Open Access Article
Open Access ArticleTwo novel potent ACEI peptides isolated from Pinctada fucata meat hydrolysates using in silico analysis: identification, screening and inhibitory mechanisms†
Jiao Liabcd,
Jilei Suabcd,
Min Chenabcd,
Jiao Chenef,
Wenping Dingabcd,
Yanqun Liabcd and
Hao Yin *abc
*abc
aCAS Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China. E-mail: yinhao@scsio.ac.cn
bGuangdong Key Laboratory of Marine Materia Medica, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China
cSouthern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou 511458, China
dCollege of Earth and Planetary Sciences, University of Chinese Academy of Sciences, Beijing 100049, China
eAffiliated Hospital of Integrated Traditional Chinese and Western Medicine, Nanjing University of Chinese Medicine, Nanjing 210028, China
fLaboratory of Cellular and Molecular Biology, Jiangsu Province Academy of Traditional Chinese Medicine, Nanjing 210028, China
First published on 25th March 2021
Abstract
The aim of this study was to discover potent angiotensin-converting enzyme (ACE) inhibitory (ACEI) peptides from Pinctada fucata (P. fucata) for treating hypertension and to characterize them using in silico analysis. The P. fucata proteins were hydrolyzed by Alcalase®, a serine endopeptidase with broad selectivity, at various times (0, 2, 4, 6, 8, 10 h). The degree of hydrolysis (DH) and ACEI activity of the different hydrolysates were measured. Considering the molecular weight and ACEI activity, the 10 h hydrolysate was purified by a series of traditional separation methods, including ultrafiltration, gel G-25 chromatography, and reversed-phase high-performance liquid chromatography (RP-HPLC), with ACEI activity as a guide. The results showed two fractions, C17 and C18, eluted by means of semi-preparative RP-HPLC, and showed the highest ACEI activities of 80.33 ± 2.70% and 81.66 ± 0.29%, respectively, at 1 mg mL−1. The two fractions were then identified using liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) and their MS/MS spectra data were subjected to de novo sequencing. Subsequently, the potential ACEI peptides were screened by in silico methods, namely, to analyze the average local confidence (ALC) value obtained from the sequencing software and the P-value from the Pepsite 2. In total, 13 potential ACEI peptide sequences were obtained and identified from the two fractions by LC-ESI-MS/MS, and two novel tetrapeptides, FRVW (607.3314 Da) and LPYY (555.2881 Da), were screened for synthesis according to the in silico analysis. The in vitro ACEI tests indicated that FRVW and LPYY had IC50 values of 18.34 and 116.26 μM, respectively. The Lineweaver–Burk plot showed that FRVW was a noncompetitive inhibitor, and LPYY was shown to be a mixed-mode type inhibitor. A stability study against ACE indicated that both peptides were hydrolyzed by ACE to some extent, the higher ACEI activity following incubation with ACE indicating that they should be classified as pro-drug substrates. Molecular docking results showed that hydrophobic amino acids (HAAs) within peptides formed vital interactions including hydrogen bonds, electrostatic forces, van der Waals forces and Pi–Pi interactions with ACE residues, which stabilized the enzyme–peptide complex. Furthermore, the docking results accorded with the inhibition kinetic mode. Our study demonstrated that FRVW and LPYY isolated from P. fucata have potential applications as antihypertensive agents.
1 Introduction
Hypertension or high blood pressure is a primary factor in the occurrence of strokes, ischemic heart disease, and arteriosclerosis, as well as end-stage renal diseases, and is one of the most prevalent cardiovascular conditions occurring worldwide.1 It is astonishing that the number of deaths in the global population reporting cardiovascular disease as the primary cause of death is approximately 17.5 million per annum, as reported by the World Health Organization (WHO).2 In the management of hypertension, angiotensin-converting enzyme (ACE) plays a vital role in vivo as it acts through the renin–angiotensin system (RAS) and the kallikrein–kinin system (KKS). In the RAS, ACE can catalyze the conversion of angiotensin I to the potent vasopressor angiotensin II, by the cleavage of the C-terminal dipeptide. In the KKS, degradation of vasodilator bradykinin takes place, with the removal of the two C-terminal dipeptides by ACE.3 Therefore, ACE has been a drug target, and its inhibition has been a key technique in developing effective agents for treating high blood pressure. Although clinically prescribed synthetic drugs including captopril, enalapril, and benazepril4 indeed work well, they cause certain adverse side effects, such as cough, insomnia, skin rashes, and fever.5 Thus, there is a trend toward finding safe and effective substitutes for these synthetic compounds. Meeting the above requirements, bioactive peptides (BPs) with important physiological regulatory functions are now regarded as the preferred substances due to their safe and effective characteristics. BPs, however, will not show any potential bioactivity unless they are liberated from their parent protein sequences by various proteases acting at specific cleavage sites. These properties of peptides are affected by the types of proteins used for hydrolysis. In general, the hydrophobic amino acid (HAA) content of proteins has been strongly correlated with the hydrophobic properties of peptides, which may support high bioactivity. In addition, the terminal amino acids of peptides are determined by the protein sequences, along with the selectivity of the enzymes employed in the hydrolysis process. Among the proteases, Alcalase® is considered to be one of the most efficient enzymes, having a broad range of selectivity and specificity.6 Usually, the utilization of Alcalase in protein hydrolysis always produces peptides with a high degree of hydrolysis as it can recognize various amino acids. In addition, the generation of BPs by proteolytic enzymes involves other factors, such as temperature, enzyme–substrate ratio (E/S), pH, and especially the time taken for hydrolysis to occur. An overlong incubation time may cause extensive hydrolysis, in which the generation of large quantities of free amino acids (FAAs) may bring about a decrease in bioactivity. Accordingly, an appropriate hydrolysis time is also critical for obtaining peptides with specific lengths that are suitable for the exertion of physiological function.In recent years, a variety of BPs, obtained from the proteins of animal-derived sources including milk, egg, and collagen and plant-derived foods such as soybean and wheat have been confirmed to have an ACEI function.7–9 Recently, the isolation and identification of ACEI peptides from other novel sources has been receiving increasing attention. Pinctada fucata (P. fucata), the Akoya pearl oyster, a highly economic marine organism found in the waters off the coast of China, possesses abundant proteins, which come not only from its meat but also exist in the pearl shell and play a regulatory role during biomineralization.10–12 Previous studies have reported that the peptides prepared from P. fucata by a series of traditional methods exhibit several potent bioactivities, such as antioxidant and ACEI activity. For example, Wu et al.13 identified an antioxidant peptide from P. fucata muscle with the sequence GAGLPGKRER, which exhibited strong antioxidant activity close to that of vitamin C and butylated hydroxytoluene. Sasaki et al.12 purified the pearl oyster shell protein hydrolysate using a gel Sephadex G-25 column and high-performance liquid chromatography (HPLC), from which the ACEI hexapeptide GVGSPY was identified, which had an IC50 value of 5.82 μg mL−1. The above traditional method, which is the method of separation guided by bioactivity, has frequently been employed in the purification and identification of peptides. However, rapid screening methods for detecting high-activity single peptides in the process of purification still need to be explored. Furthermore, information about the characterization of these identified peptides is still not available.
Current studies of ACEI peptides mainly focus on the investigation of structure–activity relationships and the discovery of peptides with potent bioactivity. Unfortunately, a general law of their structure–activity relationships has not yet been established, and further studies are needed to explore this both experimentally and theoretically. As for the discovery of BPs, previous studies have sought to determine the fraction with the greatest bioactivity, which may consist of multiple co-eluting peptides, by employing a traditional bioassay-guided method. The identification of the most potent single peptide is still a problem. To determine the most potent single bioactive peptide, the synthesis of all of the peptides is costly, time-consuming, and labor-intensive.14 In order to circumvent this, in silico methods have been applied to screen ACEI peptides. Until now, the development of bio-information, especially the establishment of certain databases, has made the prediction and discovery of BPs a lot more convenient. However, there are still some drawbacks to researches that rely on in silico methods. For example, the online databases, including BIOPEP and PeptideCutter, frequently used by researchers to predict BPs, generate different peptide sequences using virtual enzymatic hydrolysis compared with the actual sequence obtained by experimental hydrolysis as a result of several interference factors, including protein spatial structure, pH, and temperature.15 Thus, the traditional isolation and purification of peptides obtained experimentally by hydrolysis, combined with in silico screening, increase the chances of discovering the most active peptides from protein hydrolysis. Furthermore, with improvements in docking algorithms, molecular docking can be exploited to elucidate the mechanism of interaction between inhibitors and macromolecules.16
In this study, we aimed to discover the most potent ACEI peptides from P. fucata protein hydrolysates with the assistance of bioinformatic methods. Moreover, the mechanism underlying peptide inhibition was investigated by molecular docking in order to accelerate the application of the key peptides derived from P. fucata as functional foods in the management of hypertension.
2 Materials and methods
2.1 Raw materials
P. fucata meat was obtained from a shellfish market in Zhanjiang, Guangdong Province, China in September 2018. The meat was minced with a grinder, followed by defatting with isopropyl alcohol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, w/v) for 3 h. Finally, the meat was dried in air and stored at −20 °C before use.
4, w/v) for 3 h. Finally, the meat was dried in air and stored at −20 °C before use.
2.2 Enzymes and chemicals
Alcalase® (2 × 105 U g−1) was bought from Pangbo Biological Engineering Co. (Nanning, Guangxi, China). Hippuryl-histidyl-leucine (HHL) and ACE (from rabbit lung) were obtained from Sigma Chemical Co. (St. Louis, MO, USA). Ultrafiltration (UF) tubes with a molecular weight cut-off (MWCO) of 3 kDa were purchased from Millipore Co. (Bedford, MA, USA). All other reagents used were either analytical grade or HPLC grade.2.3 Preparation of hydrolysates
The defatted P. fucata meat was hydrolyzed by Alcalase at 50 °C, pH 10.0, a substrate concentration of 8%, and an enzyme/protein substrate ratio of 8000 U g−1 for 0, 2, 4, 6, 8, and 10 h. The hydrolysates were placed in boiling water for 15 min to inactivate the Alcalase, followed by centrifugation at 8000g for 20 min. The supernatant was then filtered through a 0.45 μm membrane. The filtrate, namely, the P. fucata meat hydrolysates (PFMHs), was lyophilized and stored at −20 °C before use.2.4 Degree of hydrolysis
The degree of hydrolysis (DH) was determined using the method of Zhou et al.17 The formula for calculating DH is:| DH = Nα/Nt |
2.5 Determination of amino acid composition
P. fucata meat proteins and their hydrolysates were hydrolyzed using 6 mol L−1 HCl solution at 110 °C for 22 h according to the method of Arise et al.18 An L-8900 Automatic Amino Acid Analyzer (Hitachi High-Tech Corporation, Tokyo, Japan) was employed to determine the amino acid composition. The sample was analyzed for 18 amino acids. The concentrations of the different amino acids in the hydrolysate were expressed as g/100 g.2.6 Purification of ACEI peptides from the hydrolysates
PFMHs were fractioned using ultrafiltration tubes (3 kDa MWCO), and two fractions (>3 kDa and <3 kDa) were obtained. Following the ACEI activity assay, the fraction with the higher activity was dissolved in distilled water at 100 mg mL−1 and loaded onto a Sephadex G-25 gel filtration column (3.5 × 30 cm, GE Ltd., USA), followed by elution using ultrapure water at a flow rate of 1.0 mL min−1 and evaluated at 220 nm. The eluted fractions were collected and lyophilized, and their ACEI activity was subsequently measured. Afterward, the fractions showing the highest activities were pooled and further purified on a YMC-Pack ODS-A C18 semi-preparative column (250 × 10.0 mm) using linear gradient eluting conditions at a flow rate of 2.0 mL min−1. The mobile phase consisted of eluent A (0.1% v/v trifluoroacetic acid (TFA) in ultrapure water) and eluent B (pure methanol). Following the injection of 100 μL of sample solution into the C18 column, the elution gradient was performed as follows: 99% A (0–5 min), 99–39% A (5–65 min), 39% A (65–70 min), 39–99% A (70–75 min) and 99% A (75–80 min). The eluted fractions were monitored at 220 nm and the fractions that showed the strongest ACEI activities were pooled and sequenced.2.7 Identification of purified peptide by LC-ESI-MS/MS
The two fractions (10 μL, 10 mg mL−1) with the highest ACEI activities in the RP-HPLC separation were injected into an Agilent HPLC system connected to a maXis Impact MS instrument (Bruker, Ettlingen, Germany) with an electrospray source. The separation was performed on an Agilent reversed-phase column (4.6 × 150 mm, 5 μm). Eluent A (0.1% v/v formic acid) in ultrapure water and eluent B (pure methanol) were employed to elute the sample solution under a gradient condition from 1% to 61% of solvent B over 65 min at a flow rate of 1.0 mL min−1. Mass spectrometry was conducted in the positive ion mode with m/z range of 200–3000. The parameters were set as follows: dry temperature, 180 °C; electrospray capillary voltage, 4.5 kV; and flow rate of dry gas, 4.0 L min−1. PEAKS Studio 8.5 software (Bioinformatics Solutions Inc., Waterloo, Canada) was employed to analyze the MS/MS data and perform the de novo sequencing. The parent ions and fragment ions were set at 10 ppm and 0.02 Da, respectively. Only the peptides that had high average local confidence (ALC) values (>80%) and good quality MS/MS spectra were considered.2.8 In vitro assay of ACEI activity
ACEI activity was tested based on Lan et al.’s method, with some modifications.19 Firstly, all of the reagents were dissolved in borate buffer (100 mM borate, 300 mM NaCl, pH 8.3). Next, 30 μL of 5 mM HHL (N-hippuryl-His-Leu hydrate) and 10 μL of sample solution were mixed and pre-incubated at 37 °C for 6 min. The blank was also prepared under the same conditions, with the 10 μL of sample solution substituted by 10 μL of borate buffer. Afterward, 30 μL of previously pre-incubated ACE solution (0.1 unit) was added to the blank and sample solutions, following which they were incubated at 37 °C for 30 min. The reactions were terminated by the addition of 60 μL of HCl (0.1 M) solution. The hippuric acid (HA) content was measured using RP-HPLC equipped with an Agilent Eclipse XDB-C18 column (4.6 × 150 mm, 5 μm). An isocratic elution was implemented with 25% acetonitrile (ACN) and 75% Milli-Q water (0.05% TFA) used as the mobile phase at a flow rate of 1 mL min−1 and detected at 228 nm. ACEI activity was calculated using the following equation:| ACEI activity (%) = (A1 − A2)/A1 × 100% |
2.9 In silico evaluation and synthesis of peptides
To perform the in silico analysis, Pepsite 2 (http://pepsite2.russellab.org) was employed for the prediction of peptide binding to the protein surfaces. The results of the interaction were obtained from the statistical significance (P-value), which was considered significant only when it was below 0.05.20 ToxinPred (http://crdd.osdd.net/raghava/toxinpred) was used for predicting the toxicity and hydrophobicity of the peptides. The isoelectric point (pI) and net charge were obtained using the peptide property calculator, available at https://pepcalc.com. The BIOPEP database (http://www.uwm.edu.pl/biochemia/index.php/en/biopep) was used to investigate the previously reported peptides in the identified sequence. Following the method of Zheng et al.,21 the potential ACEI peptides were selected according to the P-value and ALC, which gave the expected percentage of the correct amino acid in the sequence.22 Assisted by the in silico analysis, the key peptides were synthesized by GL Biochem Co., Ltd. (Shanghai, China) using a solid-phase method. The purity and molecular weights of the synthesized peptides were determined by HPLC and LC-MS, respectively. The ACEI activities of the synthesized peptides were assayed according to the above-mentioned method.2.10 ACEI patterns
ACEI patterns were measured based on the method of Wu et al., with some modifications.23 The HHL solutions were prepared at concentrations of 0.5, 1, 2, and 4 mM, and the concentrations of peptides were set as 0, 20, and 40 μg mL−1. The ACEI peptides (10 μL) at various concentrations were mixed with HHL solution (30 μL). The combined solution was incubated for 5 min at 37 °C after ACE (30 μL) was added, following by the addition of 0.1 M HCl solution (60 μL) to terminate the reaction. The content of HA was determined using HPLC monitored at 228 nm. The ACEI pattern was determined using the Lineweaver–Burk plot.2.11 Stability of peptides to ACE
The stability of peptides to ACE was determined using the method of Zarei et al., with modifications.24 The 4 mM peptide solutions were incubated with 150 μL of ACE (0.1 U mL−1) at 37 °C for 0, 0.5, 1, 2 or 3 h. The mixture solutions of different incubation time were taken out and stored at −40 °C until they were analyzed by HPLC. An Agilent C18 column (4.6 × 150 mm, 5 μm) was used to elute the above 20 μL mixed solution loaded at 1 mL min−1. The column was conditioned with 100% eluent A (0.1% TFA in ultrapure water) for 10 min, and a linear gradient was run as follows: eluent B (0.1% TFA in ACN) from 0 to 100% (10–50 min). The absorbance was measured at 215 nm. The 0 and 3 h incubated mixtures were tested for ACEI activity according to the method described in Section 2.8.2.12 Molecular docking simulation
Molecular docking is a simulation method used to elucidate the mechanism of interaction between a protein (receptor) and small molecules (ligands). In this study, we performed the docking between the key peptides and ACE using the Accelrys Discovery Studio software 4.1 (DS 4.1) based on a previously published method.2 The crystallographic structure of ACE (code: 1O8A) was downloaded from the Worldwide Protein Data Bank (https://www.wwpdb.org) and then was conducted with ‘prepare protein’ program to add hydrogen, remove water, and model missing loop regions. The structures of the peptides were generated by ChemOffice 2018 and the energy was minimized with the CHARMM force field. The CDOCKER protocol was used to perform docking studies. The binding sites were defined with the coordinates x: 36.189, y: 43.643 and z: 55.175 and a radius of 16 Å. The CDOCKER energy scores, interaction site, and interaction force types were analyzed to evaluate docking results.2.13 Statistical analysis
One-way analysis of variance (ANOVA) was used to carry out statistical analysis using GraphPad Prism 8.0.1 (GraphPad Software Inc., La Jolla, CA, USA). All assays were performed in triplicate. The values are expressed as the mean ± standard deviation. Significant differences between means were taken into account at P < 0.05.3 Results and discussion
3.1 DH and amino acid composition of the hydrolysates
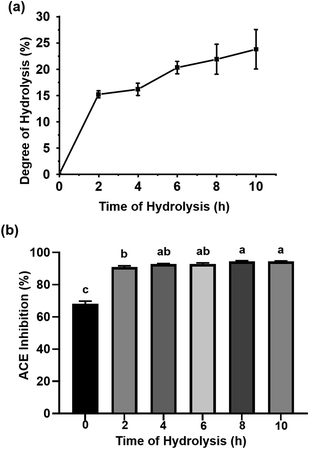
The protein in P. fucata was digested by Alcalase, a kind of serine protease widely used for generating peptides with hydrophobic C-terminal amino acids, which enhance ACEI activity.25 As shown in Fig. 1(a), the DH values during enzymatic hydrolysis showed a rapid increase during the first 2 h, indicating that numerous peptide bonds were broken in this process. Following this, the hydrolysis rate decreased, attributed to the inhibition of the reactive product, a decrease in the number of available hydrolysis sites and peptide bond concentration, and enzyme deactivation. A DH value of 23.77 ± 3.75% was obtained for a hydrolysis time of 10 h. This curve was quite similar to the result obtained by Hanafi et al., who reported the DH versus hydrolysis time results obtained from defatted green soybean meal hydrolysates produced by Alcalase following 10 h of hydrolysis.25 The hydrolysates obtained at different reaction times were assayed for ACEI capacity. As seen in Fig. 1(b), the 8 and 10 h hydrolysates achieved almost the same ACEI activities (94.46 ± 0.38% for the 8 h hydrolysate, 94.42 ± 0.35% for the 10 h hydrolysate), much higher than those of the other hydrolysates at 10 mg mL−1. In general, longer hydrolysis times were accompanied by more low molecular weight peptides, which are readily absorbed in the small intestine; thus, the 10 h hydrolysate was subjected to further study.The amino acid composition of P. fucata meat protein and its hydrolysates after 10 h of hydrolysis is listed in Table 1. It can be clearly seen that both of them had high contents of Asp, Glu, Leu, Lys, and Arg. The HAA contents in the P. fucata meat and its hydrolysates accounted for 39.52% and 39.20%, respectively, of the total amino acids. In addition, their aromatic amino acids (AAAs) accounted for 8.91% and 8.55%, respectively, of the total. The large amount of HAAs and AAAs, which are suited to bind with ACE, suggested that P. fucata could be a potential source for generating ACEI peptides. Furthermore, the high ratio of positive amino acids, including Arg, seemed to benefit the enhancement of ACEI activity as well.
| Amino acid | Meat | Hydrolysates |
|---|---|---|
| a Hydrophobic amino acids (HAAs): alanine, isoleucine, valine, leucine, tyrosine, phenylalanine, tryptophan, proline, cysteine and methionine. Positively charged amino acids (PCAAs): arginine, lysine, histidine. Aromatic amino acids (AAAs): phenylalanine, tyrosine and tryptophan. | ||
| Asp | 5.87 | 6.78 |
| Thr | 2.77 | 3.24 |
| Ser | 2.64 | 2.99 |
| Glu | 8.11 | 9.88 |
| Pro | 1.71 | 1.96 |
| Gly | 3.56 | 4.18 |
| Ala | 3.08 | 3.68 |
| Val | 2.67 | 3.11 |
| Met | 1.36 | 1.61 |
| Ile | 2.40 | 2.80 |
| Leu | 4.08 | 4.73 |
| Tyr | 1.92 | 2.13 |
| Phe | 2.36 | 2.58 |
| Lys | 4.71 | 5.23 |
| His | 1.06 | 1.17 |
| Arg | 4.07 | 4.45 |
| Cys | 1.30 | 1.23 |
| Trp | 0.55 | 0.62 |
| HAAs | 21.43 | 24.45 |
| PCAAs | 9.84 | 10.85 |
| AAAs | 4.83 | 5.33 |
3.2 Purification of peptides from P. fucata protein hydrolysates
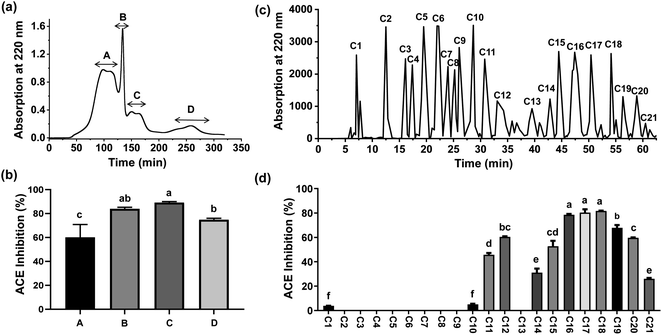
The hydrolysates were separated using 3 kDa MWCO UF tubes, which have previously been employed for preliminary isolation of bioactive peptides. Two fractions, designated PFMHs-I (MW > 3 kDa) and PFMHs-II (MW < 3 kDa), were obtained and their ACEI activities were assayed. Since PFMHs-II exhibited significantly higher ACE inhibition (88.71 ± 0.58%) than PFMHs-I (81.52 ± 1.40%) at 5 mg mL−1, it was further purified on a Sephadex G-25 column. Four peak fractions (A, B, C, and D) eluted from the Sephadex G-25 column were collected separately (Fig. 2(a)) and then subjected to an ACE inhibition test. As shown in Fig. 2(b), fraction C showed the greatest ACE inhibition rate (89.13 ± 0.84%) compared with the other fractions at the same concentration of 2 mg mL−1; thus, it was injected into the HPLC column using a semi-preparative ODS column for further separation. A total of 21 peak fractions (C1–C21) were collected based on the chromatography profile (Fig. 2(c)). Fig. 2(d) shows the ACEI capacity of each eluted fraction. It can clearly be seen that C18 exhibited the highest ACE inhibition rate (81.66 ± 0.29%), followed by C17 (80.33 ± 2.70%), at a concentration of 1 mg mL−1. Hence, they were subjected to sequencing in order to discover which peptides were responsible for ACE inhibition.3.3 Identification and in silico analysis of peptides
The two fractions C17 and C18 with the largest ACEI rates were identified by LC-ESI-MS/MS. The results listed in Table 2 show that, in total, 13 peptides were identified from fractions C17 and C18. In addition, their characteristics are also listed. The ACEI peptides from P. fucata with 10 h of hydrolysis contained four to six amino acid residues. The net charge on the peptides was forecast to be within the range −2 to 1; the isoelectric points varied between 0.62 and 10.72, and the hydrophobicity of the peptides was predicted to be in the range −0.53 to 0.31. All of the peptides were predicted to be nontoxic by ToxinPred, indicating that the peptides from P. fucata can be safely utilized in the food and pharmaceutical industries. Following a search of the BIOPEP database, the 13 identified peptides were found to be novel.26 Ten of the peptides had fragments previously reported to be ACEI peptides; for example, VW and FR were contained in the sequence of FRVW. VW was documented to be an ACEI dipeptide identified in many hydrolysates, including rapeseed, salmon protein, and tilapia proteins.27–29 Furthermore, several other ACEI peptides were reported to share VW in their sequence, such as VWP (IC50 = 4.5 μM), VWY (IC50 = 9.4 μM), and DVWY (IC50 = 581.24 μM). Likewise, the ACEI peptides that shared the common FR fragment as part of their sequences were as follows: LFRQ (IC50 = 17 μM), AFKDEDTEEVPFR (IC50 = 80.2 μM), MILLLFR (IC50 = 0.12 μM), and LFR (IC50 = 31.8 μM). For the peptide LPYY in this study, there was no reported ACEI peptide found in its sequence; however, we found that the ACEI peptide LPYPY (IC50 = 28.9 μM) shared the same three amino acids at the N-terminal and one at the C-terminal with the LPYY. We speculated that the ACEI peptides shared several common sequences that were key amino acids forming interactions with ACE.| Peptide | m/z | Fraction | ALC (%) | P-Value | Toxicity | Net charge | Hydrophobicity | Isoelectric point | Reported ACEI peptides |
|---|---|---|---|---|---|---|---|---|---|
| a Abbreviation: ALC, average local confidence. | |||||||||
| FRVW | 607.3314 | C17 | 98 | 0.0008755 | None | +1 | −0.06 | pH 10.59 | VW, FR |
| VGPGW | 515.2564 | C17 | 88 | 0.001274 | None | 0 | 0.23 | pH 3.57 | GP, GW, VG, PG, VGP |
| LPYY | 555.2881 | C18 | 87 | 0.00124 | None | 0 | 0.12 | pH 3.57 | — |
| GTPW | 460.2251 | C18 | 86 | 0.0007006 | None | 0 | 0.07 | pH 3.6 | GT, TP |
| FGFWT | 657.2963 | C17 | 86 | 0.004784 | None | 0 | 0.31 | pH 3.44 | GF, FG |
| RFTGW | 666.3347 | C17 | 86 | 0.0006071 | None | +1 | −0.16 | pH 10.55 | RF, GW, TG |
| WAPR | 529.2845 | C17 | 83 | 5.122 × 10−5 | None | +1 | −0.3 | pH 10.72 | PR, AP, WA |
| RWPN | 572.2934 | C18 | 82 | 7.924 × 10−5 | None | +1 | −0.53 | pH 10.55 | RW |
| WDDM | 566.2004 | C17 | 82 | 0.01177 | None | −2 | −0.2 | pH 0.62 | DM |
| NWLN | 546.2726 | C18 | 81 | 0.008376 | None | 0 | −0.1 | pH 3.29 | LN, WL |
| WELSKS | 749.3749 | C18 | 81 | 0.01114 | None | 0 | −0.22 | pH 6.73 | — |
| TVTW | 506.2704 | C17 | 80 | 0.00243 | None | 0 | 0.14 | pH 3.29 | — |
| PAAPW | 541.2823 | C17 | 80 | 0.0001537 | None | 0 | 0.15 | pH 4.04 | AAP, AP, AA |
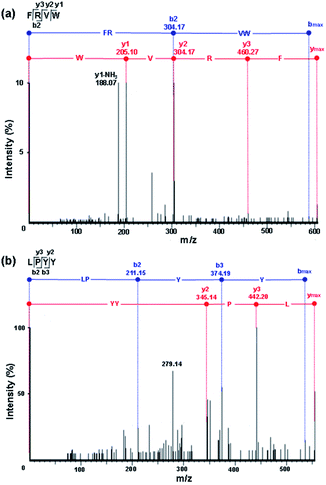
In comparison with the other peptides, the peptides FRVW, VGPGW, and LPYY displayed relatively higher ALC values (98%, 88%, and 87%, respectively), implying that they had a reliable peptide sequence. The statistical significant P-value was also obtained by Pepsite 2 which can be used to predict peptide-binding spots from protein surfaces and relies on preferred peptide binding environments calculated from a set of known protein–peptide 3D structures, combined with distance constraints derived from known peptides.20 The lower the P-value, the stronger the binding. The P-values of all of the peptides were below 0.05, illustrating that they bound strongly with ACE. Notably, the P-values of FRVW (0.0008755, shown in Table 2) and LPYY (0.00124) were lower than that of VGPGW (0.001274). According to the in silico analysis results of the ALC and P-values, the tetrapeptides FRVW and LPYY were chosen for chemical synthesis and further in vitro tests for ACE inhibition in order to confirm their activities. Their MS/MS spectra are shown in Fig. 3.
3.4 Structure–activity relationships of ACEI peptides
Recent structure–activity studies have indicated that the ACEI activity of peptides correlates with the number of amino acid residues, the ratio of HAAs in the peptide sequence, the charge characteristics, as well as the types of amino acids in the C- and N-terminals. In general, short-chained peptides with 2–12 residues tend to show higher inhibitory activity since they fit much better to the active sites of the ACE catalytic domain than larger peptides.30HAAs in the peptides contribute to inhibitory activity as they have intensive interactions with the HAAs of ACE at the active site where the conversion of Ang I to Ang II takes place. In addition, the amino acids (Lys, His, and Arg) possessing positive charges on their peptides enhance their ACEI capability, which might be due to negatively charged residues, including Glu403 and Glu162, at the S2 and S1′ active sites of the ACE domain, where the positively charged residues within the peptides may bind tightly with the negatively charged residues in ACE. Furthermore, some studies have indicated that peptides with HAAs at the C- and N-terminals might enhance ACEI activity.31,32
In this study, the IC50 value of the tetrapeptide LPYY was determined to be 116.26 μM. The inhibitory activity of LPYY may be attributed to its high ratio of HAA residues as well as its hydrophobic–hydrophilic–hydrophobic structure in the peptide sequence – the highly HAAs Leu and Tyr are located at the N- and C-terminals, respectively, whereas the hydrophilic amino acid Pro occupies the second position. Amazingly, compared to LPYY, FRVW containing three highly HAAs (Phe, Val, and Trp) and one positively charged Arg exhibited a much better ACEI activity, with an IC50 value of 18.34 μM, which is one of the best results for the tetrapeptides derived from plants, animals, and marine organisms.33 According to our study, it seems that a high ratio of HAAs is highly significant for their inhibitory activity for short-chained peptides. A similar result was reported for the peptide LIVT, which possessed three HAAs (Leu, lle, and Val) and showed an extremely low IC50 value of 0.11 μM.34
3.5 Inhibition patterns of peptides on ACE
Four inhibition patterns, including competitive, noncompetitive, uncompetitive, and mixed-competitive modes, were found for the ACEI peptides, with the noncompetitive mode being the most prevalent.In this study, the inhibitory kinetics of the two peptides were studied by means of Lineweaver–Burk plots. In the presence of three fixed peptide concentrations, the velocity of HA generation was analyzed at different concentrations of the HHL substrate. As shown in Fig. 4(a), the plots for FRVW exhibited almost the same x-axis intercepts, but different y-axis intercepts, showing it to be a noncompetitive inhibitor, which bound to essential groups outside the active part of the enzyme, leading to conformational changes and enzyme inhibition. The Vmax values decreased when the peptide concentration increased, indicating that the peptide FRVW probably prevented the HHL substrate from being converted to product. The result was similar to the ACEI peptides VGLPNSR and QAGLSPVR from tilapia skin gelatin hydrolysates and the peptides VYAP, VIIF, and MAW from cuttlefish muscle protein hydrolysates.35,36 As for the peptide LPYY, it acted in a mixed-mode type of inhibition in which the peptide formed complexes with both the active and non-active sites of ACE, thus reducing the catalytic efficiency for the substrate. As can be seen in Fig. 4(b), the Vmax values decreased as the peptide LPYY concentration increased and the Km values increased after LPYY was added to the reaction, indicating a decrease in the catalytic activity and affinity of ACE for HHL. Similar to the findings of a previous study, the peptides WVYY and WYT identified from the hydrolysate of hemp seed proteins exhibited a mixed-mode type of ACE inhibition.37
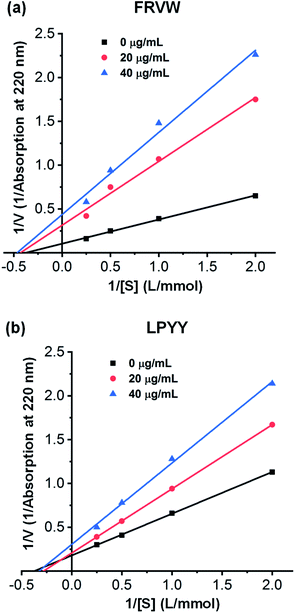 | ||
| Fig. 4 Lineweaver–Burk plots of ACE inhibition by different concentrations of synthesized peptides at various substrate concentrations: (a) FRVW; (b) LPYY. | ||
3.6 Effect of ACE on peptides
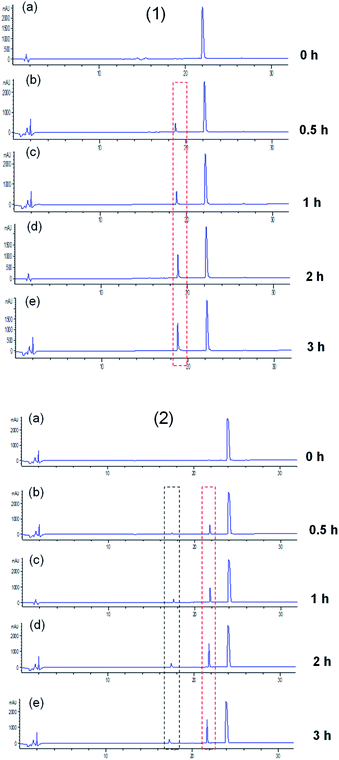
ACE exerts a physiological function due to its peptidyl dipeptidase activity, with which it can catalyze the degradation of angiotensin I and bradykinin, causing the elevation of blood pressure. Thus, it is possible that peptides will be hydrolyzed as they interact with ACE. As reported previously, the release of the C-terminal dipeptide from the oligopeptide happens when the penultimate residue is not a Pro (P) residue and the reciprocal first residue is neither Asp (D) nor Glu (E).2 In this study, the generated fragments of two key ACEI peptides, LPYY and FRVW, against ACE were visualized by eluting the mixtures solution using HPLC. From the chromatograms (Fig. 5), it can be seen that both the peptides LPYY and FRVW were hydrolyzed to some extent after incubation. LPYY showed one fragment peak, and the peak area of this fragment increased with incubation time, meaning increasing numbers of fragments released. Different from LPYY, FRVW exhibited two distinct peptide fragments, and the two peak areas also became larger over time.In general, ACEI peptides can be classified into two categories based on their interaction with ACE: (1) true inhibitors, which show resistance to ACE and are not hydrolyzed; and (2) substrates for ACE, which are further subdivided into two types: (i) real substrates, which are hydrolyzed with the release of inactive or less active fragments, and (ii) pro-drugs, which are hydrolyzed accompanied by the generation of more active fragments.38 To confirm the change in ACEI activity of peptides before and after incubation against ACE and the category they fell into, the 0 h-incubation and 3 h-incubation mixtures were tested for ACEI capacity. The results showed that LPYY exhibited a 91.30 ± 0.03% inhibitory rate before incubation and a 93.23 ± 0.21% inhibitory rate following incubation with ACE. Similarly, for the peptide FRVW, the ACEI rate increased from 90.21 ± 0.31% to 92.75 ± 0.15% (data not shown). Both of the higher ACEI activities following incubation against ACE suggested that the two peptides were pro-drug substrates. Rao et al.39 regarded both of the peptides IKPVQ and IKPVA as pro-drug substrates for ACE due to their reduced IC50 values following incubation, and found that their C-terminal dipeptides were removed by ACE, generating a common fragment IKP. Hence, theoretically at least, FRVW in this study should also have been hydrolyzed by ACE into FR and VW, which have been documented as ACEI peptides in the BIOPEP database. As anticipated, the products of FRVW hydrolyzed by ACE were identified as FR and VW by LC-MS/MS (see Fig. S1†). Similarly, the fragment LP was also detected from LPYY (see Fig. S2†).
3.7 Molecular docking simulation
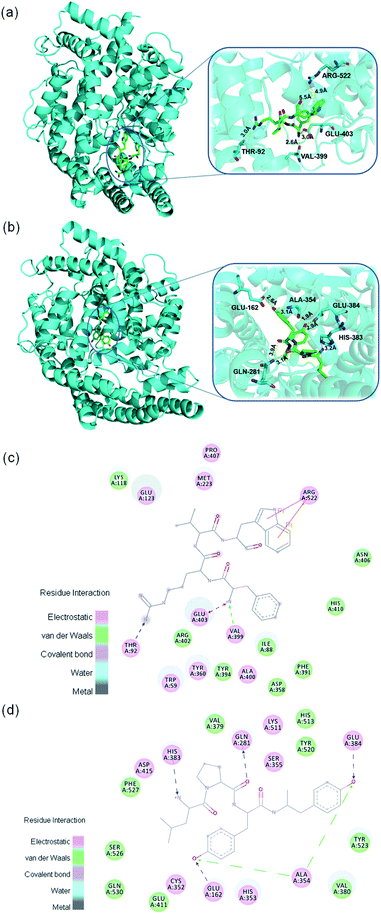
To explore the inhibitory mechanism between the identified peptides and ACE, molecular docking was performed. In this study, the two tetrapeptides, FRVW and LPYY, were docked into the crystal structure of ACE. As mentioned above, peptides with low molecular weights can adapt well to ACE; thus, FRVW (607.3314 Da) and LPYY (555.2881 Da) should dock with the active cavity well and exhibit potential binding affinity. As expected, the results showed that strong binding force between two peptides and ACE – CDOCKER_Energy of FRVW and LPYY was 128.352 kcal mol−1 and 119.652 kcal mol−1, respectively.It has been reported that HAA residues such as Phe, Trp, Leu, Pro, and Tyr are highly beneficial to ACEI activity since they participate in forming strong interactions with the ACE residues.40
As shown in Fig. 6 the Phe in FRVW formed electrostatic forces with ACE residue Glu403 and van der Waals forces with ACE residue Val399. Meanwhile, the Trp in FRVW formed Pi–Pi interactions with Arg522 of ACE. It is worth mentioning that noncompetitive peptide LSGYGP also had interaction with ACE residue Arg522,41 indicating it may be a vital amino acid for the inhibitor to bind. The Leu and Pro in LPYY bound with His383 and Gln281 residues of ACE respectively through hydrogen bonds. Furthermore, two Tyr of LPYY constructed hydrogen bonds and van der Waals forces with residues of ACE as well. The above results demonstrated that HAAs of peptides interacted with ACE through various forces, including hydrogen bonds, electrostatic forces, van der Waals forces, and Pi–Pi interactions. It is no doubt that hydrogen bonds play an important role in stabilizing the docking complex and enzyme catalytic reactions.42 However, the presence of forces other than hydrogen bonds also contributed to the stabilization of complex and great enhancement of ACEI activity.
Previous studies have shown that the active site of ACE contains three active pockets, including S1 (Ala354, Tyr523, and Glu384), S2 (Gln281, Lys511, His353, Tyr520, and His513), and S1′ (Glu162).43 Some peptides were reported to bind with these amino acid residues in active pocket. For example, VVSLSIPR bound with His353, Tyr523, and Glu384;44 SSR bound with Tyr523, Glu384, and His387;45 FHAPWK bound with Glu384 and His353.46 However, these peptides belong to competitive inhibitors. Our results showed that FRVW bound with Val399, Glu403, Thr92, and Arg522 and it didn't form any important interactions with the amino acid residues of ACE active pocket, in accordance with their ACE inhibition kinetic pattern as noncompetitive inhibitors that attach to an enzyme at an allosteric site (a site other than the active site), distorting the shape of the active site so that the enzyme can no longer take effect. LPYY formed interactions with His383, Glu281, Ala354, Glu162, and Glu384, most of which belonged to the active pocket with Glu384 and Ala354 in S1 pocket, Gln281 in S2 pocket, and Glu162 in S1′ pocket, which was consistent with the conclusion that peptide LPYY might be a mixed-type inhibitor that bound to both the active and nonactive sites. The previous reported mixed-type inhibitor WVYY formed hydrogen bonds with Gln281, His353, Asp377, Tyr520, and His513, most of which were critical components of the ACE active pocket.37 In the future, the molecular docking should be analyzed combined with the structure–activity relationships and kinetic mode which are concluded from actual test so that the simulation and in vitro experiment are verified mutually.
4 Conclusions
In this study, a combination of traditional methods and recent developments in bioinformatics has been used to discover some potential ACEI peptides. Two novel ACEI tetrapeptides, FRVW and LPYY, derived from P. fucata meat protein hydrolysates were screened and synthesized. In vitro assays of ACEI activity showed that both FRVW and LPYY had strong ACEI activities. In particular, FRVW exhibited excellent ACEI activity, with an IC50 value of 18.34 μM, which is one of the best results for tetrapeptides derived from plants, animals, and marine organisms. Lineweaver–Burk plots showed that the peptide FRVW was a noncompetitive inhibitor and that LPYY acted in a mixed-mode type of inhibition. The two peptides were unable to resist hydrolysis by ACE and showed pro-drug characteristics. Furthermore, the result of a molecular docking simulation showed that hydrophobic amino acid of peptides interacted with ACE through various forces, thus reducing catalytic activity. However, future studies should be focused on their detailed bioavailability and inhibitory mechanisms in vivo. In summary, our study shows that the peptides FRVW and LPYY derived from the hydrolysates of P. fucata are promising functional foods for the treatment of high blood pressure.Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDA13020300).Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors would like to acknowledge contributions from Zhiping Deng (Zhejiang Academy of Agricultural Sciences, Hangzhou, Zhejiang, China) for the identification of the peptide sequence.References
- R. Balti, A. Bougatef, A. Sila, D. Guillochon, P. Dhulster and N. Nedjar-Arroume, Food Chem., 2015, 170, 519–525 CrossRef CAS PubMed.
- M. L. Tu, C. Wang, C. Chen, R. Y. Zhang, H. X. Liu, W. H. Lu, L. Z. Jiang and M. Du, Food Chem., 2018, 256, 98–104 CrossRef CAS PubMed.
- P. Garcia-Mora, M. Martin-Martinez, M. A. Bonache, R. Gonzalez-Muniz, E. Penas, J. Frias and C. Martinez-Villaluenga, Food Chem., 2017, 221, 464–472 CrossRef CAS PubMed.
- H. M. Guo, Y. Q. Hao, A. Richel, N. Everaert, Y. N. Chen, M. J. Liu, X. S. Yang and G. X. Ren, J. Sci. Food Agric., 2020, 100, 5569–5576 CrossRef CAS PubMed.
- R. D. Wang, X. Lu, Q. Sun, J. H. Gao, L. Ma and J. N. Huang, Int. J. Mol. Sci., 2020, 21, 19 Search PubMed.
- V. G. Tacias-Pascacio, R. Morellon-Sterling, E. H. Siar, O. Tavano, A. Berenguer-Murcia and R. Fernandez-Lafuente, Int. J. Biol. Macromol., 2020, 165, 2143–2196 CrossRef CAS PubMed.
- H. Parmar, S. Hati, G. Panchal and A. A. Sakure, Int. J. Pept. Res. Ther., 2020, 26, 997–1011 CrossRef CAS.
- P. Puchalska, M. Concepción García and M. Luisa Marina, Food Chem., 2014, 157, 62–69 CrossRef CAS PubMed.
- C. Grootaert, B. Matthijs, S. Voorspoels, S. Possemiers, G. Smagghe and J. Van Camp, Food. Funct., 2017, 8, 3847–3855 RSC.
- L. J. You, Y. Z. Li, H. F. Zhao, J. Regenstein, M. M. Zhao and J. Y. Ren, J. Aquat. Food Prod. Technol., 2015, 24, 661–671 CrossRef CAS.
- Z. J. Ma, J. Huang, J. Sun, G. N. Wang, C. Z. Li, L. P. Xie and R. Q. Zhang, J. Biol. Chem., 2007, 282, 23253–23263 CrossRef CAS.
- C. Sasaki, S. Tamura, R. Tohse, S. Fujita, M. Kikuchi, C. Asada and Y. Nakamura, Process Biochem., 2019, 77, 137–142 CrossRef CAS.
- Y. Y. Wu, J. Wang, L. H. Li, X. Q. Yang, J. X. Wang and X. Hu, CyTA--J. Food, 2018, 16, 11–19 CrossRef CAS.
- C. C. Y. Sutopo, A. Sutrisno, L. F. Wang and J. L. Hsu, Process Biochem., 2020, 95, 204–213 CrossRef CAS.
- Y. Fu, J. F. Young, M. M. Lokke, R. Lametsch, R. E. Aluko and M. Therkildsen, J. Funct. Foods, 2016, 24, 196–206 CrossRef CAS.
- R. J. FitzGerald, M. Cermeno, M. Khalesi, T. Kleekayai and M. Amigo-Benavent, J. Funct. Foods, 2020, 64, 13 Search PubMed.
- D. Y. Zhou, Y. Tang, B. W. Zhu, L. Qin, D. M. Li, J. F. Yang, K. Lei and Y. Murata, Food Chem., 2012, 132, 815–822 CrossRef CAS.
- A. K. Arise, A. M. Alashi, I. D. Nwachukwu, S. A. Malomo, R. E. Aluko and E. O. Amonsou, J. Sci. Food Agric., 2017, 97, 2834–2841 CrossRef CAS PubMed.
- X. D. Lan, D. K. Liao, S. G. Wu, F. Wang, J. H. Sun and Z. F. Tong, Food Chem., 2015, 182, 136–142 CrossRef CAS.
- L. G. Trabuco, S. Lise, E. Petsalaki and R. B. Russell, Nucleic Acids Res., 2012, 40, W423–W427 CrossRef CAS PubMed.
- Y. J. Zheng, Y. F. Zhang and S. San, Nutrients, 2020, 12, 14 Search PubMed.
- R. Vasquez-Villanueva, J. M. Orellana, M. L. Marina and M. C. Garcia, J. Agric. Food Chem., 2019, 67, 10313–10320 CrossRef CAS PubMed.
- J. P. Wu and X. L. Ding, Food Res. Int., 2002, 35, 367–375 CrossRef CAS.
- M. Zarei, N. B. Abidin, S. M. Auwal, S. Y. Chay, Z. A. Haiyee, A. M. Sikin and N. Saari, Biomolecules, 2019, 9, 11 CrossRef PubMed.
- M. A. Hanafi, S. N. Hashim, C. S. Yea, A. Ebrahimpour, M. Zarei, K. Muhammad, A. Abdul-Hamid and N. Saari, Food Res. Int., 2018, 106, 589–597 CrossRef CAS.
- A. Iwaniak, P. Minkiewicz, M. Darewicz, K. Sieniawski and P. Starowicz, Food Res. Int., 2016, 85, 155–161 CrossRef CAS PubMed.
- E. D. Marczak, H. Usui, H. Fujita, Y. J. Yang, M. Yokoo, A. W. Lipkowski and M. Yoshikawa, Peptides, 2003, 24, 791–798 CrossRef CAS.
- M. Darewicz, J. Borawska, G. E. Vegarud, P. Minkiewicz and A. Iwaniak, Int. J. Mol. Sci., 2014, 15, 14077–14101 CrossRef CAS PubMed.
- B. H. Yesmine, B. Antoine, N. G. da Silva Ortencia Leocadia, B. W. Rogerio, A. Ingrid, B. Nicolas, M. Thierry, P. Jean-Marie, S. Frederic and B.-J. Stephanie, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2017, 1052, 43–50 CrossRef CAS PubMed.
- K. Lin, L. W. Zhang, X. Han, Z. X. Meng, J. M. Zhang, Y. F. Wu and D. Y. Cheng, J. Agric. Food Chem., 2018, 66, 3221–3228 CrossRef CAS PubMed.
- H. B. Fan, J. P. Wang, W. Liao, X. Jiang and J. P. Wu, J. Agric. Food Chem., 2019, 67, 7147–7156 CrossRef CAS PubMed.
- S. M. Auwal, N. Z. Abidin, M. Zarei, C. P. Tan and N. Saari, PLoS One, 2019, 14, 18 CrossRef PubMed.
- C. L. Liu, L. Fang, W. H. Min, J. S. Liu and H. M. Li, Food Chem., 2018, 245, 471–480 CrossRef CAS PubMed.
- V. S. Vallabha and P. K. Tiku, Int. J. Pept. Res. Ther., 2014, 20, 161–168 CrossRef.
- L. Yuan, L. P. Sun and Y. L. Zhuang, Food Funct., 2018, 9, 5251–5259 RSC.
- R. Balti, N. Nedjar-Arroume, A. Bougatef, D. Guillochon and M. Nasri, Food Res. Int., 2010, 43, 1136–1143 CrossRef CAS.
- A. T. Girgih, R. He and R. E. Aluko, J. Agric. Food Chem., 2014, 62, 4135–4144 CrossRef CAS PubMed.
- B. Forghani, M. Zarei, A. Ebrahimpour, R. Philip, J. Bakar, A. A. Hamid and N. Saari, J. Funct. Foods, 2016, 20, 276–290 CrossRef CAS.
- S. Q. Rao, S. Liu, T. Ju, W. Q. Xu, G. M. Mei, Y. S. Xu and Y. J. Yang, Biochem. Eng. J., 2012, 60, 50–55 CrossRef CAS.
- A. Moayedi, L. Mora, M. C. Aristoy, M. Safari, M. Hashemi and F. Toldra, Food Chem., 2018, 250, 180–187 CrossRef CAS PubMed.
- T. R. Zhao, B. T. Liu, L. Yuan, L. P. Sun and Y. L. Zhuang, J. Funct. Foods, 2019, 61, 9 Search PubMed.
- J. P. Li, Z. Y. Liu, Y. H. Zhao, X. J. Zhu, R. L. Yu, S. Y. Dong and H. H. Wu, Mar. Drugs, 2018, 16, 13 CrossRef PubMed.
- Y. Y. Ngoh and C. Y. Gan, Food Chem., 2018, 267, 124–131 CrossRef CAS PubMed.
- K. A. A. Nawaz, S. M. David, E. Murugesh, M. Thandeeswaran, K. G. Kiran, R. Mahendran, M. Palaniswamy and J. Angayarkanni, Phytomedicine, 2017, 36, 1–7 CrossRef CAS PubMed.
- H. J. Sun, Q. Chang, L. Liu, K. G. Chai, G. Y. Lin, Q. L. Huo, Z. X. Zhao and Z. X. Zhao, J. Agric. Food Chem., 2017, 65, 10020–10028 CrossRef CAS PubMed.
- Y. H. Shih, F. A. Chen, L. F. Wang and J. L. Hsu, J. Agric. Food Chem., 2019, 67, 7810–7820 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra10476k |
| This journal is © The Royal Society of Chemistry 2021 |