Open Access Article
Open Access ArticleEffects of granular activated carbon and Fe-modified granular activated carbon on anammox process start-up†
Guangsong Lua,
Yunqian Maac,
Lihua Zanga,
Yan Suna,
Fei Yuab and
Rong Xue *a
*a
aSchool of Environmental Science and Engineering, Qilu University of Technology (Shandong Academy of Sciences), Jinan 250353, China. E-mail: xr@qlu.edu.cn; Fax: +86-531-89631680; Tel: +86-531-89631680
bKey Laboratory of Pulp and Paper Science & Technology of Ministry of Education/Shandong Province, Qilu University of Technology, Jinan, 250353, China
cBeijing Key Laboratory of Ionic Liquids Clean Process, State Key Laboratory of Multiphase Complex Systems, Institute of Process Engineering, Chinese Academy of Sciences, Beijing, 100190, China
First published on 12th March 2021
Abstract
In this study, granular activated carbon (GAC) and Fe-modified granular activated carbon (FeGAC) prepared by ultrasonic impregnation method were added into respective up-flow anaerobic sludge blanket (UASB) reactors to explore their effects on the anammox process start-up. The results showed that the time of anammox system start-up could be reduced from 108 d in R1 (control group) to 94 d in R2 (GAC reactor) and to 83 d in R3 (FeGAC reactor). After 120 days of operation, the nitrogen removal rates (NRR) of all reactors could reach more than 0.8 kg-N m−3 d−1. Extracellular polymeric substance (EPS) amount, heme c content and the anammox bacterial functional gene copy numbers gradually increased in all reactors with the passage of culture time, and manifested the superiority in R3 especially. High throughput sequencing revealed that Candidatus Kuenenia was the dominant species in all reactors in the end. It was also demonstrated that FeGAC markedly strengthened the growth and aggregation of anammox bacteria, which is promising for the practical application of the anammox process.
1 Introduction
Nitrogen, which is determined to be the basic element of living organisms, is also responsible for the eutrophication of water bodies.1 The anaerobic ammonia oxidation (anammox) process applied to treat high-strength nitrogen and low COD content wastewater has widely interested scholars since it was first discovered.2 Ammonium is directly converted to N2 by anammox bacteria under anoxic conditions with nitrite as the terminal electron acceptor.3 Compared with the traditional nitrification-denitrification system, the anammox process has the advantages of no demand for organic carbon source, less aeration and lower sludge output.4 So far, more than 200 full-scale anammox-based wastewater treatment plants (WWTPs) have been established, and the number is still increasing.5However, the anammox bacteria in activated sludge will not show better performance in nitrogen removal until in high abundance.6 As a member of phylum Planctomycete, anammox bacteria are strictly chemolithoautotrophic and their growth rates are extremely slow, with the doubling time of 10–14 days.7,8 The characteristics of anammox bacteria, such as low cell yield and sensitivity to environmental conditions, have seriously hindered the practical application. In recent years, numerous researchers tried various strategies to shorten the start-up time of the anammox process and to improve the nitrogen removal loading capacity of biological wastewater treatment systems. Some scholars enhanced the activity of anammox bacteria by adding chemical additives, including metal ions,9 metal oxides,10 reduced graphene oxide11 and sodium glutamate,12 etc. These substances could accelerate catalysis of the key enzyme in anammox bacteria or promote cell division. On the other hand, different kinds of seed sludge have significant impacts on the anammox process start-up. Previous studies have reported that reactors inoculated with small amount of anammox sludge were more likely to achieve good performance in a short time.13 Compared with flocculent sludge, anammox granular sludge has better settleability, higher nitrogen removal rates and better resistance to loading shocks, and the high cell density of the anammox granular sludge strengthens mutual information exchange and cooperation.14,15 Therefore, anammox sludge granulation is common in the anammox process start-up. But complete granulation is difficult to achieve due to the low organic content in wastewater and the slow growth rates of anammox bacteria.
It was reported that the sludge granulation was promoted by adding carrier.16,17 Granular activated carbon (GAC) is considered as a kind of good adsorption material because of its abundant pore structure, large internal surface area and various functional groups.18 In fact, GAC could also be used as the support medium for biofilm growth in biological wastewater treatment.19 Rough and irregular surface of activated carbon provides appropriate microenvironment for bacterial adherence. Li et al.20 reported that initial activated carbon addition accelerated the formation of nitrification sludge granules. Nevertheless, sometimes the potential of functional groups on activated carbon surface obstructs microorganisms to aggregate on their surface. Metal modification could effectively change the physical and chemical properties of activated carbon, and the metal elements in moderate dose have positive effects on bacterial growth. For instance, Zhao et al.21 produced the nickel-doped magnetic carbon material to increase biohydrogen (bioH2) production of anaerobic fermentation. Song et al.22 used magnetic granular activated carbon to enhance electron transfer and methane production in the anaerobic digestion of low-strength wastewater. Qiao et al.23 demonstrated that the proper Fe2+ concentration enhanced anammox bacteria activity. However, there are few research reports on the effects of the combination of metal elements and carbon-based materials on anammox process.
In this study, the primary objectives were to (i) prepare Fe-modified granular activated carbon (FeGAC) and apply it to accelerate the anammox process start-up, (ii) evaluate the role of GAC and FeGAC on the growth of anammox bacteria by several indicators, including nitrogen removal performance of reactors, sludge EPS composition, Heme c contents and microbial community.
2 Materials and methods
2.1 Seed sludge and synthetic wastewater
The seed sludge consisted of 80% conventional activated sludge (sludge 1) obtained from the municipal WWTP (Jinan, China), and 20% anammox granular sludge (sludge 2) derived from an up-flow laboratory-scale reactor in operation for almost a year. The volatile suspended solids (VSS) concentration of concentrated sludge 1 and sludge 2 were 18.80 and 24.52 g L−1, respectively. The suspended solids (SS) concentration of the mixed sludge was about 8000 mg L−1, and the VSS concentration was about 6150 mg L−1 in all the reactors.Synthetic wastewater mainly consisted of ammonium, nitrite and inorganic carbon source. NH4+–N and NO2−–N in the synthetic medium were provided by NH4Cl and NaNO2, respectively. The composition of minerals and trace elements was as described by Van De Graaf et al.,24 as shown in Table S1.† The medium was purged with N2 to eliminate the interference of dissolved oxygen (DO) in water and the DO was controlled below 0.5 mg L−1. The pH range of influent was controlled between 7.0–7.5, adjusted by HCl (1mol L−1) and NaOH (1mol L−1).
2.2 Preparation of FeGAC
The impregnation process with ultrasonic vibration was used to prepare FeGAC in this study. The detailed steps were as follows: (1) a certain amount of GAC (20 mesh, Tian Jin Kermel Chemical Reagents CO., LTD China) was soaked in deionized water for 24 hours, and then repeatedly rinsed with deionized water to eliminate impurities until the washed-water had become clear. (2) The cleaned GAC was dried in an electric blast oven at 105 °C for 2 hours, then cooled to room temperature in a desiccator. (3) 90 g of dried GAC and 150 mL of reagent solution containing 0.01 mol L−1 of Fe2+ (FeSO4·7H2O) and 0.02 mol L−1 Fe3+ (FeCl3·6H2O), were sealed in a conical flask. (4) The flask with GAC and reagent solution was vibrated in ultrasonator for 1 h at frequency of 40 kHz, and after that the pH of the reagent solution was adjusted to 12 with 1 mol L−1 NaOH solution. (5) The mixture solution was stirred at a constant speed of 100 rpm for 24 hours in constant-temperature oscillation incubator at 25 °C. (6) The FeGAC was obtained by being dried and cooled after washed to neutral with deionized water.2.3 Reactors and experimental procedure
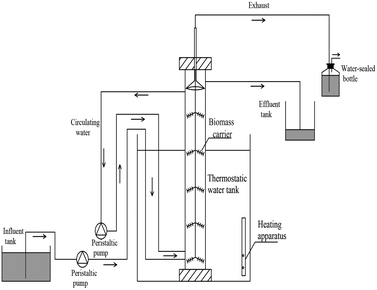
Three identical UASB reactors (Fig. 1) labeled R1, R2 and R3 were applied for anammox start-up, made of polymethyl methacrylate, with total volume of 2 L (working volume 1.8 L), inner diameter of 7 cm and height of 50 cm. In order to prevent sludge loss, the reactors were simultaneously equipped with non-woven fabrics as biomass carrier. The mixed sludge was washed for 3 times with phosphate buffer (0.13 M NaCl, 10 mM Na2HPO4 and 2.7 mM KCl) before inoculation to eliminate the effect of nitrogen background value. The virgin GAC (20 g) and FeGAC (20 g) were added into R2 and R3, respectively. R1, only with seed sludge, was control group. In the earlier stage of starting up, the NH4+–N and NO2−–N concentrations of influent were set at about 50.0 and 65.0 mg L−1 and the hydraulic retention time (HRT) was controlled at 24 h. The reactors were placed in a water bath incubator with black cloth to be in dark environment, and at about 35 ± 2 °C.2.4 Analytical methods
In order to identify the crystalline structures of GAC and FeGAC, X-ray diffraction (XRD) patterns were recorded by using X-ray diffractometer (Ultima IV, Japan). By a scanning electron microscopy (SEM, JSM-7800F, Japan) equipped with energy dispersive spectroscopy (EDS, INCA X-MAX50, Oxford, UK), the surface morphology and chemical composition of the samples were obtained. Fourier transform infrared spectrometer (FTIR, IRAffinity-1S SHIMADZU, Japan) was used to qualitatively analyze the surface functional groups of the samples. The specific surface area and average pore diameter of the particles were evaluated on a Brunauer–Emmett–Teller (BET) analyzer (Autosorb-iQ, Quantachrome, USA). The zeta potential of samples at different pH were determined by a zeta potential tester (Malvern Zetasizer Nano ZS 90, UK). Magnetic property was measured using a vibrating sample magnetometer (VSM, PPMS, SQUID). The XPS of the FeGAC was conducted on a photoelectron spectrometer (Thermo ESCALAB 250XI, USA) using Al Kα radiation (hv = 1486.6 eV) as an excitation source.Influent and effluent were sampled periodically from all reactors, and NH4+–N, NO2−–N, NO3−–N, SS, VSS and pH levels were measured according to standard methods.25 The nitrogen loading rates (NLR), nitrogen removal rates (NRR) and nitrogen removal efficiency (NRE) were calculated according to the following formulas:
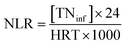 | (1) |
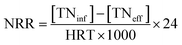 | (2) |
| NRE = NRR/NLR | (3) |
EPS of the sludge samples were extracted by the formaldehyde–NaOH method.26 The protein content of the main components in EPS was measured by the Coomassie Brilliant Blue method with bovine serum albumin used as the standard.27 The carbohydrate content of the main components in EPS was measured by the anthrone method with glucose used as the standard.28 Excitation-emission matrix (EEM) spectra of extracted EPS were evaluated by a fluorescence spectrophotometer (F-7000, Hitachi Co., Japan) at room temperature. The heme c content was calculated using the pyridine heme spectrophotometric method, based on the millimolar extinction coefficient of 23.97 for the difference in absorption between peak at 550 nm and the trough at 535 nm.29,30
2.5 Quantitative PCR (qPCR) and microbial community analysis
Primer pairs Hzocl2aF1 and Hzocl2aR1 were applied to target the anammox bacterial functional gene.31 The extraction of plasmid DNA and construction of standard curve was implemented as described in a published research.32 The amplification system was performed in 20 μL reaction substrates, which consisted of 10 μL SuperReal Color PreMix (TianGen Biotech (Beijing) CO., LTD, China), 0.6 μL each primer, 1 μL DNA template and 7.8 μL ddH2O. The accuracy of the results was tested by the melting curve.The microbial community composition in all reactors was examined with high-throughput pyrosequencing. The extraction of genomic DNA was carried out using M5635-02 Mag-Bind Soil DNA Kit (Omega Bio-Tek, USA) according to the manufacturer's manual. With regard to the PCR amplification, upstream primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and downstream primers 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used to amplify the V3 and V4 regions. Deep sequencing was implemented by Illumina Miseq platform.
2.6 Statistical analysis
The characteristics of anammox sludge were tested in triplicate. Statistical Package (SPSS 19.0, USA) was applied for statistical analysis. Analysis of variance (ANOVA) was carried out to assess the significance of experimental results, and p < 0.05 was considered to be statistically significant.3 Results and discussion
3.1 Characterization of GAC and FeGAC
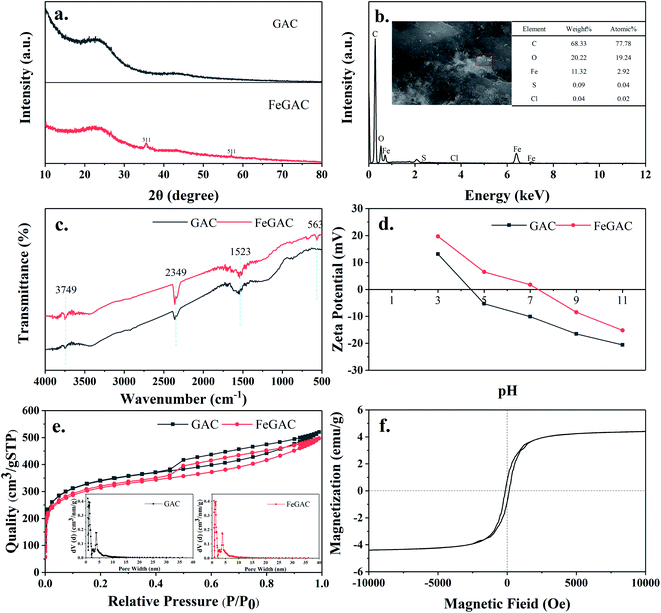
The mineral phases and crystalline structure of the GAC and FeGAC were revealed by XRD (Fig. 2a). The pattern demonstrated that GAC and FeGAC had the typical characteristics of the fluffy structured activated carbon, with a wide peak at 2θ = 26.68°, assigned to the (002) plane of the partially graphitized carbon. The reason may be that GAC and FeGAC are composed of a large amount of amorphous carbon.33 Besides, two sharp diffraction peaks on the FeGAC spectrum were observed at 35.50° and 56.98°, which correspond well to the 311 and 511 crystal planes in the Fe3O4 cubic crystal, as reported in the JCPDS card (no. 03-065-3107). It indicated that iron was successfully loaded on GAC in the form of Fe3O4 cubic crystal.As shown in Fig. 2b, the information about the elemental components of FeGAC was obtained from EDS analysis. The results demonstrated that the surface of FeGAC mainly consists of C, O, Fe, S and Cl elements, and their atomic proportion was 77.78%, 19.24%, 2.92%, 0.04% and 0.02%, respectively.
The FTIR spectra of GAC and FeGAC are shown in Fig. 2c. Two infrared spectra revealed their similar structure. A band center at 3749 cm−1 was related to the O–H stretching vibration modes of the hydroxyl group. The peaks at 1523 cm−1 and 2349 cm−1 were ascribed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C bonds stretching modes, respectively. This result was as same as previous study on carbon materials.34 However, some variations occurred in FeGAC after the modification process. The FTIR spectrum of the FeGAC exhibited a small absorption peak at 563 cm−1, corresponding to stretching vibration of Fe–O group, and this result was in compliance with the XRD result.
C bonds stretching modes, respectively. This result was as same as previous study on carbon materials.34 However, some variations occurred in FeGAC after the modification process. The FTIR spectrum of the FeGAC exhibited a small absorption peak at 563 cm−1, corresponding to stretching vibration of Fe–O group, and this result was in compliance with the XRD result.
The zeta potential means the potential difference between the electric double layer of particles and the layer of dispersing agent around them at the slipping plane.35 Fig. 2d shows the zeta potential of GAC and FeGAC in different pH solutions. Both GAC and FeGAC have positive zeta potential in strong acid environment, and negative zeta potential in alkaline solution. The pH value at which the net surface charge of the samples is zero is regarded as the point of zero charge (pHPZC). The pHPZC of the GAC sample was 4.4, while the pHPZC of the FeGAC sample increased to 7.3. It may be due to the fact that the adsorption sites on the surface of the activated carbon were occupied by metal ions. In addition, the zeta potentials of GAC and FeGAC are −10.1 mV and 1.8 mV at pH 7.0, respectively. Since the EPS secreted by microorganisms contains electronegative groups such as hydroxyl, carboxyl and azylmoieties, the slightly positive potential of FeGAC at pH 7.0 facilitates the adhesion of bacteria to reduce the loss of microorganisms.36,37
N2 adsorption–desorption isotherms and pore size distribution of FeGAC were measured, as depicted in Fig. 2e. According to the International Union of Pure and Applied Chemistry classification of adsorption isotherms, the isotherm for FeGAC exhibit type IV isotherms with a type H4 hysteresis loop, which is typical of mesoporous structure. When P/P0 was in the range of 0–0.1, the FeGAC isotherm showed uptake and the FeGAC had microporous structural properties in pore size mainly distributed between 0.7 and 5.0 nm. The BET surface area value of the GAC was 941.19 m2 g−1, which was higher than that of the FeGAC (844.58 m2 g−1). This was ascribed to the iron oxide was attached to the GAC surface and plugged pore channels.
To evaluate the magnetic properties of FeGAC, the hysteresis loop was carried out at room temperature between 10 and −10 kOe (Fig. 2f). Good ferromagnetic property of the FeGAC was testified by saturation magnetization (4.40 emu g−1), remanence (0.79 emu g−1) and coercivity (139.86 Oe). This magnetic property can produce magnetic field, which benefits the growth of microorganisms.38
The O1s XPS spectrums of GAC and FeGAC are displayed in Fig S1.† The peaks at binding energies of 533.3 and 531.5 eV likely represented C–O and –OH, respectively.39 Compared with the spectrum of GAC, a new peak of FeGAC appeared at binding energy of 530.5 eV, which attributed to Fe–O.33 This also indicated the formation of metal oxides.
3.2 The nitrogen removal performance
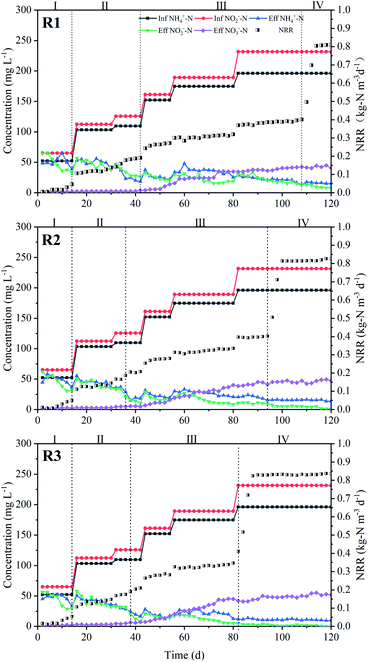
Three UASB reactors (R1, R2 and R3) were operated in parallel and initiated by increasing the influent substrates concentrations stepwise. According to NRR, four start-up phases were designated. The reactors performance and the length of each phase has been shown in Fig. 3 and Table 1.| Reactors | Time | ΔNO2−–N/ΔNH4+–N | ΔNO3−–N/ΔNH4+–N | NRR (kg-N m−3 d−1) | NRE (%) |
|---|---|---|---|---|---|
| a Note: all the data were average values during different periods. “—” indicates that the period contains negative values. | |||||
| R1 | Phase I (day 1–15) | — | — | 0.01 | 16.01 |
| Phase II (day 16–42) | 1.20 | 0.04 | 0.13 | 61.53 | |
| Phase III (day 43–108) | 1.17 | 0.17 | 0.32 | 87.55 | |
| Phase IV (day 109–120) | 1.22 | 0.23 | 0.80 | 94.26 | |
| R2 | Phase I (day 1–14) | — | — | 0.02 | 19.93 |
| Phase II (day 15–36) | 1.23 | 0.05 | 0.14 | 63.20 | |
| Phase III (day 37–94) | 1.18 | 0.16 | 0.34 | 88.81 | |
| Phase IV (day 95–120) | 1.24 | 0.25 | 0.81 | 95.34 | |
| R3 | Phase I (day 1–14) | — | — | 0.02 | 23.28 |
| Phase II (day 15–38) | 1.19 | 0.05 | 0.14 | 65.59 | |
| Phase III (day 39–82) | 1.22 | 0.17 | 0.38 | 89.90 | |
| Phase IV (day 83–120) | 1.25 | 0.26 | 0.83 | 96.96 | |
In the cell lysis phase (phase I), the similar nitrogen removal performance of the three reactors appeared and extremely low ammonium removal was observed. The effluent NH4+–N concentrations were even higher than that of influent. However, the average NO2−–N concentration removal efficiency of R1, R2 and R3 was 26.16%, 30.18% and 32.93%, respectively. This phenomenon might result from the cell lysis of aerobic bacteria, which couldn't adapt to the initial adverse conditions, and released organic nitrogen converted into ammonia.40 At the same time, the released organic matter also provided electron donors for denitrification to slightly decrease the effluent NO2−–N concentration. During this phase, anammox activity exhibited inconspicuously.
After 14 days operation, annamox activity in all reactors recovered in simultaneous decrease of NH4+–N and NO2−–N concentrations. Subsequently, although the effluent NH4+–N and NO2−–N concentrations fluctuated, they were evidently lower than the influent concentrations. The fluctuation indicated that the reactors were in the lag phase (phase II).41 In this phase, the influent NH4+–N and NO2−–N concentrations rose to 103.1 mg L−1 and 112.3 mg L−1, respectively. The NRE reached 61.53%, 63.20% and 65.59%. This phase of R1, R2, and R3 lasted for 27 days (day 16–42), 22 days (day 15–36) and 24 days (day 15–38), respectively.
The progressive enhancement of anammox performance was observed during activity elevation phase (phase III). NH4+–N and NO2−–N removal rate increased rapidly and NO3−–N accumulated apparently. Simultaneously, the three reactors showed significant differences. It took 44 days in R3 when NRR reached about 0.4 kg-N m−3 d−1 from 0.2 kg-N m−3 d−1, while 67 days in R1 and 58 days in R2, respectively. At the end of phase III, the stoichiometric ratios of nitrite consumption versus ammonium and nitrate production versus ammonium consumption were in the range of 1.22–1.25 and 0.23–0.26, which was similar to the theoretical molar ratio of 1.32 and 0.26 for anammox.3 This ratio indicated that anammox process was started up successfully. The entire start-up time could be shortened to 83 d in R3 as compared with 108 d in R1 and 94 d in R2.
During the stationary phase (Phase IV), the influent NH4+–N and NO2−–N concentrations were further increased to 196.2 mg L−1 and 231.7 mg L−1, respectively, and the HRT was gradually shortened to 12 h. High nitrogen removal performance still maintained in R3, with average NRE was 96.98%. The values of R1 and R2 were about 94.26% and 95.34%, slightly lower than that of R3. Meantime, the NRR reached 0.80, 0.81 and 0.83 kg-N m−3 d−1 in R1, R2 and R3, respectively.
During the entire sludge domestication process, the highest NRR from the three reactors was identified and counted in each time, and the weight of highest NRR was 5.00%, 8.33% and 86.67% for R1, R2, and R3, respectively (p < 0.01). This meant that there was a significant advantage in terms of nitrogen removal stability in R3. Besides, the adsorption experiments results (Fig. S2†) shown that GAC or FeGAC did not play a vital role in adsorption of NH4+–N and NO2−–N. Thus, it could be concluded that the biological reactor filled with FeGAC carrier alleviates the influence caused by increasing NLR and shortens the period of anammox process start-up.
3.3 EPS analysis
EPS dispersing on the surface of anammox bacteria plays an important role in the process of sludge aggregation and stability of microbial community structure.10 It was reported that EPS was presumed to bridge bacteria with other bacteria or particulates into aggregate to facilitate the formation of granule sludge.42 Proteins (PN) and polysaccharides (PS) are the major components in the EPS. The proportions of PN and PS has been proposed as an important indicator for the sedimentation performance of the sludge and the lower PN/PS ratio indicated the better granule strength.10 As shown in Fig. 4a, the EPS concentration of the sludge in all the three reactors increased with the domestication time throughout the experiment. When the reactors were operated for 30 days, the EPS contents in R1, R2 and R3 were 53.71 ± 4.17, 61.08 ± 3.36 and 68.36 ± 3.42 mg g−1 VSS, respectively. At the 60th day, the values in R2 and R3 were increased to 109.83 ± 2.25 and 120.43 ± 3.21 mg g−1 VSS, which were higher than the R1 (90.09 ± 4.34 mg g−1 VSS) by 17.9% and 25.2%, respectively (p < 0.01). This may be due to both the effective retention of sludge by GAC and the stimulating effect on the bacteria by iron. After 90 days, the EPS contents of sludge both in R2 and R3 were significantly higher than that in R1 (p < 0.01), but the difference between R2 and R3 was slight (p > 0.05), probably because of the decreasing concentration of iron ions in FeGAC. During the whole operation, PN/PS ratio of sludge in the three reactors varied in the range of 1.75–3.39 and the value in R3 was consistently lower than that in R1 and R2. This indicates that the sludge in R3 has better settling properties.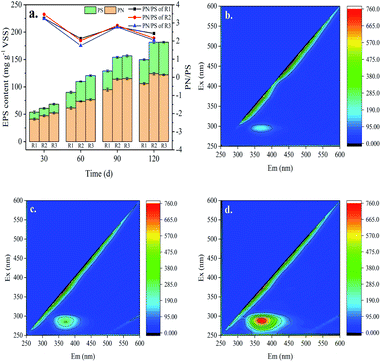 | ||
| Fig. 4 Changes in sludge EPS concentration and chemical composition. (a) Concentration of proteins and polysaccharides; (b–d) EEM fluorescence spectroscopy analysis of R1, R2 and R3. | ||
EEM fluorescence spectroscopy of anammox EPS components in the three reactors was performed after 60 days of incubation. As shown in Fig. 4b–d, only one characteristic peak was identified. According to previous studies, the peak occurred at the excitation/emission (Ex/Em) wavelength of 275–280/350–365 nm was associated with tryptophan protein-like substances.42 The fluorescence intensity of the characteristic peak indicating the PN contents increased in the order of R1 < R2 < R3. These results were consistent with the PN concentration analysis of colorimetric method. Therefore, the addition of FeGAC in the anammox sludge domestication process was conducive to bacteria secreting more EPS, which provided more cation binding sites to promote the adhesion of anammox consortia.
3.4 Heme c
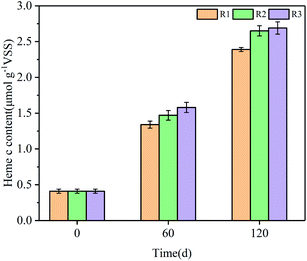
Heme proteins were detected in nitrite reductase (Nir), hydrazine synthase (HZS) and hydrazine oxidase (HAO), which were the three key catalysts involved in the anammox metabolic pathway. Especially, heme c is a pervasive constituent of redox proteins, such as cytohromes c, which serves as electron carriers to perform single electron transfers between the reduced (ferrous) and oxidized (ferric) oxidation state.43,44 Meanwhile, Heme c plays a crucial role in the energy metabolism, cell synthesis and the typical carmine color of anammox sludge.45 And the color was regarded as reflection of the activity of anammox bacteria. The redder color means the higher heme c content and the stronger cell activity. Hence, the concentration of heme c was tracked to investigate the impacts of GAC and FeGAC on the start-up of anammox.The heme c contents in all the reactors were measured on day 0, 60 and 120 (Fig. 5). During the whole operation period, the heme c contents of sludge in the three reactors were all increasing steadily, and the fastest growth rate of that was in R3. The heme c level in R3 reached 1.58 ± 0.07 μmol g−1 VSS at the 60th day. The value was about 1.18 and 1.07 times as much as those in R1 and R2. At the end of experiment, the heme c concentrations of sludge in R1, R2, and R3 were 2.39, 2.65, and 2.69 μmol g−1 VSS, respectively, and increased by 483%, 546% and 556% compared with the initial concentration (p < 0.05). It could be proposed that the more increase of heme c concentrations of sludge in R2 and R3 might be closely associated with the sustained existence of appropriate amounts of GAC and iron in the anammox reactor.
3.5 qPCR results and microbial community analysis
In order to quantify the growth of anammox bacteria, the real-time quantitative PCR (qPCR) experiments were carried out at the operational day 30, 60, 90 and 120. The results were shown in Table 2. After 30 days incubation, the anammox bacterial functional gene copy numbers in R3 was slightly higher than that in R1 and R2. However, there was significant increase in the copy numbers of anammox bacteria functional gene in all rectors after 60 days operation and the numbers for R1, R2 and R3 were 8.67 ± 0.17 × 107, 9.90 ± 0.22 × 107, 1.01 ± 0.10 × 108 copies per g biomass, respectively. In addition, obvious difference between R2 and R3 was not observed after 90 days operation. The trends of the anammox bacteria abundance were all consistent with the changes in EPS during the long-term experiments.| Reactors | Bacteria abundance of different periods (copies per g biomass) | |||
|---|---|---|---|---|
| 30 d | 60 d | 90 d | 120 d | |
| R1 | 3.06 ± 0.18 × 107 | 8.67 ± 0.17 × 107 | 1.82 ± 0.10 × 108 | 4.39 ± 0.09 × 108 |
| R2 | 3.37 ± 0.14 × 107 | 9.90 ± 0.22 × 107 | 2.04 ± 0.10 × 108 | 5.03 ± 0.12 × 108 |
| R3 | 3.54 ± 0.29 × 107 | 1.01 ± 0.10 × 108 | 2.03 ± 0.11 × 108 | 5.12 ± 0.09 × 108 |
The 16S rRNA-based high-throughput sequencing technology was employed to study the microbial community evolutions of the three reactors before and after the experiment. Microbial composition at the phylum level in different reactors could be found in Fig. 6a. Proteobacteria, Planctomycetes, Chloroflexi, Bacteroidetes, Acidobacteria, Patescibacteria, Actinobacteria and Firmicutes, which accounted for more than 95%, were the major microorganisms in all sludge samples. Previous studies reported that nearly all known anammox genera belong to phyla Planctomycetes.46 The relative abundance of Planctomycetes phylum accounted for 11.05% in the seed sludge and rose to 17.61%, 22.85% and 30.58% in R1, R2 and R3, respectively, at the end of the experiment. The results indicate that anammox bacteria were enriched in all reactors. Proteobacteria related to NH4+–N removal also dominated in all the samples, but the relative abundance of it decreased obviously. The third dominant phylum was chloroflexi accounted for 13.46% in seed sludge, 22.98% in R1, 23.41% in R2 and 19.32% in R3, which is universally detected in the biological nitrogen removal systems.
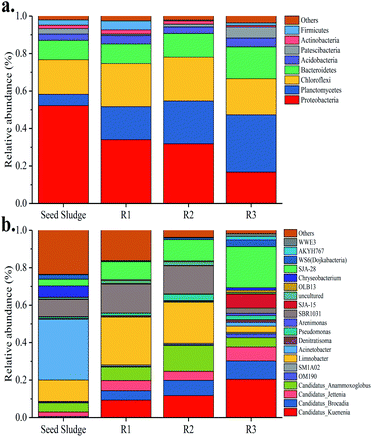 | ||
| Fig. 6 Microbial community structure. (a) Distribution of dominant bacteria in phylum; (b) distribution of dominant bacteria in genus level. | ||
At the genus level (Fig. 6b), twenty dominant genera were found in all sludge samples, containing Candidatus Kuenenia, Candidatus Brocadia, Candidatus Jettenia, Candidatus Anammoxoglobus, SM1A02, OM190, Limnobacter, Acinetobacter, Denitratisoma, Pseudomonas, Arenimonas, SBR1031, SJA-15, uncultured, OLB13, Chryseobacterium, SJA-28, WS6 (Dojkabacteria), AKYH767 and WWE3. Compared with the seed sludge sample, Candidatus Kuenenia, Candidatus Brocadia, Candidatus Jettenia and Candidatus Anammoxoglobus, which were mainly responsible for anammox function, became the dominant species in all the three reactors at the end of experiment. Especially, the relative abundance of Candidatus Kuenenia increased sharply to 9.30%, 11.71% and 20.45% in R1, R2 and R3, respectively. By contrast, there were almost no Candidatus Kuenenia and Candidatus Brocadia in the seed sludge. Furthermore, SM1A02 belonging to the Planctomycete phylum was detected in all reactors and it was considered to be a novel anammox strain.47 Regarding R3, the relative abundance of heterotrophic denitrifying bacteria, such as Denitratisoma, increased gradually and was higher than that of seed sludge and other two reactors. This phenomenon might be related to higher EPS content of the sludge in R3, which promoted the heterotrophic denitrifying bacteria growth. Based on the above discussion, it could be concluded that the addition of GAC and FeGAC changed the microbial community composition.
3.6 Morphological changes of carrier and iron analysis
The typical appearance of GAC before and after modification, and the attachment of microorganisms on the surfaces of different carbon carriers from R2 and R3 were investigated by SEM at the end of experiments. In Fig. 7a, amorphous and fragmented structures and porous textures of GAC could be observed. The surface of the GAC was relatively smooth and flat. Some acicular crystalline particles (Fig. 7b) dispersing evenly on the surface made the surface of FeGAC relatively rough. It indicated that the iron oxides were successfully agglomerated on the GAC. Fig. 7c and d showed the terminal conditions of microorganism attachment on the surface of GAC and FeGAC. It could be observed that the biomass on the surface of FeGAC was more than that of GAC. Because there is the inherence of quorum sensing (QS) in anammox bacteria, microbial aggregation will be apt to occur on the FeGAC. These results confirmed that activated carbon was a suitable carrier and that FeGAC was conducive to bacterial aggregation and adhesion.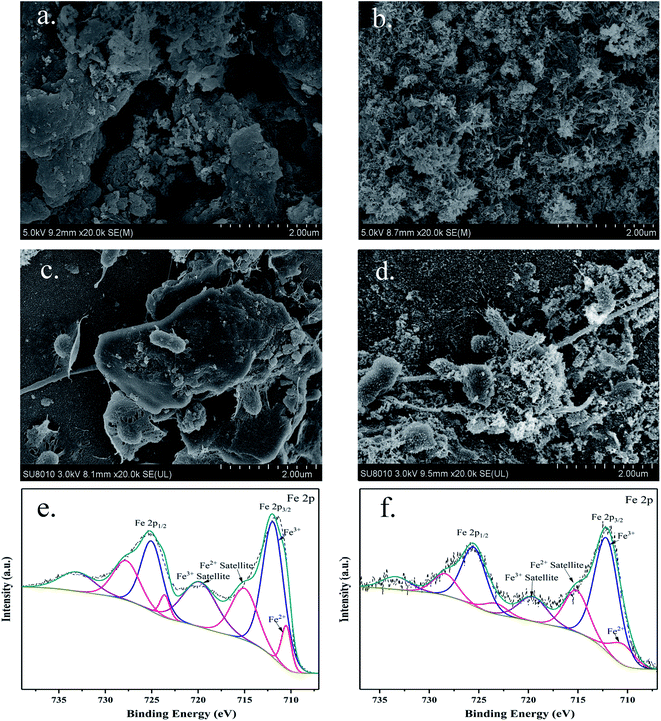 | ||
| Fig. 7 SEM images of (a) granular activated carbon (GAC); (b) Fe-modified granular activated carbon (FeGAC); (c) recovered GAC; (d) recovered FeGAC; and XPS spectra of (e) FeGAC; (f) recovered FeGAC. | ||
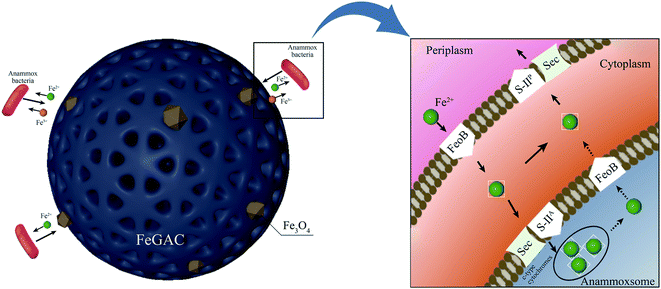
To examine the original and final iron content of FeGAC added into the anammox system, XPS measurements were performed. According to Fig. 7e and f, the Fe 2p peaks of initial FeGAC and recovered FeGAC centered at binding energies of 712.1 and 725.4 eV with two usual shakeup satellites at 715.0 and 719.8 eV, which can be attributed to the Fe3O4.48 The conclusion was consistent with the aforementioned XRD result. Furthermore, compared with the content of iron in initial FeGAC, that in recovered FeGAC decreased from 2.92% to 1.06%. And the content ratio of Fe2+/Fe3+ was 48.88/51.12 in initial FeGAC and 30.26/69.74 in recovered FeGAC. The possible explanation was that Fe2+ or Fe3+ from Fe3O4 crystals dissolved gradually, some of the released substances may be in drainage, while the other was assimilated by microorganisms. The FeGAC served as the supporting carrier and iron provider, providing considerable and favorable sites for microbial aggregation, reducing sludge loss and enhancing bacterial activity. According to previous studies, anammox bacteria are seriously rely on iron-containing proteins contained in the anammoxosome, especially cytochromes c. Anammoxosome is not only responsible for the energy metabolism, but also serves as an iron storage facility for heme-containing enzymes.49 The mechanism for the role of Fe on anammox bacteria was proposed in Fig. 8. The divalent ferrous was transported across the membrane to the cytoplasm via Fe(II)-specific FeoB system, and then made cytochrome c maturation through the Sec-translocon (Sec) and cytochrome c maturation systems S-IIA and S-IIP.50,51 In addition, the presence of iron made the EPS content significantly increasing with the PN/PS decreasing. More EPS could enhance the hydrophobicity of the sludge and promote the sludge to aggregate into granular sludge.52 However, further investigation is needed for the optimal iron load on activated carbon and the detailed mechanism of iron on anammox bacteria.
4 Conclusions
GAC and FeGAC introduced into the different reactors could effectively reduce the anammox process start-up time, and R3 with FeGAC manifested more prominent results. The NRE of anammox systems via GAC and FeGAC assistance reached 95.34% and 96.96% in the stationary phase, respectively, and were higher than that of the control group. Besides, R3 with FeGAC had superior resistance to the shocks by increasing NLR. The changes of EPS amount, heme c content and qPCR results in all reactors were consistent with the nitrogen removal performance. The combination of porous carbon material and Fe facilitated the anammox bacteria proliferation and aggregation.Author contributions
Guangsong Lu: conceptualization, methodology, investigation, writing – original draft; Yunqian Ma: validation, writing – review & editing; Lihua Zang: writing – review & editing, supervision; Yan Sun: investigation; Fei Yu: writing – review & editing, funding acquisition; Rong Xue: conceptualization, writing – review & editing, funding acquisition, supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Natural Science Foundation Projects of Shandong Province in China (ZR2019BD050) and the Foundation of Key Laboratory of Pulp and Paper Science and Technology of Ministry of Education/Shandong Province of China (No. KF201717).References
- Z. Zhao, Y. Cao, Y. Fan, H. Yang, X. Feng, L. Li, H. Zhang, L. Xing and M. Zhao, Water Res., 2019, 156, 297–304 CrossRef CAS PubMed.
- B. Kartal, J. G. Kuenen and M. C. M. Van Loosdrecht, Sci, 2010, 328, 702–703 CrossRef CAS PubMed.
- M. Strous, J. J. Heijnen, J. G. Kuenen and M. S. M. Jetten, Appl. Microbiol. Biotechnol., 1998, 50, 589–596 CrossRef CAS.
- M. Ibrahim, N. Yusof, M. Z. Mohd Yusoff and M. A. Hassan, Desalin. Water Treat., 2016, 57, 13958–13978 CrossRef CAS.
- Y. Cao, M. C. M. van Loosdrecht and G. T. Daigger, Appl. Microbiol. Biotechnol., 2017, 101, 1365–1383 CrossRef CAS PubMed.
- J. Chen, Q. Ji, P. Zheng, T. Chen, C. Wang and Q. Mahmood, Water Res., 2010, 44, 3321–3328 CrossRef CAS PubMed.
- T. Wang, H. Zhang, D. Gao, F. Yang, S. Yang, T. Jiang and G. Zhang, Desalination, 2011, 271, 193–198 CrossRef CAS.
- B. Ma, S. Wang, S. Cao, Y. Miao, F. Jia, R. Du and Y. Peng, Bioresour. Technol., 2016, 200, 981–990 CrossRef CAS PubMed.
- X. Zhang, Z. Chen, Y. Zhou, Y. Ma, C. Ma, Y. Li, Y. Liang and J. Jia, Sci. Total Environ., 2019, 648, 798–804 CrossRef CAS PubMed.
- J. J. Xu, X. L. Zhu, Q. Q. Zhang, Y. F. Cheng, L. Z. J. Xu, Y. H. Zhu, Z. Q. Ji and R. C. Jin, Sci. Total Environ., 2018, 633, 848–856 CrossRef CAS PubMed.
- X. Yin, S. Qiao, J. Zhou and X. Tang, Chem. Eng. J., 2016, 283, 160–166 CrossRef CAS.
- X. Xu, G. hua Liu, D. Zhao, J. Chen, Y. Shao, J. Wang, K. Zhou, Q. Li, Q. Wei and H. Wang, Chemosphere, 2020, 244, 125570 CrossRef CAS PubMed.
- Y. Miao, J. Zhang, Y. Peng and S. Wang, J. Hazard. Mater., 2020, 384, 121325 CrossRef CAS PubMed.
- B. Kartal, W. J. Maalcke, N. M. De Almeida, I. Cirpus, J. Gloerich, W. Geerts, H. J. M. Op Den Camp, H. R. Harhangi, E. M. Janssen-Megens, K. J. Francoijs, H. G. Stunnenberg, J. T. Keltjens, M. S. M. Jetten and M. Strous, Nature, 2011, 479, 127–130 CrossRef CAS PubMed.
- X. Lin and Y. Wang, Water Res., 2017, 120, 22–31 CrossRef CAS PubMed.
- H. A. Ahmad, S. Q. Ni, S. Ahmad, J. Zhang, M. Ali, H. H. Ngo, W. Guo, Z. Tan and Q. Wang, Bioresour. Technol., 2020, 313, 123642 CrossRef CAS PubMed.
- D. chul Shin, J. suk Kim and C. hwi Park, Journal of Water Process Engineering, 2019, 29, 100784 CrossRef.
- H. S. Hsieh and J. J. Pignatello, Appl. Catal., B, 2018, 233, 281–288 CrossRef CAS.
- S. W. Maloney, N. R. Adrian, R. F. Hickey and R. L. Heine, J. Hazard. Mater., 2002, 92, 77–88 CrossRef CAS PubMed.
- A. J. Li, X. Y. Li and H. Q. Yu, Chem. Eng. J., 2013, 218, 253–259 CrossRef CAS.
- W. Zhao, J. Zhang, H. Zhang, M. Yang and L. Zang, J. Cleaner Prod., 2020, 270, 122730 CrossRef CAS.
- X. Song, J. Liu, Q. Jiang, P. Zhang, Y. Shao, W. He and Y. Feng, Chem. Eng. J., 2019, 374, 1344–1352 CrossRef CAS.
- S. Qiao, Z. Bi, J. Zhou, Y. Cheng and J. Zhang, Bioresour. Technol., 2013, 142, 490–497 CrossRef CAS PubMed.
- A. A. Van De Graaf, P. De Bruijn, L. A. Robertson, M. S. M. Jetten and J. G. Kuenen, Microbiology, 1996, 142, 2187–2196 CrossRef CAS.
- APHA (American Public Health Association), Standard Methods for the Examination of Water and Wastewater, Washington DC, USA, 19th edn, 2012 Search PubMed.
- H. Liu and H. H. P. Fang, J. Biotechnol., 2002, 95, 249–256 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- C. Le and D. C. Stuckey, Water Res., 2016, 94, 280–287 CrossRef CAS PubMed.
- E. A. Berry and B. L. Trumpower, Anal. Biochem., 1987, 161, 1–15 CrossRef CAS PubMed.
- H. Ma, Y. Zhang, Y. Xue, Y. Zhang and Y. Y. Li, Sci. Total Environ., 2019, 659, 568–577 CrossRef CAS PubMed.
- M. C. Schmid, A. B. Hooper, M. G. Klotz, D. Woebken, P. Lam, M. M. M. Kuypers, A. Pommerening-Roeser, H. J. M. Op Den Camp and M. S. M. Jetten, Environ. Microbiol., 2008, 10, 3140–3149 CrossRef CAS PubMed.
- X. Xu, G. hua Liu, Q. Fan, J. Chen, Y. Wang, Y. Zhang, Y. Yang, J. Wang, Y. Zhang, H. Jiang, L. Qi and H. Wang, J. Environ. Manage., 2018, 225, 104–111 CrossRef CAS PubMed.
- Z. Feng, H. Chen, H. Li, R. Yuan, F. Wang, Z. Chen and B. Zhou, Sci. Total Environ., 2020, 713, 136423 CrossRef CAS PubMed.
- İ. Demiral, C. Samdan and H. Demiral, Surf. Interfaces, 2020, 22, 100873 CrossRef.
- S. Bhattacharjee, J. Controlled Release, 2016, 235, 337–351 CrossRef CAS PubMed.
- G. P. Sheng, J. Xu, H. W. Luo, W. W. Li, W. H. Li, H. Q. Yu, Z. Xie, S. Q. Wei and F. C. Hu, Water Res., 2013, 47, 607–614 CrossRef CAS PubMed.
- J. Wang, J. Liang, L. Sun, G. Li, H. Temmink and H. H. M. Rijnaarts, Bioresour. Technol., 2020, 309, 123448 CrossRef CAS PubMed.
- H. Li, Z. Chi and B. Yan, Environ. Sci. Pollut. Res., 2018, 25, 29584–29592 CrossRef CAS PubMed.
- S. Feng, W. Yang and Z. Wang, Mater. Sci. Eng., B, 2011, 176, 1509–1512 CrossRef CAS.
- N. Chamchoi and S. Nitisoravut, Chemosphere, 2007, 66, 2225–2232 CrossRef CAS PubMed.
- Z. Bi, S. Qiao, J. Zhou, X. Tang and J. Zhang, Bioresour. Technol., 2014, 170, 506–512 CrossRef CAS PubMed.
- W. Wang, Y. Yan, Y. Zhao, Q. Shi and Y. Wang, Water Res., 2020, 169, 115223 CrossRef CAS PubMed.
- S. E. J. Bowman and K. L. Bren, Nat. Prod. Rep., 2008, 25, 1118–1130 RSC.
- B. Kartal and J. T. Keltjens, Trends Biochem. Sci., 2016, 41, 998–1011 CrossRef CAS PubMed.
- D. Wu, Q. Zhang, W. J. Xia, Z. J. Shi, B. C. Huang, N. S. Fan and R. C. Jin, Chemosphere, 2019, 226, 934–944 CrossRef CAS PubMed.
- A. Gonzalez-Martinez, F. Osorio, J. A. Morillo, A. Rodriguez-Sanchez, J. Gonzalez-Lopez, B. A. Abbas and M. C. M. van Loosdrecht, Biotechnol. Prog., 2015, 31, 1464–1472 CrossRef CAS PubMed.
- F. Xie, X. Ma, B. Zhao, Y. Cui, X. Zhang and X. Yue, Bioresour. Technol., 2020, 297, 122429 CrossRef CAS PubMed.
- R. Zhang, X. Zheng, B. Chen, J. Ma, X. Niu, D. Zhang, Z. Lin, M. Fu and S. Zhou, J. Cleaner Prod., 2020, 256, 120662 CrossRef CAS.
- L. Lv, L. F. Ren, S. Q. Ni, B. Y. Gao and Y. N. Wang, RSC Adv., 2016, 6, 99989–99996 RSC.
- D. Shu, Y. He, H. Yue and S. Yang, RSC Adv., 2016, 6, 68005–68016 RSC.
- C. Ferousi, S. Lindhoud, F. Baymann, B. Kartal, M. S. Jetten and J. Reimann, Curr. Opin. Chem. Biol., 2017, 37, 129–136 CrossRef CAS PubMed.
- S. min Tang, Z. hao Xu, Y. lei Liu, G. feng Yang, J. Mu, R. cun Jin, Q. Yang and X. ling Zhang, Biodegradation, 2020, 31, 223–234 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra00384d |
| This journal is © The Royal Society of Chemistry 2021 |