
Hui Zhanga,
Xue Lib,
Ailan Huanga,
Zhifeng Yana,
Yang Chen ab and
Zijun Bie
ab and
Zijun Bie *ab
*ab
aDepartment of Chemistry, Bengbu Medical University, 2600 Donghai Avenue, Bengbu 233000, China. E-mail: biezijun@bbmc.edu.cn
bSchool of Pharmacy, Bengbu Medical University, 2600 Donghai Avenue, Bengbu 233000, China
First published on 25th February 2021
Abstract
Saponin is a large family of important natural products with various pharmacological activities. Selective enrichment of saponin from complex biological samples is a key step for analysis of saponin. Despite that aptamers have been widely used for selective enrichment, aptamers that can specifically recognize saponins have never been reported. In this study, a facile and efficient SELEX approach was developed for in vitro evolution of saponin-binding aptamers, using PEI-assisted boronate affinity magnetic nanoparticles (p-BA-MNPs) that exhibit highly favorable binding properties as a general affinity platform. As a proof of the principle, ginsenoside Re and Rb1 were employed as two target saponins. Two aptamers towards each target saponin, with dissociation constant at the 10−5 M level, were selected within 6 rounds. An affinity magnetic nanoparticle was constructed by using the selected aptamer as a affinity ligand. The resulting material allowed for the quantitative analysis of ginsenoside Re in real samples with high reliability. The p-BA-MNPs based SELEX is straightforward and generally applicable for a wide range of target saponins, providing a promising aptamer evolution approach for aptamer-based research and pharmaceutical analysis.
Introduction
Saponins, a large family of important natural products with either steroidal or terpenoidal backbone and glycosides, have found various pharmacological activities including anti-cancer activity, immune system regulatory functions, therapeutic effects in leukemia, and so on.1–4 However, due to endogenous interference, matrix effects and trace amounts of exogenous compounds, a simple and comprehensive analytical approach for analysis of saponins remains challenging. Thus, the selective separation of saponins is the key to eliminate all the possible interference before further analysis.Due to the rapid development of separation science, different techniques have been employed for separation of saponins, including TLC,5,6 reverse phase chromatography (RPLC),7–9 HILIC,10 CE11 and immunoaffinity chromatography (IAC).12,13 However, the existing approaches are associated with several drawbacks. For example, the strongly polar saponins are not suitable for RPLC. Furthermore, the applications of IAC are also very limited due to the low immunogenicity of the glycosyl chains of saponins.14 As a consequence, a novel selective enrichment approach for saponins is still of great importance.
Aptamers are single-stranded oligonucleotides (DNA or RNA) that have specific and complex three-dimensional shapes, characterized by stems, loops, hairpins, bulges, pseudoknots, triplexes, or quadruplexes.15 Because of the spatial structures and multiple functional groups, aptamers can bind with a large variety of chemical or biological species selectively.16–19 By employing the aptamers as specific recognition elements, numerous analytical approaches have been developed for different purposes.20–23 Thus, the aptamers that can specifically bind with saponins will provide new access to develop novel selective analytical approaches for saponins. However, such aptamers have never been reported.
Aptamers are usually evolved from random oligonucleotide libraries through systematic evolution of ligands by exponential enrichment (SELEX).24,25 During SELEX, specific isolation of the target-binding single stranded DNA (ssDNA) from the random pool is the key step. Widely used SELEX mainly rely on several isolation strategies, including affinity chromatography,24,26 surface plasmon resonance (SPR)27 and capillary electrophoresis (CE).28,29 Despite CE is the most commonly used isolation strategy, the CE-SELEX is not suitable for the evolution of saponin-binding aptamers. This is because the employment of organic solvent during separation process (to improve the solubility of the saponins) could have negative effect on the formation of spatial structure of ssDNA. Therefore, new efficient evolution methods are much needed. A possible solution to the above dilemma is the immobilization of target onto certain matrices, which allows the employment of different buffers for target immobilization and ssDNA/target incubation. Recently, our group has demonstrated that boronate affinity materials can be used as efficient sorbents for saponins.30–32 Besides, boronate affinity materials have been proven to be a rapid and efficient platform for screening of aptamers for biomolecules.33–36 Thus, all these findings inspired us to further explore the applications of boronate affinity materials based affinity platform for saponin-binding aptamers evolution.
Herein, we present a PEI-assisted boronate affinity magnetic nanoparticle (p-BA-MNPs)-based SELEX approach, in which p-BA-MNPs were used as an affinity interface for efficient evolution of saponin-specific aptamers. The p-BA-MNPs could specifically bind the target saponins at relatively high pH (≥8.5), yet the bound target saponins could be released from the MNPs when the surrounding pH is acidic (pH ≤ 3.0). The principle of the p-BA-MNPs-based SELEX approach is illustrated in Fig. 1. First, the target saponin was incubated with the p-BA-MNPs for a certain period of time, ensuring the immobilization of the target. Afterward, the saponins bounded p-BA-MNPs was incubated with a fluorescein-labeled ssDNA library for certain time. After removing the unbound ssDNA during the washing step, only the ssDNA that can bind with the target saponins were remained on the p-BA-MNPs due to the ssDNA–saponin complexes. Subsequently, the ssDNA–saponin complexes were released from the MNPs by acidic solution (0.1 M HAc). The presence of the aptamer candidate–saponin complexes were verified by CE. Finally, the aptamer candidate–saponin complexes were used as templates for amplification by polymerase chain reaction (PCR) to finish a single round of evolution. During each evolution cycle, the binding affinity toward the target of the enriched ssDNA library was estimated by the fluorescence intensity of ssDNA–saponin complexes. No more evolution round was needed when the affinity met the requirement; otherwise, the PCR products were used as a new library for a new round of evolution. The whole evolution procedure was not complete until the affinity of the obtained DNA library met with the requirement or not significantly improved as compared with previous evolution round. The obtained aptamer candidates were sent to cloning and sequencing for further applications.
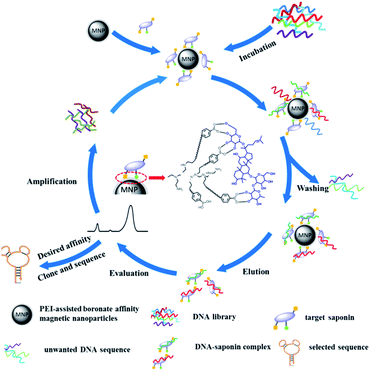 | ||
| Fig. 1 PEI-assisted boronate affinity magnetic nanoparticle-based SELEX for efficient in vitro evolution of saponin-binding aptamers. | ||
The p-BA-MNPs-based SELEX exhibited several advantages. The pH on/off controlled capture and release of ssDNA–target complexes and the easy manipulation of the magnetic cores enable the simple implementation of all the steps involved in the SELEX. Besides, more boronic acids moieties derived from the branched scaffold of PEI will not only leads to larger binding capacity but also stronger affinity toward the saponin.37–41 Moreover, because of the high affinity of the p-BA-MNPs toward the saponin target, the existence of competing cis-diol compounds such as monosaccharides will not interfere the evolution, ensuring efficient screening. Benefit from these, ssDNA that bind with the target with weak affinity could be washed away in each round of evolution. In addition, the hydrophilic nature of the PEI will greatly eliminate nonspecific adsorption and negative selection is not necessary. As a proof of the principle, typical saponins (ginsenoside Re and Rb1) were employed as examples, aptamers which can bind these two target saponins with high affinity and selectivity were successfully screened out within 6 rounds. For real-world application, a ginsenoside Re-binding aptamer based affinity material was prepared and used for selective enrichment of ginsenoside Re real sample.
Experimental
Materials and reagents
Adenosine, deoxyadenosine, branched polyethyleneimine (PEI) (Mw = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), sodium cyanoborohydride (NaBH3CN) and 2,4-difluoro-3-formyl-phenylboronic acid (DFFPBA) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Standard ginsenosides (Re, Rb1 and Rb2) were purchased from Yuanye bio-technology Co., Ltd (Shanghai, China). Ginseng-based oral liquid was purchased from the local pharmacy (AnTai, Bengbu, China). Glycine was purchased from Bio-Rad (Hercules, CA, USA). Sodium cyanoborohydride was purchased from J&K Scientific (Shanghai, China). Ferric trichloride hexahydrate, 1,6-hexanediamine, anhydrous sodium acetate, glycol, ammonium bicarbonate, and acetic acid were ordered from Nanjing Reagent Company (Nanjing, China). The DNA library, primers, FAM-modified, and HS-modified ssDNA sequences were purchased from Sangon (Shanghai, China). The DNA library was composed of 39 random nucleotides in the central and two constant primer regions at both ends. Hot start Taq enzyme and dNTPs were obtained from Takara Biotechnology (Dalian, China). The other chemical reagents were all of analytical grade or higher. For all the evolution cycles, the DNA library were dissolved in 0.05 M ammonium bicarbonate buffer solution containing 0.05 M sodium chloride (NaCl), 0.001 M magnesium chloride (MgCl2), and pH 8.5. Before the start of each evolution round, the ssDNA library and the collected aptamer at each cycle of screening were denatured by heating at 95 °C for 10 min and then cooled in an ice bath for another 10 min. Ultrapure water gained by a Millipore Milli-Q Advantage A10 system (Milford, MA, USA) was used throughout this work. The samples and buffer used for CE were filtered by membrane filter (0.45 μm) before use. Fused-silica capillary of 75 μm ID × 365 μm OD was acquired from Ruifeng Chromatographic Devices (Hebei, China).
000), sodium cyanoborohydride (NaBH3CN) and 2,4-difluoro-3-formyl-phenylboronic acid (DFFPBA) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Standard ginsenosides (Re, Rb1 and Rb2) were purchased from Yuanye bio-technology Co., Ltd (Shanghai, China). Ginseng-based oral liquid was purchased from the local pharmacy (AnTai, Bengbu, China). Glycine was purchased from Bio-Rad (Hercules, CA, USA). Sodium cyanoborohydride was purchased from J&K Scientific (Shanghai, China). Ferric trichloride hexahydrate, 1,6-hexanediamine, anhydrous sodium acetate, glycol, ammonium bicarbonate, and acetic acid were ordered from Nanjing Reagent Company (Nanjing, China). The DNA library, primers, FAM-modified, and HS-modified ssDNA sequences were purchased from Sangon (Shanghai, China). The DNA library was composed of 39 random nucleotides in the central and two constant primer regions at both ends. Hot start Taq enzyme and dNTPs were obtained from Takara Biotechnology (Dalian, China). The other chemical reagents were all of analytical grade or higher. For all the evolution cycles, the DNA library were dissolved in 0.05 M ammonium bicarbonate buffer solution containing 0.05 M sodium chloride (NaCl), 0.001 M magnesium chloride (MgCl2), and pH 8.5. Before the start of each evolution round, the ssDNA library and the collected aptamer at each cycle of screening were denatured by heating at 95 °C for 10 min and then cooled in an ice bath for another 10 min. Ultrapure water gained by a Millipore Milli-Q Advantage A10 system (Milford, MA, USA) was used throughout this work. The samples and buffer used for CE were filtered by membrane filter (0.45 μm) before use. Fused-silica capillary of 75 μm ID × 365 μm OD was acquired from Ruifeng Chromatographic Devices (Hebei, China).
Instruments
CE evaluation were carried out on a Beckman Coulter P/ACE MDQ instrument with UV detector (Fullerton, CA, USA). The wavelength for UV detection was set at 260 nm for ssDNA and 203 nm for saponins. PCR amplification was implemented on an ABI GeneAmp 9700 instrument (Shanghai, China). Transmission electron microscopy (TEM) characterization was carried out on a JEOL JEM-1010 instrument (Tokyo, Japan). The ultraviolet (UV) spectra were recorded by a Thermo Fisher NanoDrop 2000/2000C spectrophotometer (MA, USA).Methods
Synthesis of p-BA-MNPs
The procedure for the synthesis of p-BA-MNPs includes three major parts: (1) synthesis of amino-functionalized magnetic nanoparticles (AMNPs), (2) preparation of PEI modified MNPs (PEI-MNPs), and (3) boronic acid functionalization. The AMNPs were synthesized based on a previously reported approach.39 Briefly, ferric trichloride hexahydrate (2.0 g) was first dissolved in 60 mL ethylene glycol to form a clear orange yellow color solution. Then, 4.0 g anhydrous sodium acetate and 13.0 g 1,6-hexanediamine were added to the above solution. Finally, the mixture was stirred vigorously for 30 min and sealed in a PTFE-lined autoclave and reacted at 198 °C for 6 h. The resulting MNPs were rinsed with water and ethanol 3 times each, and then dried at 60 °C under vacuum overnight. After 200 mg AMNPs were added to 40 mL anhydrous methanol containing 5% glutaraldehyde, the mixture was mechanically stirred for 12 h at room temperature. The resulting glutaraldehyde-activated MNPs were washed 3 times with anhydrous methanol and then dispersed in 20 mL anhydrous methanol containing 0.5 g PEI by using ultrasound. The mixture was mechanically stirred for 12 h at room temperature. After that, 20 mg mL−1 sodium cyanoborohydride was added during the course of the reaction for 12 h. The PEI-MNPs were collected using a magnet, washed with ethanol 3 times each, and then dried at 60 °C overnight. 200 mg PEI-MNPs were dispersed in 50 mL anhydrous methanol containing 20 mg mL−1 DFFPBA and mechanically stirred for 12 h at room temperature. Then, 20 mg mL−1 sodium cyanoborohydride was added for another 12 h-reaction. The obtained MNPs were collected by magnet, washed with water and ethanol and dried at 60 °C. The obtained p-BA-MNPs were stored in a dry chamber for further use.Evolution of aptamers by the p-BA-MNPs-based SELEX
To screen aptamers against saponin, 5 mg of p-BA-MNPs were first dispersed in 1 mL of 1 mg mL−1 saponin solution (0.05 M ammonium bicarbonate buffer solution containing 10% methanol, pH 8.5). Then, 1 h of shaking at 25 °C was undertaken. Unbound saponin was washed away with 1 mL buffer solution (50 μM ammonium bicarbonate buffer containing 50 μM NaCl, 1 μM MgCl2, pH 8.5) for 3 times. After that, 500 μL of 1 μM ssDNA library was incubated with the saponin bound p-BA-MNPs for 1 h. Then, the p-BA-MNPs were rinsed with 500 μL buffer solution for 3 times to get rid of the unbound ssDNA. Ultimately, the bound species which were in the form of aptamer candidates–saponin complexes were eluted using 100 μL of 0.1 M acetic acid solution. In addition, the collected elutes containing aptamer candidates–saponin complexes were used as templates for amplification by PCR. The PCR products were used as a new library for subsequent round of evolution.Affinity evaluation
The affinity of the aptamer against target saponin was evaluated by dissociation constants (Kd). Different concentrations of each FAM-modified aptamer solution (from 0 to 200 nM) were prepared with 10 mM Tris–HCl buffer (pH 8.5). Firstly, 500 μL of 5 mg mL−1 target saponin was added to 500 μL of 5 mg mL−1 of p-BA-MNPs and shaken at room temperature for 2 h. Then, 200 μL aliquots of FAM-modified aptamer solution were added to the above mentioned saponin bound p-BA-MNPs and shaken at room temperature for 1 h. After rinsing with buffer (50 μM ammonium bicarbonate buffer containing 50 μM NaCl, 1 μM MgCl2, pH 8.5) 3 times, the aptamer–saponin complexes were eluted using 100 μL of 0.1 M acetic acid solution. The fluorescence intensity of elution was recorded. To estimate the Kd values, nonlinear least squares regression analysis was performed using the Prism software (Ver. 4.00, GraphPad Software, San Diego, CA).Selective enrichment of ginsenoside Re with Apt Re-3@MNPs
The aptamer functionalized MNPs (Apt Re-3@MNPs) was first synthesized. Briefly, 100 μL of NH2-modified selected aptamers (5 OD) were immobilized onto AMNPs through linking with glutaraldehyde. The nonbinding sequence-functionalized MNPs were obtained with the same process, expect the NH2-modified aptamer was replaced by an NH2-modified nonbinding sequence. For enrichment process, Apt Re-3@MNPs (2 mg) was added to 200 μL of 20-fold diluted human serum solution. The mixture was shaken on a rotator for 2 h at 25 °C. After that, the MNPs were collected by a magnet and rinsed with 200 μL of 0.1 M PBS containing 0.5 M NaCl (pH 7.4) and 0.1 M phosphate buffer (pH 7.4) for three times. For elution, 20 μL of 0.1 M HAc solution was employed. The elution process was carried out by 1 h shaking and 10 min 95 °C heating respectively. Finally, the mixture was subjected to magnetic separation to collect the eluates, and then sent for further analysis.Result and discussions
Characterization of PEI-assisted boronate affinity magnetic nanoparticles (p-BA-MNPs)
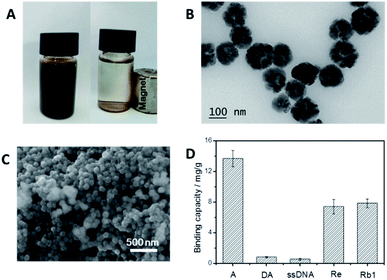
As shown in Fig. 2B and C, the average diameter of the p-BA-MNPs was determined by TEM and SEM to be 100 nm. Table S1† shows the element analysis of the BA-MNPs and p-BA-MNPs, the content of N of the p-BA-MNPs is significantly higher than that of BA-MNPs indicating more reactive sites for boronic acid functionalization. This is further confirmed capacity of the nanoparticles (Fig. S2†). Fig. 2A shows that the p-BA-MNPs dispersed well in buffer and are able to be attracted to the wall of the glass container by an external magnet, which greatly favours the following experiments. The selectivity of the p-BA-MNPs was the first priority to the aptamer evolution in this study. As shown in Fig. 2D, the p-BA-MNPs selectively captured adenosine (a typical cis-diol molecule) but excluded deoxyadenosine (a typical non cis-diol molecule), indicating successful functionalization of boronic acid. As shown in Fig. 2D, typical saponins (ginsenoside Re, Rb1) were extracted by the p-BA-MNPs under alkaline environment (pH 8.5), but released at an acidic environment (pH 3.0). Because of the high affinity towards the saponins, intensive washing could be adopted to prevent the non-specific adsorption of DNA library. Because of this desired selectivity, the p-BA-MNPs allowed for selective immobilization of the target saponins as well as evolution of target-specific ssDNA.In vitro evolution of saponin-binding aptamers using p-boronate affinity MNPs
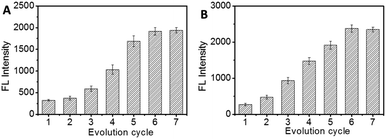
In vitro evolution of saponin-binding aptamers were carried out by p-BA-MNPs based SELEX. In order to eliminate the possible interference of some weak binding ssDNA, washing step was introduced after incubation in every evolution round. As shown in Fig. S3,† certain UV absorbance of washing buffer could be recorded. Considering the limited UV absorbance of the saponin in the corresponding region, the UV absorbance could be ascribed to either ssDNA or saponin–ssDNA complex. Based on the UV absorbance of the washing buffer (Fig. S3†), we can conclude that certain amount of weak binding ssDNA was washed away in the washing step. Furthermore, the UV absorbance of washing buffer was decreased as the rounds of evolution increased, which indicates the increased affinity between ssDNA and target saponin. In addition, the presence of saponin–ssDNA complex was investigated by CE (Fig. S4†). By using a widely used CE running buffer (25 μM glycine, 25 μM Na2B4O7, 5 μM KH2PO4, and pH 9.0) for aptamer selection, the unbound ssDNA, free target saponin and saponin–ssDNA complexes were evaluated by CE with UV detection. The ssDNA library with a pI of 4–5, showed one broad peak around 10 min. Considering the poor aqueous solubility of the saponin, no apparent peaks for saponins could be observed. After the first round of evolution cycle, a series of CE peaks was observed at 5–8 min. This could be ascribed to the formation of saponin–ssDNA complex, which provided a solid basis for the following evolution process.In order to characterize the evolution process, the aptamers obtained at different evolution cycles were evaluated. During the evaluation, the target saponin were first immobilized onto the p-BA-MNPs and then incubated with 5′-FAM-modified ssDNA obtained at each evolution round; after washing away unbound ssDNA, the fluorescence intensity of ssDNA–protein complex was recorded to estimate the affinity of the selected aptamers towards target saponins. As shown in Fig. 3, the fluorescence intensity increased fast as the evolution cycle increased. The intensity value of the 6th round and the 7th round was almost the same, indicating that the selection reached saturation. Therefore, the evolution process was terminated.
Characterization of selected aptamers
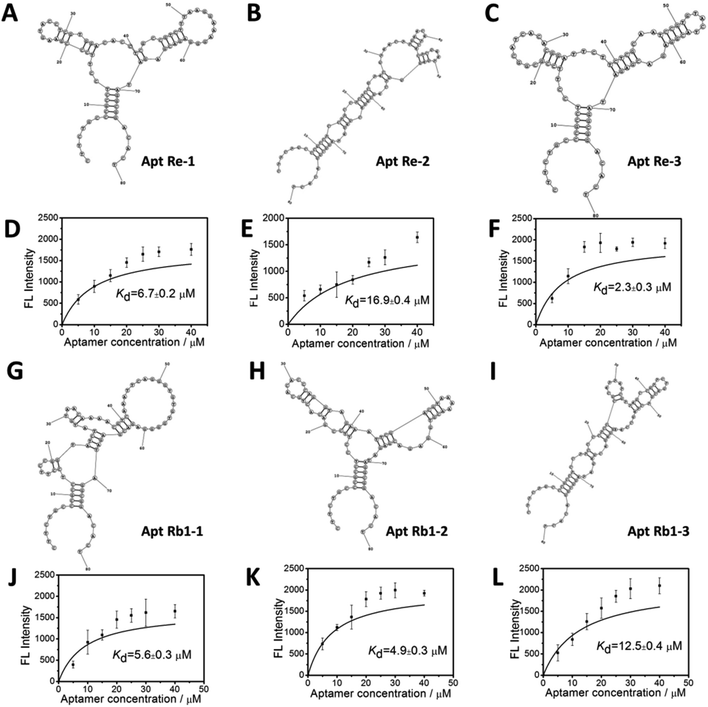
After 6 rounds of evolution, the selected aptamer candidates were cloned and sequenced. Among the obtained sequences, three aptamers for each specific saponin (ginsenoside Re and Rb1) were chosen due to their lowest Gibbs free energy.34–36 The secondary structures and binding curve for the selected aptamers are shown in Fig. 4. The primary structures and detailed values of the measured Kd values are summarized in Table 1. As shown in the above results, the selected aptamers exhibited high affinity to target saponin, giving Kd values within the 10−6 to 10−5 M range (2.3–16.9 μM).| Aptamer | Sequence of random region (5′–3′) | Kd (μM) |
|---|---|---|
| a The sequence of the 5′-end constant region: 5′-CTTCTGCCCGCCTCCTTCC; the sequence of the 3′-end constant region: GGAGACGAGATAGGCGGACACT-3′. | ||
| Re-1 | CGAAGTACCTCGTGGACACCTCGCCGTCTTTAAGGAAAG | 6.7 ± 0.2 μM |
| Re-2 | TCTCCGTACGTAACTTGAACATCACTCGAATGAGGGTCA | 16.9 ± 0.4 μM |
| Re-3 | GCGGCATCACACGGATTCTTCTCGCAATCCGTGTACTAC | 2.3 ± 0.3 μM |
| Rb1-1 | GGTATCTATGTAACAAAACGTCAAATTCAGGGTGTGTGG | 5.6 ± 0.3 μM |
| Rb1-2 | CCTCCCGCATTATCGTTAGCCGTTCTGACGTCTAGCCAA | 4.9 ± 0.3 μM |
| Rb1-3 | GACACTGCACACCCAGGTAGAAATTCGACGTAAAAACGT | 12.5 ± 0.4 μM |
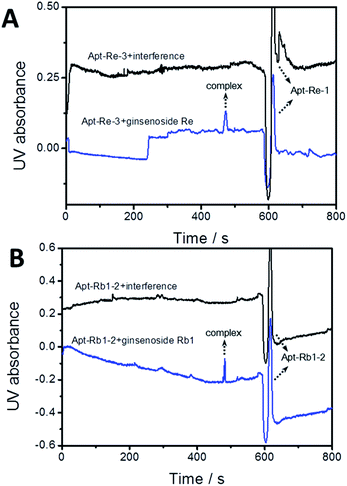
Furthermore, the selectivity of the obtained aptamers toward the saponin target with a series of structural analogues was investigated. Apt Re-3 and Rb1-2 were selected as representatives of the selected sequences for ginsenoside Re and Rb1, respectively. As shown in Fig. 5A, an apparent peak was observed for apt Re-3–ginsenoside Re complex, whereas nearly nothing was recorded for the mixture of apt Re-3 with other structural analogues. Similar results could also be observed for aptamer Rb1-2 (Fig. 5B). These results suggest that both apt Re-3 and Rb1-2 exhibited specificity toward their target saponins. The high affinity and good selectivity of screened aptamers verified the high efficiency of the p-BA-MNPs based SELEX approach.
Extraction of ginsenoside Re from complex sample
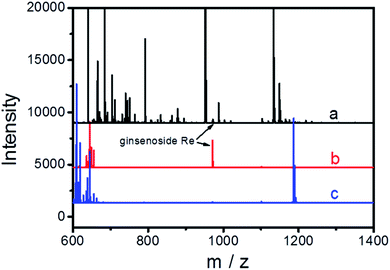
The extraction of ginsenoside Re from spiked human serum was employed to demonstrate the potential of the selected aptamers for real-world applications. The ginsenoside Re-binding aptamer (apt Re-3) was used as an affinity ligand and modified onto amino-functionalized MNPs to form the aptamer-functionalized MNPs (Apt Re-3@MNPs, Fig. S5†). The obtained magnetic nanoparticles were then employed as the selective sorbent to enrich ginsenoside Re from a spiked human serum sample. In the meantime, nonbinding sequence-modified MNPs were used as a control. Considering the spiked level of ginsenoside Re (100 ng mL−1) is relatively low, meanwhile there a lots of interfering compounds, direct analysis of the spike sample is of great difficulty. As shown in Fig. 6, the MS peak for ginsenoside Re is barely observable during the direct analysis and the result is greatly improved after enrichment with Apt Re-3@MNPs. As a comparison, no ginsenoside Re was extracted by the nonbinding sequence-modified MNPs, indicating the high specificity of the selected aptamers. Furthermore, HPLC-UV was also employed to evaluate the performance of Apt Re-3@MNPs for quantitative analysis. As demonstrated in Fig. S6,† an enhanced and clean chromatograph could be obtained after the enrichment. For different spiked levels, the recoveries are 63–75%, and RSDs are 8.3–14.1% (Table S2†). Thus, all these results reveal that the aptamer used was of excellent performance and held great potential in real-world applications.Conclusion
We have established a p-BA-MNPs-based SELEX for efficient in vitro evolution of saponin-binding aptamers. This is the first example of evolution of the saponin-binding aptamers. p-BA-MNPs, as an advanced functional material, permitted facile capture/release of saponins as well as easy magnetic manipulation. By the introduction of PEI scaffold, both binding capacity and affinity were improved, which ensured the efficiency of the SELEX approach. Besides, the hydrophilic nature of the PEI could greatly eliminated sequences with nonspecific binding. The effectiveness of this SELEX method was verified with the successful evolution of aptamers which can bind specifically two representative target saponin, ginsenoside Re and ginsenoside Rb1. The selected aptamers exhibited high affinity and high specificity. By the employment of one aptamer against ginsenoside Re (Apt Re-3) as affinity ligand, an affinity MNPs was synthesized for selective enrichment of ginsenoside Re in real sample. Because most saponin contains glycosides, the aptamers that is specific toward other saponins can also be efficiently selected by this developed new SELEX method. Therefore, the p-BA-MNPs based SELEX can hold a great promised for the efficient evolution of aptamers.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the financial support of the grants from the Natural Science Foundation of Anhui Province (1908085QH349), Natural Science Research Project of Anhui Educational Committee (KJ2019A0390).Notes and references
- M. J. Kim, H. Yun, D. H. Kim, I. Kang, W. Choe, S. S. Kim and J. Ha, J. Ginseng Res., 2014, 38, 16 CrossRef.
- K. R. Vinoth, T. W. Oh and Y. K. Park, Neurochem. Res., 2016, 41, 951 CrossRef.
- J. Tian, F. Fu, M. Geng, Y. Jiang, J. Yang, W. Jiang, C. Wang and K. Liu, Neurosci. Lett., 2005, 374, 92 CrossRef CAS.
- H. J. Jang, I. H. Han, Y. J. Kim, N. Yamabe, D. Lee, G. S. Hwang, M. Oh, K. C. Choi, S. N. Kim, J. Ham, D. W. Eom and K. S. Kang, J. Agric. Food Chem., 2014, 62, 2830 CrossRef CAS.
- Y. Jiang, B. David, P. Tu and Y. Barbin, Anal. Chim. Acta, 2010, 657, 9 CrossRef CAS.
- R. J. Vanhaelen-Fastré, M. L. Faes and M. H. Vanhaelen, J. Chromatogr. A, 2000, 868, 269 CrossRef.
- S. Y. Wang, L. Z. Qiao, X. Z. Shi, C. X. Hu, H. W. Kong and G. W. Xu, Anal. Bioanal. Chem., 2015, 407, 331 CrossRef CAS.
- Z. T. Liang, Y. J. Chen, L. Xu, M. J. Qian, T. Yi, H. B. Chen and Z. Z. Zhao, J. Pharm. Biomed. Anal., 2015, 105, 121 CrossRef CAS.
- Y. J. Chen, Z. Z. Zhao, H. B. Chen, E. Brand, T. Yi, M. J. Qin and Z. T. Liang, J. Ginseng Res., 2017, 41, 10 CrossRef.
- Y. Wang, J. S. Wang, M. Yao, X. J. Zhao, J. Fritsche, P. Schmitt-Kopplin, Z. W. Cai, D. F. Wan, X. Lu, S. L. Yang, J. R. Gu, H. U. Häring, E. D. Schleicher, R. Lehmann and G. W. Xu, Anal. Chem., 2008, 80, 4680 CrossRef CAS.
- Y. L. Tian, Y. Y. Lu, J. Xie, Y. Cheng, R. B. Qi, Y. J. Wu and S. S. Zhang, Anal. Methods, 2009, 1, 203 RSC.
- B. Stritularak, O. Morinaga, C. S. Yuan, Y. Shaoyama and H. Tanaka, J. Nat. Med., 2009, 63, 360 CrossRef.
- O. Morinaga, H. Tanaka and Y. Shaoyama, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2006, 830, 100 CrossRef CAS.
- E. Sterner, N. Flanagan, J. C. Gildersleeve, E. Sterner, N. Flanagan and J. C. Gildersleeve, ACS Chem. Biol., 2016, 11, 1773 CrossRef CAS.
- R. Stoltenburg, C. Reinemann and B. Strehlitz, Biomol. Eng., 2007, 24, 381 CrossRef CAS.
- B. A. Williams, L. Lin, S. M. Lindsay and J. C. Chaput, J. Am. Chem. Soc., 2009, 131, 6330 CrossRef CAS.
- Z. Xi, R. Huang, Z. Li, N. He, T. Wang, E. Su and Y. Deng, ACS Appl. Mater. Interfaces, 2015, 7, 11215 CrossRef CAS.
- R. H. Wang and Y. B. Li, Biosens. Bioelectron., 2013, 42, 148 CrossRef CAS.
- L. J. Wang, R. H. Wang, F. Chen, T. S. Jiang, H. Wang, M. Slavik, H. Wei and Y. B. Li, Food Chem., 2017, 221, 776 CrossRef CAS.
- S. Tombelli, M. Minunni and A. Mascini, Biosens. Bioelectron., 2005, 20, 2424 CrossRef CAS.
- B. Deng, Y. W. Lin, C. Wang, F. Li, Z. X. Wang, H. Q. Zhang, X. F. Li and X. C. Le, Anal. Chim. Acta, 2014, 837, 1 CrossRef CAS.
- K. Maehashi, T. Katsura, K. Kerman, Y. Takamura, K. Matsumoto and E. Tamiya, Anal. Chem., 2007, 79, 782 CrossRef CAS.
- W. H. Tan, M. J. Donovan and J. H. Jiang, Chem. Rev., 2013, 113, 2842 CrossRef CAS.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818 CrossRef CAS.
- C. Tuerk and L. Gold, Science, 1990, 249, 505 CrossRef CAS.
- J. J. Tang, J. W. Xie, N. S. Shao and Y. Yan, Electrophoresis, 2006, 27, 1303 CrossRef CAS.
- T. S. Misono and P. K. R. Kumar, Anal. Biochem., 2005, 342, 312 CrossRef CAS.
- S. D. Mendonsa and M. T. Bowser, J. Am. Chem. Soc., 2005, 127, 9382 CrossRef CAS.
- J. Yang and M. T. Bowser, Anal. Chem., 2013, 85, 1525 CrossRef CAS.
- W. M. Zhao, A. L. Huang, Z. F. Yan, Z. J. Bie and Y. Chen, Spectrochim. Acta, Part A, 2020, 234, 118258 CrossRef CAS.
- H. Zhang, Y. N. Zhang, H. L. Wang, H. Wen, Z. F. Yan, A. L. Huang, Z. J. Bie and Y. Chen, J. Sep. Sci., 2020, 43, 2162 CrossRef CAS.
- Y. Chen, W. M. Zhao, Q. Chen, W. Wei, S. Y. Chen and Z. J. Bie, Anal. Methods, 2019, 11, 5673 RSC.
- H. Y. Nie, Y. Chen, C. C. Lü and Z. Liu, Anal. Chem., 2013, 85, 8277 CrossRef CAS.
- X. L. Li, Y. J. He, Y. Y. Ma, Z. J. Bie, B. R. Liu and Z. Liu, Anal. Chem., 2016, 88, 9805 CrossRef CAS.
- Y. Y. Ma, X. L. Li, W. Li and Z. Liu, ACS Appl. Mater. Interfaces, 2018, 10, 40918 CrossRef CAS.
- Y. Y. Ma, W. Li, R. R. Xing, P. F. Li and Z. Liu, ACS Sens., 2020, 5, 2537 CrossRef CAS.
- N. N. Zhao, Z. Q. Liu, J. P. Xing, Z. Zheng, F. R. Song and S. Liu, Chem. Eng. J., 2020, 383, 123079 CrossRef CAS.
- Y. Chen, A. L. Huang, Y. N. Zhang and Z. J. Bie, Anal. Chim. Acta, 2019, 1076, 1 CrossRef CAS.
- H. Y. Wang, Z. J. Bie, C. C. Lü and Z. Liu, Chem. Sci., 2013, 4, 4298–4303 RSC.
- D. J. Li, H. J. Xia and L. Wang, Talanta, 2018, 184, 235 CrossRef CAS.
- D. J. Li and Z. J. Bie, Analyst, 2017, 142, 4494 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: All the supplementary figures and tables. See DOI: 10.1039/d1ra00889g |
| This journal is © The Royal Society of Chemistry 2021 |