Open Access Article
Open Access ArticleUltra-specific genotyping of single nucleotide variants by ligase-based loop-mediated isothermal amplification coupled with a modified ligation probe†
Yuanyuan Sun *ac,
Bingjie Han
*ac,
Bingjie Han a and
Fangfang Sun
a and
Fangfang Sun *b
*b
aDepartment of Translational Medicine Center, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan Province, P. R. China. E-mail: sunyy@zzu.edu.cn
bDepartment of Gynecology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan Province, P. R. China. E-mail: fccsunff@zzu.edu.cn
cSchool of Chemistry and Chemical Engineering, Shaanxi Normal University, Xi'an 710062, Shaanxi Province, P. R. China
First published on 10th May 2021
Abstract
Specific and accurate detection of single nucleotide variants (SNVs) plays significant roles in pathogenic gene research and clinical applications. However, the sensitive but ultra-specific detection of rare variants in biological samples still remains challenging. Herein, we report a novel, robust and practical SNV assay by integrating the outstanding features of high selectivity of an artificial mismatched probe, and the powerful loop-mediated isothermal amplification. In this strategy, we rationally introduce artificial mismatched bases into the 3′-terminal regions of the probe located in the ligation region to reduce the risk of nonspecific ligation, which can dramatically improve the specificity for the SNV assay. The proposed method can discern as little as 0.01% mutant DNA in the high background of wild-type DNA with high sensitivity (10 aM). In virtue of its outstanding performance, the artificial mismatched probe may also be employed and expanded in various DNA and RNA genetic analyses with ligase-assisted approaches, showing great potential in biomedical research, clinical diagnostics, and bioanalysis.
Introduction
Accurate detection of inherited genetic variation and sequence-specific nucleic acids at single-base resolution is vitally important for research on disease pathogenesis, genetic studies and personalized medicine.1–4 In real biological samples, the detection of single nucleotide variants (SNVs) in the presence of a high background of wild-type DNA (wtDNA) requires an analytical method that possesses powerful amplification and highly accurate single-base discrimination ability. Nucleic acid amplification techniques are undoubtedly powerful tools that can enhance the sensitivity of the sequence-specific target DNA detection, which is necessary for accurate SNV genotyping.5–7Various methods have been applied in site-specific SNVs genotyping. These methods can mainly be divided into the polymerase chain reaction (PCR),8–13 and the ligation-based nucleic acid amplifications.14–18 However, due to its low specificity, the allele-specific PCR based on primer extension catalyzed by DNA polymerase cannot accurately recognize single-base variation, and this can lead to false-positive results caused by amplification artifacts (such as primer-dimers).19 Owing to the superior performance of ligase to distinguish a single-base difference, the ligation reaction is generally combined with multiple nucleic acid amplification techniques, such as ligase chain reaction (LCR),20,21 ligation-assisted PCR,14 strand displacement amplification (SDA),16,22 rolling circle amplification (RCA)17,23 and isothermally exponential amplification (LIEXA),24 to improve the efficiency of the genotyping assay. The ligation probe with complementary base of the mutation site at the 3′-end is specially designed to detect mutant DNA (mutDNA). Therefore, the ligation product cannot be generated from the wtDNA template, and thus, subsequent amplification will not be initiated. Although the ligation reaction is generally available in single-base discrimination, it has a fundamental drawback that specificity is limited due to non-specific ligation produced by some ligation probes with the template of wtDNA. The ligation-based approaches have sufficient specificity for conventional SNVs genotyping analysis (1% or even 0.1%),20–24 but there is still a bottleneck for analysis of rare variants in clinical applications. Furthermore, the ligation reaction is used not only in SNVs genotyping analysis, but also in the detection of various gene biomarkers, such as microRNA,25–27 messenger RNA,28 circular RNA,29 DNA and RNA methylation,30–33 by the ligase-assisted approaches mentioned above. To our knowledge, research focusing on effective reduction of non-specific ligation to improve the analysis specificity has not been reported.
Loop-mediated isothermal amplification (LAMP) is an outstanding strategy that can rapidly detect the target genes with only a few copies, which can be completed by only using one type of DNA polymerase at a constant temperature within an hour.34 Nevertheless, the primer extension-based LAMP has poor specificity for single-base variations analysis (1%).35 In our previous study, we employed the ligase-based isothermally exponential amplification (LIEXA) as a general strategy to achieve highly sensitive detection of genetic biomarkers at a single-base resolution,24 which can accurately detect 10 aM mutant DNA (mutDNA) and 0.1% mutDNA in the presence of a large amount of wtDNA. However, due to the influence of non-specific ligation, the specificity may not meet the requirement for the accurate analysis of rare variants.
To address this issue, in this study, we report a novel SNVs genotyping assay based on the artificial mismatched ligation probe combined with the ligase-assisted LAMP amplification (AML-LAMP). The artificial mismatched base is elegantly introduced at the third position from the 3′-terminus of Probe mB-3 located in ligation region. Due to the mismatch between wtDNA and Probe mB-3 by synergy of terminal base and artificial mismatched base, three overhanging bases are generated at the 3′-end of Probe mB-3, and these overhanging bases can effectively prevent the ligation of wtDNA as the template. The proposed genotyping assay is able to unequivocally detect 10 aM mutDNA (equal to 60 copies) with ultrahigh specificity to discriminate as low as 0.01% mutDNA in a high background of wtDNA.
Experimental
Reagents and instruments
Thermostable Ampligase was purchased from Epicenter Technologies (USA). Bst DNA polymerase large fragment was obtained from New England Biolabs (USA). HPLC-purified DNA targets, probes and primers used in this study were purchased from Integrated DNA Technologies (USA). The betaine was obtained from Sigma-Aldrich (Shanghai, China). The dNTPs were purchased from Takara Biomedical Technology Co., Ltd. (Beijing, China). SYBR Green I (20 ng μL−1 stock solution in DMSO) was obtained from Zhishan Biotechnology Co., Ltd. (Xiamen, China). All the solutions were prepared with sterilized and deionized water. The sequences of oligonucleotides were listed in Table S1 (see in ESI†).The ligation reaction was carried out in a T100 Thermal Cycler (Bio-Rad, USA). The real-time fluorescence signal was monitored with the StepOne Real-Time PCR System (Applied Biosystems, USA).
Human genomic DNA extraction
The human genomic DNA was extracted from the human whole blood by using the TIANamp Genomic DNA Kit (TIANGEN Biotechnologies) following the guidelines of manipulate instructions. The extracted genomic DNA was quantified with the Nanodrop One (Thermo Fisher Scientific, USA). Notably, before with the genomic DNA as the template of ligation reaction, it should be heat-denatured at 95 °C for 3 min and then placed on ice immediately.Standard protocol for SNVs genotyping with the AML-LAMP assay
The ligation reaction was carried out in a 10 μL mixture containing reaction buffer (20 mM Tris–HCl, 25 mM KCl, 10 mM MgCl2, 0.5 mM NAD and 0.01% Triton X-100, pH 8.3 @ 25 °C), 1 U Ampligase, 2 nM Probe A, 2 nM Probe mB-3, and different amounts of DNA targets or genomic DNA samples. The mixture was firstly heated to 95 °C for 3 min and then incubated at 60 °C for 30 min. After the ligation reaction, the ligation products were immediately placed on ice.A volume of 2 μL of the ligation product was employed for subsequent LAMP amplification. The LAMP reaction was proceeded in a 10 μL mixture including 0.8 μM forward primer (FP), 0.8 μM backward primer (BP), 0.2 mM dNTPs, 1 M betaine, SYBR Green I, 4 U Bst DNA polymerase large fragment and ThermoPol Reaction Buffer (20 mM Tris–HCl, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100, pH 8.8 @ 25 °C). Then the final mixture solution was immediately put into the StepOne Real-Time PCR System to perform the LAMP amplification at 60 °C and the real-time fluorescence intensity was simultaneously monitored at intervals of one minute.
Results and discussion
Principle of the proposed method for genotyping assay
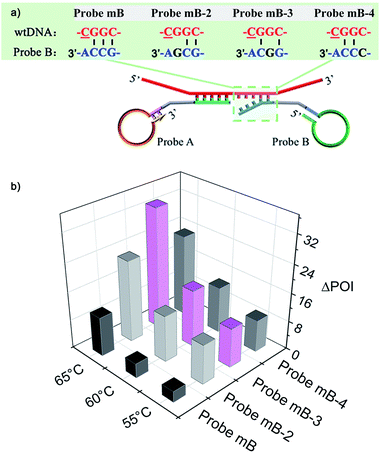
The mechanism of the novel AML-LAMP assay is schematically illustrated in Fig. 1. Since mutations in the p53 tumor-suppressor gene are found at high frequency in a wide range of human cancers, the SNV site located in exon 8 of p53 gene (Arg282Trp) is selected as the model gene in this study, in which base C in the wild-type (wtDNA) is replaced with base T in the mutant type (mutDNA).3 Probe A and Probe mB-3 are firstly designed. To obtain high specificity for the detecting of SNV, on the one hand, the melting temperatures between the probes and mutDNA should be similar. On the other hand, the bases at 5′ end of Probe A and 3′ end of Probe mB-3 are designed as the anti-target sequences complementary to mutDNA at the SNV site, and the 3′ terminus base of Probe mB-3 is A complementary to mutDNA (T) at the SNV site. Probe A is modified with a phosphate group at its 5′-terminus. Moreover, the artificial mismatched base (G) is introduced at the third position from the 3′-terminus of Probe mB-3. The stem-loop DNA sequences in Probe A and Probe mB-3 can be used to perform the LAMP amplification with a pair of primers (FP and BP). In the presence of mutDNA target, Probe A and Probe mB-3 will adjacently hybridize with mutDNA and subsequently be ligated by Ampligase to form a double stem-loop DNA, which is the starting material for LAMP (Fig. 1A). Due to the mismatch between wtDNA and Probe mB-3 by synergy of terminal base and artificial mismatched base, three overhanging bases are generated at the Probe mB-3 3′-terminus and these overhanging bases can effectively prevent the ligation of wtDNA as the template, thus no subsequent amplification occurs.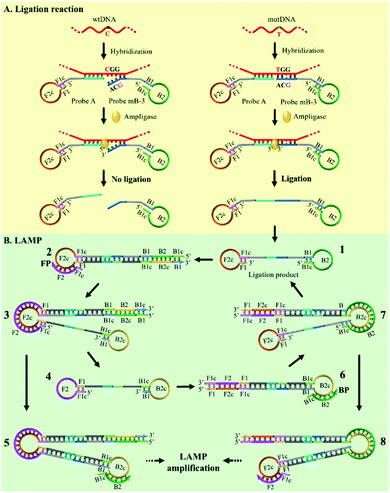 | ||
| Fig. 1 Schematic representation of the AML-LAMP for genotyping assay. (A) Ligation reaction with the template of mutDNA and wtDNA. (B) LAMP amplification with the ligation products of mutDNA. | ||
The ligation product of double stem-loop DNA (structure 1, Fig. 1B) is the essential starting material for LAMP. The rapid auto-cycling strand displacement amplification is carried out using Bst DNA polymerase, FP and BP. As demonstrated in Fig. 1B, self-primed DNA synthesis of structure 1 is performed from the 3′ end to form a single stem-loop structure (structure 2). The FP containing the F1c and F2 sequences (c denotes the complementary sequence) is annealed to structure 2 and then elongates along the upper strand to produce a stem-loop DNA at its 3′-terminus (structure 3). Meanwhile, the self-primed extension can take place to generate a stem-loop structure with a longer stem dsDNA (structure 5) and to release another double stem-loop DNA (structure 4). With the 3′-end self-extension of structure 4 and the participation of BP in hybridization with the single-strand loop (where BP is made up of B1c and B2), another round of extension and displacement can be initiated, resulting in structures 8 and 1. As a result, the recycling amplification of structures 1 and 4 or of structures 5 or 8 will be established and this allows the rapid exponential DNA amplification to occur at a constant temperature. The real-time amplifications are monitored by fluorescence curves using dsDNA-specific SYBR Green I dye.
Critical roles of the modified ligation probe in discriminating single base variation
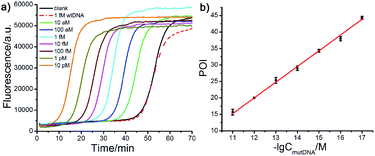
The performance of the modified Probe B containing artificial mismatched bases was firstly investigated. A series of artificial mismatched bases were rationally introduced into different positions of the 3′-terminal region of Probe mB, which were defined as Probe mB-2, Probe mB-3 and Probe mB-4 according to the position of mismatched base from their 3′ terminus. The hybridization between modified Probe B and wtDNA could generate several overhanging bases at the probe 3′-end due to the synergy of terminal base and artificial mismatched base, and these overhanging bases could prevent the ligation of wtDNA as the template.The terminal overhanging bases between modified Probe B and wtDNA were highlighted in Fig. 2a. The ligation temperature could affect the specificity of the single-base discrimination, thus mutDNA and wtDNA assay using the different probes (Probe mB, mB-2, mB-3 and mB-4) were carried out at different temperatures (55 °C, 60 °C and 65 °C). Various concentrations of mutDNA (10 aM, 100 aM and 1 fM) and 1 fM wtDNA were simultaneously detected. The effects of the ligation temperature and modified Probe B on single-base discrimination were displayed as a three-dimensional histogram. As depicted in Fig. 2b, with increasing ligation temperature, the difference between the point of inflection (ΔPOI, POI means the time at which the slope of the fluorescence curve of the target DNA reaches the maximum value) of 1 fM mutDNA and that of wtDNA for all of the Probe B increased. Nevertheless, 10 aM mutDNA could not be accurately detected by modified Probe mB-2, mB-3 and mB-4, which should be attributed to their low hybridization efficiency at high temperature, causing the loss of their analysis sensitivity. Considering the specificity and sensitivity, the optimal ligation temperature of 60 °C and Probe mB-3 were selected for SNP genotyping assay, in which the best position in the modified probe for such artificial mismatched bases was the third position from the probe 3′-terminus.
Sensitivity and detection range of the proposed assay
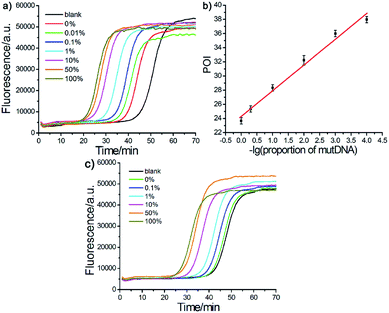
Other important parameters affecting the performance of the AML-LAMP strategy, including the amount of FP and BP and the concentration of Probe A and Probe mB-3, were optimized (see Fig. S1 and S2, ESI†). The sensitivity and detection range of the proposed method in detecting mutDNA were evaluated under the optimal conditions. As displayed in Fig. 3a, well-defined real-time fluorescence curves at different mutDNA concentrations ranging from 10 aM to 10 pM were obtained. With increasing concentrations of mutDNA, the time of the corresponding POI value gradually decreased. When the POI values were plotted against the negative logarithm (−lg) of mutDNA concentrations, as shown in Fig. 3b, an excellent linear relationship was obtained in the range of 10 aM to 10 pM. The linear correlation equation was POI = −37.27 − 4.77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CmutDNA/M with the correlation coefficient R2 = 0.9995. These results indicated the ultrahigh sensitivity of the proposed strategy that could clearly distinguish as low as 10 aM (∼60 copies) mutDNA from the blank control, indicating that the artificial mismatched base in Probe mB-3 had no effect on the analytical sensitivity that did not lead to the reduction of sensitivity. Furthermore, the fluorescence response aroused by 1 fM wtDNA almost overlapped with that of the blank control, further demonstrating the high selectivity of the proposed method for mutDNA analysis.
CmutDNA/M with the correlation coefficient R2 = 0.9995. These results indicated the ultrahigh sensitivity of the proposed strategy that could clearly distinguish as low as 10 aM (∼60 copies) mutDNA from the blank control, indicating that the artificial mismatched base in Probe mB-3 had no effect on the analytical sensitivity that did not lead to the reduction of sensitivity. Furthermore, the fluorescence response aroused by 1 fM wtDNA almost overlapped with that of the blank control, further demonstrating the high selectivity of the proposed method for mutDNA analysis.
Evaluation of specificity of the proposed assay
The effect of the Probe mB-3 with artificial mismatched base on genotyping assay specificity was further investigated. It is worth noting that a practical SNVs assay should have high specificity to accurately detect low levels of mutDNA in complex clinical biosamples. To verify the specificity of the proposed AML-LAMP-based SNVs genotyping assay, mutDNA was mixed with wtDNA at a ratio from 0% to 100%, relative to the total DNA concentration of 100 fM. As depicted in Fig. 4a, as the mutDNA concentration in the mixtures increased, the fluorescence signals more rapidly appeared. The POI values were linearly proportional to −lg of proportion of mutDNA in the range of 0.01–100%. Fascinatingly, one could see from Fig. 4a and b that the AML-LAMP assay could clearly discern as low as 0.01% mutDNA target in a large pool of wtDNA, indicating an ultrahigh specificity at single-base resolution. In addition, as shown in Fig. 4a and c, the specificity of genotyping assay with Probe mB-3 was 10-fold higher than that of by unmodified Probe mB (0.1%). Furthermore, the specificity of the proposed method is also higher than that of other approaches, such as allele-specific PCR, ligase-mediated RCA, SDA, LIEXA and LCR, while is comparable to that of very few ligation-based PCR, which requires TaqMan probe, precise thermal cycling and not suitable for point-of-care (POC) diagnostics. The detailed comparison of the proposed method with other most widely used SNPs assays are summarized in Table S2 (ESI†). Overall, these results indicate that the proposed assay can accurately detect rare variants in a large number of DNA samples, thus can suitably be employed in early diagnosis of cancer and other human diseases.Detection of genomic DNA in blood
Sharing the distinct advantages of high sensitivity and ultrahigh specificity to detect as low as 0.01% mutation frequency, the proposed AML-LAMP method may offer a powerful and practical tool for the accurate quantification of mutated DNA in complex biosamples. To validate its robustness and practicability in detection of real biological samples, the AML-LAMP-based genotyping assay was employed to directly detect genomic DNA extracted from human whole blood samples with mutant probes (Probe A and Probe mB-3) or wild-type probes (Probe A and Probe wB-3). As demonstrated in Fig. S3a,† no effective fluorescence response of genomic DNA incubated with Probe A and Probe mB-3 could be observed by all the patients and healthy people, indicating that mutDNA target could not be detected in the genomic extracts. In contrast, genomic DNA of all the samples could produce the well-defined real-time fluorescence signals by using Probe A and Probe wB-3 (w represented wild-type – the 3′ terminal base in Probe wB-3 was complementary to wtDNA) to perform the assay (Fig. S3b†). Moreover, as little as 100 pg genomic DNA could be obviously detected, and a good linear relationship between the POI values and the −lg of genomic DNA amounts (ranging from 100 pg to 100 ng) was obtained in Fig. S3c and d.† The above results suggested that all the extracted genomic DNA were wild-type, which were also confirmed by DNA sequencing (see Fig. S5†).To further verify its potential clinical applicability, the proposed assay was also applied to detect spiked mutDNA in 10 ng genomic DNA, and as illustrated in Fig. S4,† it could successfully detect mutDNA (at various concentrations) in the vast wtDNA matrixes. These results suggest that the proposed AML-LAMP-based genotyping assay has the potential to be applied for detecting SNVs, particularly for rare variants, in complex biological samples.
Conclusions
In summary, a facile artificial mismatched probe-assisted ligation strategy is proposed for SNVs genotyping assay. The modified ligation probe with artificial mismatched base located in ligation region is employed to prevent the non-specific ligation of wtDNA as the template, thus could effectively improve the specificity of single-base resolution. We have developed the AML-LAMP-based genotyping assay that by combining the artificial mismatched ligation probe with the LAMP reaction for sensitive and ultra-specific genotyping of SNVs. As ligation-assisted approaches have been widely applied in various fields, such as detection of SNPs, DNA and RNA methylation, messenger RNA, microRNA, circular RNA, and even cancer cells, in virtue of the outstanding performance of the artificial mismatched probe, we believe that the proposed assay can provide a useful tool for biomedical research and molecular diagnostics.Compliance with ethical standards
All experiments were performed in accordance with the Guidelines of International Ethics Code of Biomedical Research. Experiments were approved by the ethics committee of The First Affiliated Hospital of Zhengzhou University. Informed consents were obtained from human participants of this study.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21335005), and Research Initiation Fund for Young Teachers of Zhengzhou University.Notes and references
- S. H. Katsanis and N. Katsanis, Nat. Rev. Genet., 2013, 14, 415–426 CrossRef CAS PubMed.
- G. L. Bond, W. Hu, E. E. Bond, H. Robins, S. G. Lutzker, N. C. Arva, J. Bargonetti, F. Bartel, H. Taubert, P. Wuerl, K. Onel, L. Yip, S. J. Hwang, L. C. Strong, G. Lozano and A. J. Levine, Cell, 2004, 119, 591–602 CrossRef CAS PubMed.
- G. L. Bond and A. J. Levine, Oncogene, 2007, 26, 1317–1323 CrossRef CAS PubMed.
- M. S. Lawrence, P. Stojanov, C. H. Mermel, J. T. Robinson, L. A. Garraway, T. R. Golub, M. Meyerson, S. B. Gabriel, E. S. Lander and G. Getz, Nature, 2014, 505, 495–501 CrossRef CAS PubMed.
- S. Kim and A. Misra, Annu. Rev. Biomed. Eng., 2007, 9, 289–320 CrossRef CAS PubMed.
- Y. Mitani, A. Lezhava, Y. Kawai, T. Kikuchi, A. Oguchikatayama, Y. Kogo, M. Itoh, T. Miyagi, H. Takakura and K. Hoshi, Nat. Methods, 2007, 4, 257–262 CrossRef CAS PubMed.
- A. C. Syvänen, Nat. Genet., 2005, 37(suppl. 6), S5–S10 CrossRef PubMed.
- X. Duan, Z. Li, F. He and S. Wang, J. Am. Chem. Soc., 2007, 129, 4154–4155 CrossRef CAS PubMed.
- G. Zhou, M. Kamahori, K. Okano, G. Chuan, K. Harada and H. Kambara, Nucleic Acids Res., 2001, 29, e93 CrossRef CAS PubMed.
- K. L. Gunderson, F. J. Steemers, G. Lee, L. G. Mendoza and M. S. Chee, Nat. Genet., 2005, 37, 549–554 CrossRef CAS PubMed.
- S. Zhang, Y. Cai, J. Zhang, X. Liu, L. He, L. Cheng, K. Hua, W. Hui, J. Zhu, Y. Wan and Y. Cui, Nanoscale, 2020, 12, 10098–10105 RSC.
- Y. Lu, S. Chen, L. Wei, L. Sun, H. Liu and Y. Xu, Anal. Chem., 2019, 91, 6111–6117 CrossRef CAS PubMed.
- M. G. Mohsen, D. Ji and E. T. Kool, Chem. Sci., 2019, 10, 3264–3270 RSC.
- K. Zhang, R. Deng, H. Gao, X. Teng and J. Li, Chem. Soc. Rev., 2020, 49, 1932–1954 RSC.
- C. Shi, S. H. Eshleman, D. Jones, N. Fukushima, L. Hua, A. R. Parker, C. J. Yeo, R. H. Hruban, M. G. Goggins and J. R. Eshleman, Nat. Methods, 2004, 1, 1–7 CrossRef PubMed.
- Z. Chen, L. Miao, Y. Liu, T. Dong, X. Ma, X. Guan, G. Zhou and B. Zou, Chem. Commun., 2017, 53, 12922–12925 RSC.
- J. H. Parka, H. Janga, Y. K. Jung, Y. L. Junga, I. Shin, D. Y. Cho and H. G. Park, Biosens. Bioelectron., 2017, 91, 122–127 CrossRef PubMed.
- K. Zhang, R. Deng, X. Teng, Y. Li, Y. Sun, X. Ren and J. Li, J. Am. Chem. Soc., 2018, 140, 11293–11301 CrossRef CAS PubMed.
- T. D. Harris, P. R. Buzby, H. Babcock, E. Beer, J. Bowers, I. Braslavsky, M. Causey, J. Colonell, J. DiMeo, J. W. Efcavitch, E. Giladi, J. Gill, J. Healy, M. Jarosz, D. Lapen, K. Moulton, S. R. Quake, K. Steinmann, E. Thayer, A. Tyurina, R. Ward, H. Weiss and Z. Xie, Science, 2008, 320, 106–109 CrossRef CAS PubMed.
- Y. Cheng, Q. Du, L. Wang, H. Jia and Z. Li, Anal. Chem., 2012, 84, 3739–3744 CrossRef CAS PubMed.
- Y. Sun, X. Lu, F. Su, L. Wang, C. Liu, X. Duan and Z. Li, Biosens. Bioelectron., 2015, 74, 705–710 CrossRef CAS PubMed.
- H. Wang, W. Liu, Z. Wu, L. Tang, X. Xu, R. Yu and J. Jiang, Anal. Chem., 2011, 83, 1883–1889 CrossRef CAS PubMed.
- H. Zhou, H. Wang, C. Liu, H. Wang, X. Duan and Z. Li, Chem. Commun., 2015, 51, 11556–11559 RSC.
- H. Wang, H. Wang, Y. Sun, X. Liu, Y. Liu, C. Wang, P. Zhang and Z. Li, Talanta, 2020, 212, 120754 CrossRef CAS PubMed.
- H. Tian, Y. Sun, C. Liu, X. Duan, W. Tang and Z. Li, Anal. Chem., 2016, 88, 11384–11389 CrossRef CAS PubMed.
- Z. Hu, F. Xu, G. Sun, S. Zhang and X. Zhang, Chem. Commun., 2020, 56, 5409–5412 RSC.
- G. Wang, W. Tian, X. Liu, W. Ren and C. Liu, Anal. Chem., 2020, 92, 6702–6708 CrossRef CAS PubMed.
- Y. Tang, X. Zhang, L. Tang, R. Yu and J. Jiang, Anal. Chem., 2017, 89, 3445–3451 CrossRef CAS PubMed.
- P. Zhang, N. Guo, K. Gao, F. Su, F. Wang and Z. Li, Org. Biomol. Chem., 2020, 18, 3269–3273 RSC.
- Z. Zhang, D. Yang, W. Tian, Y. Qi, W. Ren, Z. Li and C. Liu, Anal. Chem., 2020, 92, 3477–3482 CrossRef CAS PubMed.
- Y. Wang, Z. Zhang, C. Sepich-Poore, L. Zhang, Y. Xiao and C. He, Angew. Chem., Int. Ed., 2021, 60, 873–880 CrossRef CAS PubMed.
- J. Ding, C. Ma, M. Chen, B. Chen, B. Yuan and Y. Feng, Anal. Chem., 2020, 92, 2612–2619 CrossRef CAS PubMed.
- W. Liu, J. Yan, Z. Zhang, H. Pian, C. Liu and Z. Li, Chem. Sci., 2018, 9, 3354–3359 RSC.
- N. Tomita, Y. Mori, H. Kanda and T. Notomi, Nat. Protoc., 2008, 3, 877–882 CrossRef CAS PubMed.
- Y. S. Jiang, S. Bhadra, B. Li, Y. R. Wu, J. N. Milligan and A. D. Ellington, Anal. Chem., 2015, 87, 3314–3320 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra00851j |
| This journal is © The Royal Society of Chemistry 2021 |