Open Access Article
Open Access ArticleIsolation of 5′-O-sulfamyladenosine and related 3′-O-β-glucosylated adenosines from the nucleocidin producer Streptomyces calvus†
Xuan Feng,
Davide Bello and
David O'Hagan *
*
School of Chemistry, University of St Andrews, North Haugh, St Andrews, Fife, KY16 9ST, UK. E-mail: do1@st-andrews.ac.uk
First published on 28th January 2021
Abstract
The isolation of three adenosine based metabolites 6–8 from Streptomyces calvus is reported. The metabolites are structurally related to the fluorine containing antibiotic nucleocidin 1 and two recently identified glycosylated fluoroadenosines 2 and 3, however in this case the three metabolites do not contain a fluorine, suggesting that the biosynthetic enzymes to the fluorometabolites also process their non-fluorinated counterparts.
Introduction
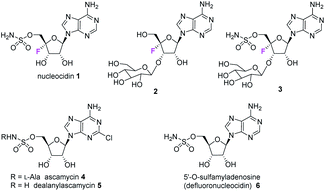
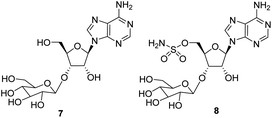
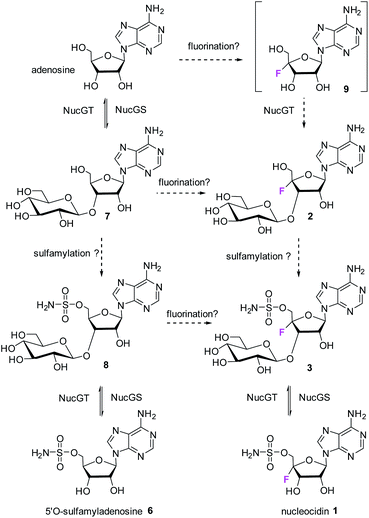
The antibiotic nucleocidin 1 is unusual in two respects.1 Firstly it contains a fluorine atom at C-4′ of the ribose ring and is among the very rare fluorine containing metabolites.2 There has been a recent interest in exploring the biosynthesis of nucleocidin 1 in the soil bacterium Streptomyces calvus,3 and during those investigations the two 4′-fluoro-3′-O-β-glucosylated adenosines 2 and 3 have been identified.4 These are the most recently identified fluorine containing metabolites and they are co-produced in the fermentation immediately prior to the production of nucleocidin 1. Their presence early in the biosynthesis timeline suggests that they are early formed fluorometabolites on the nucleocidin 1 pathway.Secondly, nucleocidin 1 has a sulfamyl ester moiety, which is attached to the O-5′ oxygen. Sulfamyl esters are also an exceedingly rare motif in natural products. There are only a few reported natural products of sulfamyl class5 and the collection is entirely restricted to O-5′-sulfamyladenosine derivatives as illustrated in Fig. 1. Nucleocidin 1 is the most well known,1 and then there are the ascamycins 4 and 5 isolated from Streptomyces sp. JCM9888, which have a chlorine at C-2 of the adenine ring, but no fluorine on the ribose.6 There is one instance of the isolation of O-5′-sulfamyladenosine (defluoronucleocidin) 6 (ref. 7) as a natural product, also from a Streptomyces, however there is an extensive literature associated with the bioactivity of O-5′-sulfamyladenosine 6 coming almost entirely from medicinal chemistry, as it is readily prepared by synthesis.8 Its bioactivity has been widely explored and it is demonstrated for example to be an antiparasitic agent9a–c as well as displaying platelet aggregation activity9d and inhibiting protein biosynthesis9e where it was suggested to interfere with adenosine monophosphate enzymology. In order to gain a better insight into the timing of the fluorination event during nucleocidin 1 biosynthesis, it became of interest to establish if sulfamyladenosine 6 and the defluoro-3′-O-β-glucosylated analogues 7 and 8 of the previously identified 4′-fluoro-metabolites 2 and 3 are also found as natural products in the S. calvus fermentation. The identification of such compounds suggest candidate substrates for the unknown fluorination enzyme in nucleocidin biosynthesis (Fig. 2).
Results and discussion
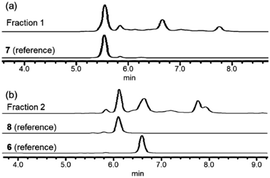
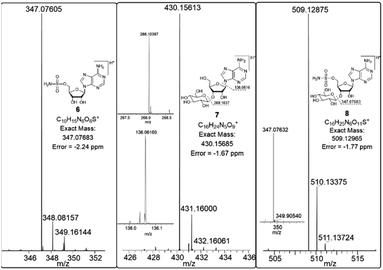
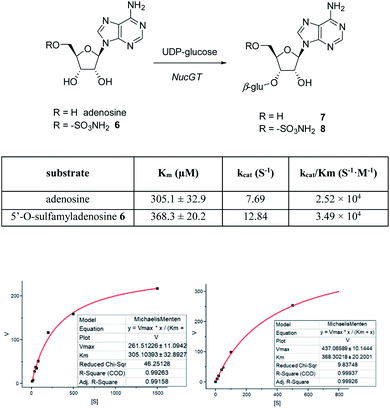
In order to investigate the presence of any non-fluorinated adenosine based metabolites from the nucleocidin producing bacterium, seed cultures of S. calvus were grown in a TSB liquid medium and cells were used to inoculate a defined medium as has been described previously.3 The cultures were then incubated on a rotary shaker at 32 °C for 6–11 days. A crude metabolite extract was recovered, usually around day 8 following the method used to isolate nucleocidin 1 and 4′-fluoro-3′-O-β-glucosylated metabolites 2 and 3. This involved spinning down the cells of the bacterial culture by centrifugation to obtain a supernatant, and then the organics were adsorbed onto activated charcoal, washed with water and then the adsorbed organic residue was released from the charcoal by washing with acetone.3c Concentration and then HPLC guided MS-MS analysis was used to search for metabolites with molecular weights associated with target metabolites 6–8. In the event elution peaks (see Fig. 3) with the correct masses for all three of these metabolites were identified and each was isolated in low milligram amounts by preparative HPLC. High resolution mass spectrometry (HRMS) gave accurate masses (see Fig. 4) and therefore elemental constitutions for 7 [Met I + H]+ = 430.15613 amu; C16H24N5O9+, 8 [Met II + H]+= 509.12875 amu; and C16H25N6O11S+ and 6 [defluoronucleocidin + H]+ = 347.07605 amu; C10H15N6O6S+. Although the titres were low, sufficient material was isolated to be able to record 1H-NMR in each case. The 1H-NMR data for 6–8 are collated in Table 1. O-5′-Sulfamyladenosine 6 is well known8 and a sample was prepared by synthesis for comparison of the 1H-NMR, which demonstrated they were identical compounds. The 1H-NMR spectrum of 7 could be compared with an authentic sample of 7 prepared previously by a de novo synthesis in our lab.4 Additionally reference samples of 7 and 8 were also prepared enzymatically using a novel glucosyl transferase (NucGT) which was reported recently4 as having the ability to form fluorometabolite 3 from nucleocidin 1. This 3′-O-adenosine-β-glucosyl transferase is encoded by the nucGT gene identified within the putative nucleocidin gene cluster in Streptomyces calvus. Over-expression of this gene in E. coli has demonstrated its ability to transfer the β-glucosyl moiety of UDP-glucose to 3′-O-hydroxyl group of adenosines. Accordingly incubation of adenosine and 5′-O-sulfamyl adenosine with NucGT and UDP-glucose generated 7 and 8 respectively, and these products were purified by HPLC and used as reference compounds. They had identical 1H-NMR spectra to those isolated from S. calvus. The ability of NucGT to catalyse these transformations in vitro offers a very obvious rationale as to how they are biosynthesised in vitro by S. calvus. A study of the reaction kinetics for NucGT with adenosine and 5′-O-sulfamyladenosine 6 indicates that the enzyme has a slightly greater affinity (Km) for adenosine, but it is more efficient overall (kcat/Km) with 5′-O-sulfamyladenosine 6 as illustrated in Fig. 5. Given that 5′-O-sulfamyladenosine 6 is known to display a wide spectrum of bioactivity,8,9 the observed 3′-O-β-glycosylation with NucGT and UDP-glucose may offer a detoxification strategy by S. calvus, including tagging for extra cellular transport. A range of natural products including antibiotics, appear also to use glycosylation as an in vivo resistance mechanism, and then where the active component is released extracellularly by the action of glucosidase enzymes.10 A candidate glucosidases (NucGS) has been biochemically validated from S. calvus, with its gene (nucGS) located adjacent to nucGT on the S. clavus genome.4| Assignment | 7 (isolated) | 7 (synthetic) | 8 (isolated) | 8 (enzymatic) | 6 (isolated) | 6 (synthetic) |
|---|---|---|---|---|---|---|
| δH ppm | δH ppm | δH ppm | δH ppm | δH ppm | δH ppm | |
| 2 | 8.34 (s) | 8.33 (s) | 8.24 (s) | 8.22 (s) | 8.23 (s) | 8.27 (s) |
| 8 | 8.44 (s) | 8.44 (s) | 8.24 (s) | 8.21 (s) | 8.23 (s) | 8.28 (s) |
| 1′ | 6.09 (d, 4.8) | 6.09 (d, 4.7) | 6.09 (d, 3.6) | 6.08 (d, 3.6) | 6.09 (d, 4.9) | 6.10 (d, 4.8) |
| 2′ | 4.78 (t, 5.0) | 4.78 (t, 4.9) | 4.81 (m, overlapped) | 4.81 (m, overlapped) | 4.81 (t, 5.0) | 4.79 (t, 5.0) |
| 3′ | 4.51 (t, 5.2) | 4.52 (t, 5.0) | 4.79 (t, 5.1) | 4.80 (t, 5.0) | 4.52 (t, 4.8) | 4.51 (t, 4.9) |
| 4′ | 4.29 (m) | 4.30 (m) | 4.47 (m, overlapped) | 4.46 (m, overlapped) | 4.33 (q, 4.4) | 4.34 (td, 4.7, 3.5) |
| 5′ Ha | 3.75 (m) | 3.75 (d, 4.2) | 4.47 (dd, 4.8, 2.5) | 4.46 (m, overlapped) | 4.41 (dd, 11.0, 4.9) | 4.42 (dd, 11.0, 4.9) |
| 5′ Hb | 3.82 (d, 3.0) | 3.83 (d, 2.9) | 4.51 (dd, 12.5, 5.1) | 4.50 (m) | 4.47 (dd, 11.0, 3.6) | 4.47 (dd, 11.0, 3.5) |
| 1′′ | 4.50 (d, 7.9) | 4.49 (d, 7.9) | 4.64 (d, 7.8) | 4.64 (d, 7.8) | — | — |
| 2′′ | 3.31–3.27 (m) | 3.30 (t, J 8.7) | 3.35–3.45 (m) | 3.35–3.5 (m) | — | — |
| 3′′ | 3.41 (t, J 9.2) | 3.40 (t, 8.9) | — | — | ||
| 4′′ | 3.35–3.32 (m) | 3.33 (t, 9.4) | — | — | ||
| 5′′ | 3.67 (m) | 3.60–3.64 (m) | 3.57 (m) | Overlapped | — | — |
| 6′′ Ha | 3.73 (d, 2.1) | 3.73 (d, 4.1) | 3.69 (dd, 11.7, 4.7) | 3.68 (d, 8.9) | — | — |
| 6′′ Hb | 3.84 (d, 3.2) | 3.84 (d, 2.9) | 3.86 (m) | 3.86 (d, 11.8) |
The average ratios of the fluoro to non-fluoro metabolites 1 to 6, 3 to 8, and 2 to 7 were calculated from the LC-MS ion intensities at their specific molecular weights. Measurements were taken and averaged from the total ion chromatographs (TICs) of up to ten different S. calvus fermentation extracts, harvested between days 6–11. Although there was considerable variation between the relative amounts of the metabolite classes with time, the fluorometabolites dominate by several fold within each metabolite class as summarised in Table 2.
| Compounds | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 6 |
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 7 |
| Ratio | 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Conclusions
The known bioactive O-5′-sulfamyladenosine 6 and the two 4′-defluoro-3′-O-β-glucosylated adenosines 7 and 8 have been identified as natural products from cultures of S. calvus. These non-fluorinated compounds map nucleocidin 1 and the recently identified 4′-fluoroadenosine metabolites 2 and 3 suggesting that the biosynthetic enzymes to the fluorometabolites also process their non-fluorinated counterparts.4The presence of 7 and 8 is also consistent with the in vitro activity of the glucosylation enzyme NucGT. A network of putative biochemical relationships between the non-fluoro and the 4′-fluoro metabolites is illustrated in Fig. 6. The network contains redundancy which may be refined when the specificity of sulfamylation and fluorination enzymes become known.
Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
We thank the Royal Society Wolfson Laboratory Refurbishment Scheme for support, the EPSRC for a Grant (EP/R013799/1) and the University of St Andrews for a Studentship (XF). We also thank Pfizer Ltd., (Groton, USA) for supplying a nucleocidin producing strain of S. calvus T-3018.Notes and references
- (a) S. O. Thomas, V. L. Singleton, J. A. Lowery, R. W. Sharpe, L. M. Pruess, J. N. Porter, J. H. Mowat and N. Bohonos, Antibiot. Annu., 1956, 1956–1957 Search PubMed; (b) I. D. Jenkins, J. P. H. Verheyden and J. G. Moffatt, J. Am. Chem. Soc., 1976, 98, 3346 CrossRef CAS PubMed.
- (a) M. F. Carvalho and R. S. Oliveira, Crit. Rev. Biotechnol., 2017, 37, 880–897 CrossRef CAS PubMed; (b) D. O'Hagan and H. Deng, Chem. Rev., 2015, 115, 634–649 CrossRef PubMed.
- (a) S. Zhang, D. Klementz, J. Zhu, R. Makirtynskyy, A. R. O. Pasternak, S. Günther, D. L. Zechel and A. Bechthold, J. Biotechnol., 2019, 292, 23–31 CrossRef CAS PubMed; (b) X. Feng, N. Al Maharik, A. Bartholomé, J. E. Janso, U. Reilly and D. O'Hagan, Org. Biomol. Chem., 2017, 15, 8006–8008 RSC; (c) A. Bartholomé, J. E. Janso, U. Reilly and D. O'Hagan, Org. Biomol. Chem., 2017, 15, 61–64 RSC; (d) X. M. Zhu, S. Hackl, M. N. Thaker, L. Kalan, C. Weber, D. S. Urgast, E. M. Krupp, A. Brewer, S. Vanner, A. Szawiola, G. Yim, J. Feldmann, A. Bechthold, G. D. Wright and D. L. Zechel, ChemBioChem, 2015, 16, 2498–2506 CrossRef CAS PubMed; (e) L. Kalan, A. Gessner, M. N. Thaker, N. Waglechner, X. M. Zhu, A. Szawiola, A. Bechthold, G. D. Wright and D. L. Zechel, Chem. Biol., 2013, 20, 1214–1224 CrossRef CAS PubMed.
- X. Feng, D. Bello, P. T. Lowe, J. Clark and D. O'Hagan, Chem. Sci., 2019, 10, 9501 RSC.
- P. Mujumdar, S. Bua, C. T. Supuran, T. S. Peat and S.-A. Poulsen, Bioorg. Med. Chem. Lett., 2018, 28, 3009–3013 CrossRef CAS PubMed.
- (a) C. Zhao, J. Qi, W. Tao, L. He, W. Xu, J. Chen and Z. Deng, PLoS One, 2014, 9, e114722 CrossRef PubMed; (b) K. Isono, M. Uramoto, H. Kusakabe, N. Miyata, T. Koyama, M. Ubukata, S. K. Sethi and J. A. McCloskey, J. Antibiot., 1984, 37, 670–672 CrossRef CAS PubMed.
- S. Rengaraju, S. Narayanan, P. L. Ganju, M. A. Amin, M. R. S. Iyengar, T. Sasaki, S. Miyadoh, T. Shomura, M. Sezaki and M. Kojima, Meiji Seika Kenkyu Nenpo, 1986, 25, 49–55 Search PubMed.
- (a) M. L. Bovee, M. A. Pierce and C. S. Francklyn, Biochemistry, 2003, 42, 15102–15113 CrossRef CAS PubMed; (b) D. A. Shuman, M. J. Robins and R. K. Robins, J. Am. Chem. Soc., 1970, 92, 3434–3440 CrossRef CAS PubMed.
- (a) S. Dixit, R. S. Upadhayaya and J. Chattopadhyaya, Org. Biomol. Chem., 2012, 10, 6121 RSC; (b) K. Isono, M. Uramoto, H. Kusakabe, N. Miyata, T. Koyama, M. Ubukata, S. K. Sethi and J. A. McCloskey, Jpn. J. Antibiot., 1984, 37, 670–672 CrossRef CAS PubMed; (c) J. J. Jaffe, J. J. McCormack and E. Meymerian, Exp. Parasitol., 1970, 28, 535–543 CrossRef CAS PubMed; (d) G. R. Gough, D. M. Nobbs, J. C. Middleton, F. Penglis-Caredes and M. H. Maguire, J. Med. Chem., 1978, 21(6), 520–525 CrossRef CAS PubMed; (e) A. Bloch and C. Coutsogeorgopoulos, Biochemistry, 1971, 10, 4394–4398 CrossRef CAS PubMed.
- (a) D. M. Liang, J.-H. Liu, H. Wu, B.-B. Wang, H.-J. Zhu and J.-J. Qiao, Chem. Soc. Rev., 2015, 44, 8350–8374 RSC; (b) L. Zhao, N. J. Beyer, S. A. Borisova and H. W. Liu, Biochemistry, 2003, 42, 14794–14804 CrossRef CAS PubMed; (c) C. Vilches, C. Hernandez, C. Mendez and J. A. Salas, J. Bacteriol., 1992, 174, 161–165 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra00235j |
| This journal is © The Royal Society of Chemistry 2021 |