Open Access Article
Open Access ArticleRegenerating heavily biofouled dissolved oxygen sensors using bacterial viruses†
Fereshteh Bayat‡
 a,
Dhanyasri Maddiboina‡b,
Tohid F. Didar
a,
Dhanyasri Maddiboina‡b,
Tohid F. Didar acd and
Zeinab Hosseinidoust
acd and
Zeinab Hosseinidoust *abd
*abd
aSchool of Biomedical Engineering, McMaster University, 1280 Main Street West, ETB 406, Hamilton, Ontario L8S 4K1, Canada. E-mail: doust@mcmaster.ca; Tel: +1 (905) 525 9140
bDepartment of Chemical Engineering, McMaster University, Hamilton, Ontario L9S 8L7, Canada
cDepartment of Mechanical Engineering, McMaster University, 1280 Main Street West, JHE 310, Hamilton, Ontario L8S 4L7, Canada
dInstitute for Infectious Disease Research, McMaster University, 1280 Main Street West, MDCL 2235, Hamilton, Ontario L8S 4K1, Canada
First published on 23rd February 2021
Abstract
Bacterial biofilms are aggregates of bacterial cells embedded in a self-produced extracellular polymeric matrix. Biofilm formation has always been considered a major challenge for sensors used in underwater measurements, and is a primary source of measurement error, especially when it comes to long-term in situ monitoring. We demonstrate the utility of lytic bacteriophages (bacterial viruses) as a non-invasive strategy for removing bacterial biofilms formed on the gas permeable membrane of electrochemical dissolved oxygen sensors. Our results show that a 4 day Pseudomonas aeruginosa biofilm with a fully developed matrix significantly affected the sensor signal and response time, decreasing the signal by 32% and increasing the response time by 94%. In addition, measurements with the biofouled membrane had a very low signal to nose ratio compared to a clean sensor membrane. A single dose of overnight phage treatment effectively removed the biofilm (as indicated by scanning electron micrographs and fluorescence images of the membrane), without the need for repeated treatments. Furthermore, the sensor signal that had plummeted by 32% for a fully biofouled membrane, was returned to the original value (7.96 ± 0.27 mg L−1) after phage treatment and the signal to noise ratio (calculated as the ratio of mean to standard deviation) increased 8 folds for a phage-treated membrane compared to a biofouled membrane. Our data indicate near complete regeneration and signal recovery for the dissolved oxygen sensor, making the biofouled sensor reusable without the use of harsh chemicals that could destroy the fragile sensor membrane.
Introduction
Biofouling, or biological fouling, is an unwanted deposition of microorganisms on biotic and abiotic surfaces.1 Bacteria are major contributors to biofouling events, and they can readily form surface-bound, 3-dimensional communities. These communities are known as bacterial biofilms and are usually encased in a matrix of biopolymers, called the extracellular polymeric substance (EPS), which is secreted by the bacterial cells themselves.4 From the point of view of a bacterial cell, biofilms provide a means for survival and that is why bacterial biofilms are the most common form of bacterial existence in nature. Bacteria form biofilms on infected catheters and implants,2 on surfaces in the food industry5 and membranes in water treatment plants,6 on ship hulls,7 and on marine sensors.8,9 Biofouling has always been considered a major challenge for underwater measurements, especially when it comes to long-term in situ monitoring (e.g., and environmental monitoring devices installed in lakes and rivers).10 Biofouling of sensor surfaces is a major concern in all physico-chemical sensors, including pH and redox sensors, optical sensors, and electrochemical sensors, and could result in unreliable measurements.11,12 Biofilms start to grow as soon as a sensor is immersed in water, and it has been reported that the quality of measurements decreases in less than a week.10 Although many potential solutions have been proposed to combat biofouling in underwater measurements and monitoring, there is no single universally accepted method applicable to underwater environmental sensors.Various methods and technologies have been used to prevent biofouling or remove biofilms from biofouled surfaces, such as mechanical treatment (wipers, water jetting, ultrasonic or electrical surface excitation),11,13 chemical detergents and biocides,14 UV light exposure (which may kill bacteria but will not completely remove the biofilm),15,16 chlorination,8 chemical modifications of surfaces (e.g., using zwitterions, carboxylation, and charged hydrogels), and coating surfaces with antimicrobials17 or repellent coatings.18–20 For sensing surfaces, however, cleaning methods should be chosen with care. Cleaning methods that involve harsh physical or chemical treatments could cause structural damage and harm the sensitive and sometimes delicate sensing surfaces.5 In addition, improved intercellular interactions in biofilms leads to enhanced metabolic capacity of cells and antimicrobial resistance.21 As a result, biofilms have shown to be more resistant to chemical disinfectants and biocides compared to planktonic (free swimming) bacteria. Antimicrobial or repellent surface coatings are not as invasive as mechanical scrubbing or chemical chlorination, but require changing the sensing surface, which may interfere with measurements and sensor sensitivity due to altered bio-interface properties. Besides, once the coating is depleted, the sensor will start to foul and will thus have to be removed and changed.
Bacteriophages are bacterial viruses that were discovered over a hundred years ago, well before penicillin, and were almost immediately used to treat bacterial infections.22,23 Bacteriophages (or phages, for short) are natural bacteria devourers, and their job in nature is to keep bacterial populations in check. They have been proposed and explored as potential biological tools to control or mitigate biofilm formation in different settings.24 These human-friendly viruses are capable of infecting bacteria very specifically, thus not harming the delicate microbial balance in the surrounding environment, a characteristic that has made them attractive for different applications as therapeutics, biocontrol agents, prophylactics and even sensing agents.25–27 After binding to bacterial host cell surface receptors, phages insert their genome into the host bacterium. A class of phages known as lytic or virulent phages immediately take control of the host cell machinery and turn the bacteria cell into a phage propagation factory. At the end of this replication process, known as the lytic cycle, the bacterial cell is lysed, and hundreds of new phages are released to infect other bacteria. Temperate or mild phages are another class of phage. Those integrate their genome with that of the host bacteria and lay dormant until activated by an external stimulus such as UV light, temperature change, or a sub-lethal concentration of a toxic chemical, which in turn activate the lytic cycle and ultimately lyse the bacterial cell.28,29 Lytic phages are the group of phages that are mostly explored for controlling bacterial populations, because they offer an immediate antibacterial action.
Phages are currently heavily investigated for their use as therapeutics and for biomedical applications24,30,31 but have remained obscure to most environmental engineers. With only a few reports published on mitigation of membrane fouling32,33 and no prior report on regenerating biofouled sensors, there is a gap in knowledge regarding the performance of phage for sensor regeneration, which calls for more investigations in this field. It is noteworthy that chemical antimicrobial agents in general have limited activity against biofilms, because they could be neutralized or diluted to sublethal concentrations by binding to the biofilm matrix before reaching all the individual bacterial cells within the biofilm.34 In addition to propagating on-site, lytic phages have enzymes that can degrade the biofilm matrix, which is a major advantage over chemical treatment.24 Bacteriophages, however, are not the silver bullet for fighting bacterial biofilms. Even in demonstrated biomedical applications of phage against bacterial biofilms, bacteriophages have been demonstrated to work along with the host immune system to completely eradicate the biofilm, a phenomenon absent in environmental applications. Despite the imperfection, what makes bacteriophages ideal for biomedical applications over antibiotics (mild and non-destructive to non-target bacteria, and evolution of resistant mutants that are less virulent13,35) also makes them important options of investigations for environmental applications. However, for regeneration of submerged sensors, it is important to adequately clean the sensing substrate so as to regenerate a sensing signal without damaging or disintegrating the surface, and so effectiveness of phage treatment is not immediately clear and must be investigated with a correct choice of phage. A wrong choice of phage could be completely ineffective or even result in increased biofilm formation.36
In this work, we use a lytic bacteriophage cocktail to remove a fully developed biofilm of Pseudomonas aeruginosa PAO1 (PA) from the membrane of a dissolved oxygen (DO) sensor. The cocktail consists of vB_Pae-Kakheti25 and vB_Pae-TbilisiM32, which are well-characterized Pseudomonas phages that we have determined are capable in dispersing established biofilms of PA.37 Pseudomonas aeruginosa is one of the most prevalent opportunistic microorganisms, inhabiting soil, seawater, freshwater, sewage, plants, and animals.38 This microorganism is a strong biofilm former and is responsible for difficult to treat hospital acquired infections,39 persistent food contamination,39 and marine biofouling, due to its high capacity to colonize and form biofilms on surfaces submerged in aquatic environments.40 It has been shown that other microorganisms are stimulated to settle and colonize the submerged surfaces in marine environments in the presence of the EPS secreted by biofilm forming species such as Pseudomonas aeruginosa.3 In addition, studying the biofilms formed on nanofiltration membranes in water treatment plants has confirmed that P. aeruginosa is one of the important species dominating the biofilm communities.9 As a result, in this proof of concept study, P. aeruginosa is used for biofilm formation on the surface of DO sensor membranes as a representative for biofilm forming species in aquatic environments. We demonstrate the effectiveness of a single dose of lytic phage treatment on biofouled sensor membranes in recovering DO sensor signals to the original level, increasing the signal to noise ratio, and decreasing the response time.
Methods
Materials
The ExStik II DO600 dissolved oxygen meter and DO603 membrane kits were purchased from Extech Instruments (Nashua, USA). Phosphate buffered saline (PBS) (0.1 M, pH 7.4) was prepared using tablets from VWR (Mississauga, Canada). Clear, flat-bottom, sterilized, 96-well tissue culture plates and crystal violet were also purchased from VWR. LB broth, Miller, sterile 50 mL conical centrifuge tubes, and glutaraldehyde (50%) were purchased from Fisher Scientific (Mississauga, Canada). Hoechst 33342 nucleic acid stain (ex/em 350/461 nm) was purchased from Thermo Fisher Scientific (Massachusetts, USA). Formaldehyde was purchased from Caledon Laboratories (Georgetown, Canada).Bacterial strain, phage, and culture conditions
The bacterial strain used in this study was Pseudomonas aeruginosa PAO1 (PA) and the phages used were vB_Pae-Kakheti25 and vB_Pae-Tbilisi32 (DSMZ, Germany). Bacterial overnights of PA were prepared by inoculating LB broth with a frozen glycerol stock and incubating at 37 °C and 180 rpm for 16–18 h. Phage suspensions were propagated by diluting the overnight culture 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 into 50 mL of LB media in a baffled Erlenmeyer flask and incubating at 37 °C and 180 rpm. When the optical density of the culture at 600 nm (OD600) reached 0.5, 10 μL of either phage vB_Pae-Kakheti25 or vB_Pae-Tbilisi32 was added and the culture and incubated for 6 h, or overnight. The resulting phage lysate was centrifuged for 10 min at 7000 × g, then the supernatant was filtered through a 0.2 μm syringe filter and stored in the fridge. The concentration of the phage suspension (number of plaque forming units per mL, PFU mL−1) was determined using the agar overlay technique.41
1000 into 50 mL of LB media in a baffled Erlenmeyer flask and incubating at 37 °C and 180 rpm. When the optical density of the culture at 600 nm (OD600) reached 0.5, 10 μL of either phage vB_Pae-Kakheti25 or vB_Pae-Tbilisi32 was added and the culture and incubated for 6 h, or overnight. The resulting phage lysate was centrifuged for 10 min at 7000 × g, then the supernatant was filtered through a 0.2 μm syringe filter and stored in the fridge. The concentration of the phage suspension (number of plaque forming units per mL, PFU mL−1) was determined using the agar overlay technique.41
Biofilm formation and quantification
Biofilms were formed on the membranes by placing the DO membrane cassette face down in a 50 mL conical tube containing 2 mL of an overnight PA culture, diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in LB broth. The membranes were incubated at 37 °C and 80 rpm for 1–4 days. Biofilm was then quantified through fluorescence microscopy followed by image analysis. For biofilms formed in 96-well plates, we diluted an overnight culture 1
1000 in LB broth. The membranes were incubated at 37 °C and 80 rpm for 1–4 days. Biofilm was then quantified through fluorescence microscopy followed by image analysis. For biofilms formed in 96-well plates, we diluted an overnight culture 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in LB broth, added 200 μL of diluted suspension to each well, and incubated the plate at 37 °C and 80 rpm for 1–4 days. Biofilms were then quantified using the crystal violet method, as described elsewhere.42 Briefly, biofilms were washed three times with PBS buffer, stained with crystal violet, and then quantified by dissolving the crystal violet in acetone and reading the absorbance at 590 nm with a BioTek plate reader.
1000 in LB broth, added 200 μL of diluted suspension to each well, and incubated the plate at 37 °C and 80 rpm for 1–4 days. Biofilms were then quantified using the crystal violet method, as described elsewhere.42 Briefly, biofilms were washed three times with PBS buffer, stained with crystal violet, and then quantified by dissolving the crystal violet in acetone and reading the absorbance at 590 nm with a BioTek plate reader.
Phage treatment
Membranes with biofilm were rinsed in PBS three times and then incubated in 2 mL of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of vB_Pae-Kakheti25 and vB_Pae-Tbilisi32 (109 PFU mL−1 each) in a 50 mL conical tube at 37 °C and 80 rpm for 6 h or overnight. The control fouled membranes were treated the same way, but incubated in LB broth instead of a phage suspension.
1 mixture of vB_Pae-Kakheti25 and vB_Pae-Tbilisi32 (109 PFU mL−1 each) in a 50 mL conical tube at 37 °C and 80 rpm for 6 h or overnight. The control fouled membranes were treated the same way, but incubated in LB broth instead of a phage suspension.
Bleach treatment
Same as phage treatment, membranes with biofilm were rinsed in PBS three times, and then incubated in 2 mL of a 10% bleach solution in a 50 mL conical tube at 37 °C and 80 rpm overnight.Sensitivity readings
DO concentration of stirred deionized (DI) water was measured using the DO sensor with clean, biofouled, phage-treated and bleached membranes installed. Two membranes were used per condition and the measurements were repeated three times with each membrane. Each experiment was performed in triplicate. The response time for each membrane was recorded as the time the sensor readings showed fluctuations <±0.01 mg L−1 for 30 seconds.Fluorescence microscopy
Biofouled and phage-treated membranes were fixed with 4% formaldehyde in PBS for 30 min and rinsed with DI water. Fixed samples were stained with 400 μL of 25 μg mL−1 Hoechst 33342 in DI water for 1 h and rinsed lightly with DI water afterwards to remove unbound fluorescence dye. Images were recorded using Nikon Eclipse Ti2-E motorized inverted fluorescence microscope, and the results were quantified using ImageJ image processing software. For quantification, two membranes were used, and seven frames of each membrane were quantified.Scanning electron microscopy (SEM)
Membranes were cut out of their cassette and rinsed with PBS three times. Following fixation with 30% glutaraldehyde for 30 min at 4 °C, the membranes were dehydrated in a gradient of ethanol solutions (10%, 30%, 50%, 70%, 90%, 100%) for 10 min each, then dried using a Leica EM CPD300 critical point dryer (Leica Mikrosysteme GmbH, Austria). Those were then coated with 20 nm of gold using a Polaron Model E5100 sputter coater (Polaron Equipment Ltd., UK). SEM images were obtained using a Tecan VEGA II LSU SEM (Tecan USA, PA, USA) at 10 kV and 4700× magnification.Statistical analysis
Data are presented as means ± standard deviation. Statistical significance was assessed using analysis of variance and post-hoc analysis with Tukey's test, where significant difference was considered as P < 0.05. All experiments were performed in triplicates unless otherwise specified.Results and discussion
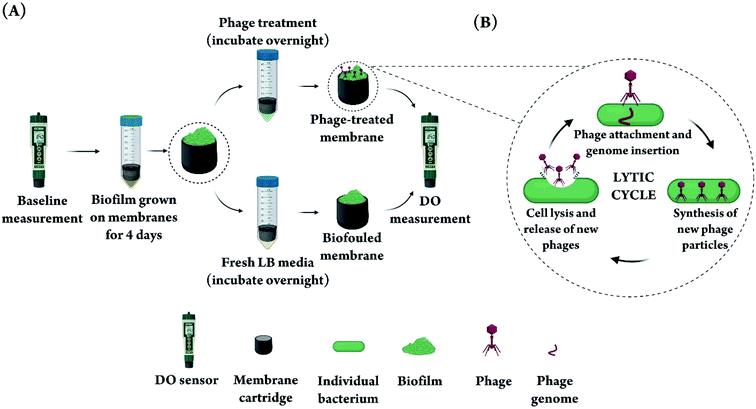
In this study, PA biofilms were grown on DO sensor membranes following baseline measurements (Fig. 1A). Biofouled membranes were then subjected to overnight incubation in a phage suspension consisting of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of phages vB_Pae-Kakheti25 and vB_Pae-Tbilisi32 in LB media. The choice of phage was based on previous experience in our lab with a library of PA phages and their performance against PA biofilms in a stationary microtiter plate model. We chose two phages for treating the biofilm instead of one phage, because it has been shown that using more than one phage decreases the frequency of evolution of phage-resistant bacteria,43–45 thus increasing the efficiency of phage treatment. To justify the choice of a phage mixture, we demonstrated that the two phages used lead to a significantly higher biofilm reduction (in a microtiter biofilm model) when used in combination, compared to the case when each phage is used alone (Fig. S1†). Fig. 1B shows a simplified schematic of the phage lytic cycle that leads to the destruction of bacterial cells in a biofilm as a result of phage action. Control biofilms were incubated overnight in fresh LB media without any phage.
1 mixture of phages vB_Pae-Kakheti25 and vB_Pae-Tbilisi32 in LB media. The choice of phage was based on previous experience in our lab with a library of PA phages and their performance against PA biofilms in a stationary microtiter plate model. We chose two phages for treating the biofilm instead of one phage, because it has been shown that using more than one phage decreases the frequency of evolution of phage-resistant bacteria,43–45 thus increasing the efficiency of phage treatment. To justify the choice of a phage mixture, we demonstrated that the two phages used lead to a significantly higher biofilm reduction (in a microtiter biofilm model) when used in combination, compared to the case when each phage is used alone (Fig. S1†). Fig. 1B shows a simplified schematic of the phage lytic cycle that leads to the destruction of bacterial cells in a biofilm as a result of phage action. Control biofilms were incubated overnight in fresh LB media without any phage.
Biofilm removal with phage
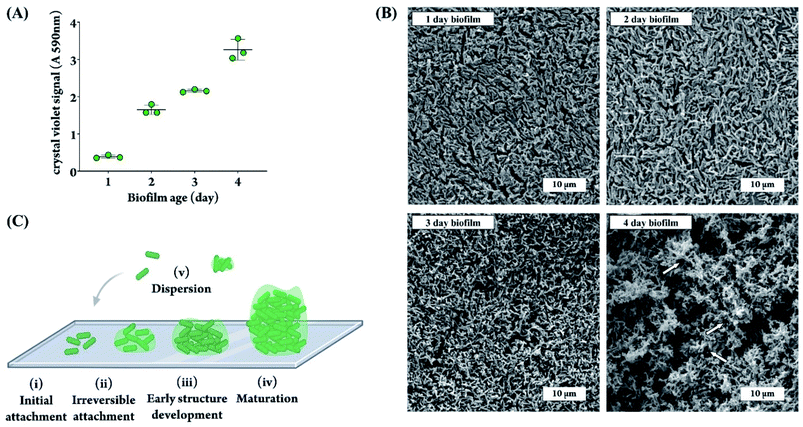
To ensure a thick biofilm forms on the sensor membranes, we first assessed the appropriate length of time needed to allow for biofilm formation on the membrane by quantifying biofilm growth in a 96 well plate for the duration of four days. The biofilm level was quantified using crystal violet, a commonly used dye that binds and stains both bacteria and the surrounding matrix.42 As shown in Fig. 2A, the level of total biomass steadily increased over the four day period. The SEM images of PA biofilm grown on DO sensor membranes in Fig. 2B confirm the trend shown in Fig. 2A and correlate with the well-documented stages of a PA biofilm lifecycle.46–48 As depicted in Fig. 2C, the biofilm developmental process begins with (i) attachment of PA cells to a surface. This is followed by (ii) the cells spreading across the surface instead of upward, as can be seen in our 1 day and 2 day biofilm SEM images by visibility of the membrane underneath the cells, and the beginning of EPS production. A consistent increase in cell clustering and matrix production can be seen between day 1–3 images as the biofilm matures. The next stage of growth (iii) is the development of water channel structures and an increase in biofilm height, exhibited in the 3 day and 4 day SEM images were the membrane surface is not visible anymore (Fig. 2B). The 4 day biofilm SEM image shows full maturation (this is stage iv in Fig. 2C) as evidenced by encasement of the cells in EPS, which appears as the glue-like substance between the bacterial cells (Fig. 2B). Mature biofilms are very difficult to eradicate, even with harsh chemical and/or mechanical treatments.49,50 Additionally, clusters from a mature biofilm commonly detach,51 allowing the bacterial cells to spread and establish biofilms elsewhere (this is stage v in Fig. 2C). Based on these results, we chose 4 days of incubation as the time point that would result in a strong, well-developed biofilm and a thick matrix for all subsequent experiments, in an effort to properly demonstrate the biofilm-fighting ability of our phage treatment. We have used a microtiter plate model to quantify PA biofilm formation over time using a well-accepted and simple biomass quantification method, namely the crystal violet method. This method stains all biomass and will thus provide an overall value for the amount of bacteria plus the extracellular matrix at any given time. The crystal violet method, however, could not be applied directly to the DO sensor membranes because of non-specific membrane staining. Therefore, we quantified the biofilm using a microtiter plate model to demonstrate the increase in biomass over time, and used electron microscopy of biofilms grown on the DO sensor membranes to qualitatively confirm the quantitative data obtained using crystal violet method.Next, we determined the required length of time for phage treatment to reduce biofilm levels. We chose 6 h and overnight (16–18 h) as these reflect timepoints commonly found to be effective in our lab for various phage-host pairs. The amount of biofilm remaining on the sensor membranes was quantified before and after phage treatment by staining the biofilm with Hoechst, a deoxyribonucleic acid (DNA)-binding fluorescent dye that can penetrate the bacterial cell wall and stain the bacterial genome.52 As shown in Fig. 3A, a 6 h phage treatment was capable of diminishing the fluorescence signal from stained bacteria, but an overnight treatment was the most effective at reducing the amount of biofilm on the DO membrane.
The fluorescence images from treated and non-treated membranes reflect the quantitative measurements, with an overnight phage treatment resulting in the least amount of staining on the membrane (Fig. 3B). It should be noted that the biofilm EPS is composed of exopolysaccharides, proteins, and extracellular DNA (eDNA),4 and therefore the DNA-binding fluorescent dye can bind to both the eDNA and the DNA inside the bacterial cells. To confirm the trends observed with fluorescence imaging of the membranes, SEM was used to visualize a biofouled membrane following phage treatment. The SEM images show that the phages were able to clear the dense matrix structure and no intact bacterial cells were visible (Fig. 3C). It is noteworthy that we chose a phage concentration of 109 PFU mL−1 mainly to speed up the biofilm removal process. With these particular phages, we have observed in our lab that lower phage concentrations are also effective (106–108 PFU mL−1), but it takes a longer time to achieve similar levels of biofilm dispersal with lower concentration. Should the reader chose to adapt this method to in situ treatment, we recommend that the phage treatment be applied by submerging the sensor in a closed container with phage, regardless of the phage concentration used because the dispersal and thus reduction of phage concentration will prolong treatment. As shown in Fig. 1B, lytic phages self-propagate by lysing bacterial cells and propagating themselves in the process, on site, which results in biofilm reduction over time. There are, however, traces of what appears to be cellular debris on the phage-treated membrane, which is expected to be a result of phage-mediated bacterial lysis. The full removal of the biofilm matrix is an interesting observation because it suggests the involvement of matrix-degrading enzymes. Phages coevolved with bacteria over millions of years and have developed specific strategies to combat biofilms. Some phages carry or express enzymes to degrade extracellular proteins and polysaccharides, key components of a biofilm matrix, and reach the bacterial cells embedded inside.53 Alternatively, some phages can induce such enzymes from within the bacterial genome.53 We would like to highlight the fact that genomes for the two selected phages have been sequenced and reported in the literature (NCBI: txid1141525 and NCBI: txid1141526), but only a fraction of those genes have a known function, with many classified as hypothetical proteins of unknown function. This is a well-known gap in knowledge in the phage field. A more comprehensive bioinformatic analysis would have to be performed on the phage genomes to uncover putative enzymes involved with biofilm degradation. We cannot, therefore, comment on the presence of genes encoding for matrix-degrading enzymes. However, determining appropriate phages for treating environmental biofilms does not necessarily require identification of anti-biofilm genes, but can be done with simple high-throughput assays which assess biofilm reduction in response to phage addition, which we have performed in our lab using our in-house phage library to choose the appropriate phage for this work.
Sensor signal recovery
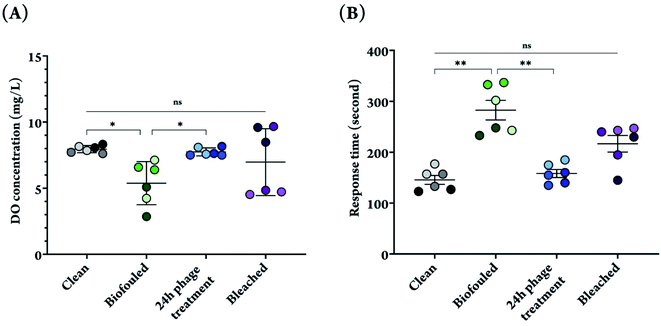
As is evident from the fluorescence microscopy images of phage treated samples, overnight phage treatment led to a significant decrease in the amount of biomass, and successfully removed biofilms from the surface of DO sensor membranes. As a result, these membranes were used for further analysis to measure DO concentrations in DI water, and the resulting sensor signals were compared with clean and biofouled membranes. The sensor used in this study is an electrochemical sensor that contains an electrode, electrolyte and a selectively permeable membrane. This polytetrafluoroethylene (PTFE)-based membrane allows dissolved oxygen in water to pass through and hinders the diffusion of water and other large components, thus playing a central role in the generation of an accurate electrochemical DO signal.54 Once passed through the membrane, diffused oxygen undergoes an electrochemical reaction within the electrolyte solution, producing an electrical signal that is shown in the sensor display.The DO sensor signal decreased from 7.96 ± 0.27 mg L−1 for clean membranes to 5.39 ± 1.63 mg L−1 after growing biofilms for four days on those membranes (32% decrease in signal after biofouling). Biofilm formation prevents proper oxygen diffusion through the small pores on the surface of permeable membranes, therefore significantly decreasing the signal and increasing the noise (note the increase in standard deviation). After incubating the biofouled membranes in the phage suspension overnight, which allowed for biofilm removal from the surface of the membranes, the signal to noise ratio increased significantly (8 fold increase) and allowed for recovery of the absolute DO sensor measurement (7.75 ± 0.30 mg L−1, Fig. 4A). Although some cellular and matrix debris still remained on the surface, results from DO measurements indicate that this did not affect the measurements, and the drastic decrease in amount of biomass following phage treatment worked well to enhance oxygen diffusion through the permeable membrane. In addition, the sensor response time that had increased following the biofilm formation on the membrane was also recovered after phage treatment, decreasing from 282.7 ± 47.12 s for a biofouled membrane to 158.3 ± 19.41 s for a phage treated membrane, which was comparable to the response time of a clean sensor 145.7 ± 21.27 s (Fig. 4B).
On the other hand, when a 10% bleach solution, a common bactericidal chemical, was used to remove biofilms from the surface of DO sensor membranes, a high variability in DO measurement and response time were observed. Although the results do not show significant difference from phage treated membranes, it is not a hidden fact that using chemicals such as bleach have adverse consequences including but not limited to corrosion of metals and painted surfaces, reaction with other chemicals and release of toxic gases and products, which can be extremely harmful to both humans and the environment.55
Dissolved oxygen is one of the pivotal parameters defining the health of water bodies and marine environments. Aquatic creatures rely on dissolved oxygen and they require at least 5 mg L−1 to survive.54 Electrochemical DO sensors, such as the one used for our investigation, are known for accuracy and fast response time, both of which have been significantly affected by biofouling of the sensor membrane, as indicated by our data, clearly demonstrating how biofouling could pose a major challenge for the use of DO sensors in marine environment monitoring. A single phage treatment, however, regenerated the sensor surface and completely recovered the signal to its original value, as well as lowering the response time to that of a clean sensor. This demonstrates the utility of phage for non-invasive cleaning of DO sensors and suggests potential for cleaning of other sensor surfaces that have been subjected to biofouling.
Conclusion
We present an effective method for cleaning biofouled sensors and completely restoring the sensor signal to its original value with no harsh chemicals or mechanical treatments required, which is beneficial for use on delicate membranes and sensitive sensing surfaces. We achieved these results with only a single phage treatment, and without the need for repeated treatments, owing to the self-propagating nature of bacteriophages.56 Phages can replicate on the sensor membrane, as long as they have access to viable host bacteria, allowing for a continuous cycle of infection until the biofilm is cleared and resulting in recovery of the DO sensor function. Aside from being polymicrobial, which would require the use of multiple phages, environmental biofilms typically develop a strong matrix, which would require the use of phage with matrix degrading enzymes. This means that biofilm control using bacteriophages requires a thorough understanding of the candidate bacteriophages. Not all bacteriophages perform equally for biofilm removal and this is an area of active research in our lab and others.The specificity of phages in being able to only infect a few closely related strains of bacteria means they are non-destructive towards beneficial microbes present in the environment in which the sensor is installed,57 unlike harsh cleaning agents. The lack of environmental toxicity means phage treatments could, in principle, be readily applied on site, without the need to remove the sensor from the point at which it was installed. In that case, the phage treatment can be applied by submerging the sensor in a closed container with phage.
It is worth noting that phage-host specificity would require the real-life application of our proposed method to involve a pre-screening for specific lytic phages that can infect the bacteria comprising the biofilm. In this study, we grew monospecies biofilms using a lab strain of bacteria known to be a strong biofilm former as a demonstration of the effectiveness of phage treatment against sensor biofouling. Environmental biofilms are typically a mixture of multiple species,58 which would require the use of more than one type of phage. Finding phages that infect the multispecies environmental biofilm is usually as easy as screening the same aquatic environment for phage, because phages reside in the same niche as their host. In fact, even for therapeutic use in humans, phages are routinely isolated from environmental reservoirs such as ponds, lakes, streams, or even sewage.19 This means that with seasonal changes in the composition of environmental biofilms formed in aquatic environments, we have access to an endless supply of phages that change and evolve with changes in the host population. With time, this would lead to a library of phages that can be used for a certain niche to tackle environmental biofilms on sensors and can be easily screened against new target bacteria. Generating phage preparations for environmental applications is relatively low cost because, unlike therapeutic applications, extensive purification may not be required.
In summary, with the correct selection of phage, bacteriophage treatment could find broad utility in regenerating not only DO or membrane-based sensors, but any sensor surface that is subject to extensive biofouling.
Abbreviations
| DO | Dissolved oxygen |
| EPS | Extracellular polymeric substance |
| PA | Pseudomonas aeruginosa PAO1 |
| DNA | Deoxyribonucleic acid |
| eDNA | Extracellular DNA |
| PBS | Phosphate buffered saline |
| DI water | Deionized water |
| PTFE | Polytetrafluoroethylene |
| SEM | Scanning electron microscopy |
| OD | Optical density |
| PFU mL−1 | Number of plaque forming units per mL |
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. DM and FB contributed equally to the manuscript.Funding sources
ZH and TFD each acknowledge support from NSERC Discovery Grant and McMaster start up funds.Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
Electron microscopy was performed at the Canadian Centre for Electron Microscopy (also supported by NSERC and other government agencies). All illustrations were created with Biorender (https://BioRender.com).References
- H. Flemming, Biofouling and Me: My Stockholm Syndrome with Biofilms, Water Res., 2020, 173, 115576, DOI:10.1016/j.watres.2020.115576.
- J. Malheiro and M. Simões, Antimicrobial Resistance of Biofilms in Medical Devices, in Biofilms and Implantable Medical Devices, Elsevier, 2017, pp. 97–113, 10.1016/B978-0-08-100382-4.00004-6 Search PubMed.
- M. Salta, J. A. Wharton, Y. Blache, K. R. Stokes and J. F. Briand, Marine Biofilms on Artificial Surfaces: Structure and Dynamics, Environ. Microbiol., 2013, 15(11), 2879–2893, DOI:10.1111/1462-2920.12186.
- N. Rabin, Y. Zheng, C. Opoku-Temeng, Y. Du, E. Bonsu and H. O. Sintim, Biofilm Formation Mechanisms and Targets for Developing Anti biofilm Agents, Future Med. Chem., 2015, 7(4), 493–512, DOI:10.4155/fmc.15.6.
- J. M. R. Moreira, R. Fulgêncio, P. Alves, I. Machado, I. Bialuch, L. F. Melo, M. Simões and F. J. Mergulhão, Evaluation of SICAN Performance for Biofouling Mitigation in the Food Industry, Food Control, 2016, 62, 201–207, DOI:10.1016/j.foodcont.2015.10.023.
- T. Nguyen, F. A. Roddick and L. Fan, Biofouling of Water Treatment Membranes: A Review of the Underlying Causes, Monitoring Techniques and Control Measures, Membranes, 2012, 2(4), 804–840, DOI:10.3390/membranes2040804.
- S. Cao, J. D. Wang, H. S. Chen and D. R. Chen, Progress of Marine Biofouling and Antifouling Technologies, Chin. Sci. Bull., 2011, 56(7), 598–612, DOI:10.1007/s11434-010-4158-4.
- A. Whelan and F. Regan, Antifouling Strategies for Marine and Riverine Sensors, J. Environ. Monit., 2006, 8(9), 880–886, 10.1039/b603289c.
- P. Henderson, Fouling and Antifouling in Other Industries-Power Stations, Desalination Plants-Drinking Water Supplies and Sensors, in Biofouling, Wiley-Blackwell, Oxford, UK, 2010, pp. 288–305. DOI:10.1002/9781444315462.ch20.
- L. Delauney, C. Compare and M. Lehaitre, Biofouling Protection for Marine Environmental Sensors, Ocean Sci., 2010, 6(2), 503–511, DOI:10.5194/os-6-503-2010.
- M. Lehaitre, C. Compère, Biofouling and Underwater Measurements. In Real-Time Coastal Observing Systems for Marine Ecosystem Dynamics and Harmful Algal Blooms: Theory, instrumentation and modelling, 2008, pp. 463–493 Search PubMed.
- M. Grzegorczyk, S. J. Pogorzelski, A. Pospiech and K. Boniewicz-Szmyt, Monitoring of Marine Biofilm Formation Dynamics at Submerged Solid Surfaces with Multitechnique Sensors, Front. Mar. Sci., 2018, 5, 1–16, DOI:10.3389/fmars.2018.00363.
- M. Dargahi, Z. Hosseinidoust, N. Tufenkji and S. Omanovic, Investigating Electrochemical Removal of Bacterial Biofilms from Stainless Steel Substrates, Colloids Surf., B, 2014, 117, 152–157, DOI:10.1016/j.colsurfb.2014.02.021.
- C. Whittaker, H. Ridgway and B. H. Olson, Evaluation of Cleaning Strategies for Removal of Biofilms from Reverse-Osmosis Membranes, Appl. Environ. Microbiol., 1984, 48(2), 395–403, DOI:10.1128/aem.48.2.395-403.1984.
- J. Li, K. Hirota, H. Yumoto, T. Matsuo, Y. Miyake and T. Ichikawa, Enhanced Germicidal Effects of Pulsed UV-LED Irradiation on Biofilms, J. Appl. Microbiol., 2010, 109(6), 2183–2190, DOI:10.1111/j.1365-2672.2010.04850.x.
- S. Gora, K. Rauch, C. Ontiveros and G. Gagnon, Ultraviolet Light Emitting Diodes (UV LEDs) for Biofilm Control, 16th Int. Environ. Spec. Conf. 2018, Held as Part Can. Soc. Civ. Eng. Annu. Conf. 2018, 2019, vol. 2017, pp. 226–232 Search PubMed.
- D. Pichardo-Romero, Z. P. Garcia-Arce, A. Zavala-Ramírez and R. Castro-Muñoz, Current Advances in Biofouling Mitigation in Membranes for Water Treatment: An Overview, Processes, 2020, 8(2), 1–22, DOI:10.3390/pr8020182.
- M. Osborne, A. Aryasomayajula, A. Shakeri, P. R. Selvaganapathy and T. F. Didar, Suppression of Biofouling on a Permeable Membrane for Dissolved Oxygen Sensing Using a Lubricant-Infused Coating, ACS Sens., 2019, 4(3), 687–693, DOI:10.1021/acssensors.8b01541.
- R. T. Schooley, B. Biswas, J. J. Gill, A. Hernandez-Morales, J. Lancaster, L. Lessor, J. J. Barr, S. L. Reed, F. Rohwer, S. Benler, A. M. Segall, R. Taplitz, D. M. Smith, K. Kerr, M. Kumaraswamy, V. Nizet, L. Lin, M. D. McCauley, S. A. Strathdee, C. A. Benson, R. K. Pope, B. M. Leroux, A. C. Picel, A. J. Mateczun, K. E. Cilwa, J. M. Regeimbal, L. A. Estrella, D. M. Wolfe, M. S. Henry, J. Quinones, S. Salka, K. A. Bishop-Lilly, R. Young and T. Hamilton, Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails to Treat a Patient with a Disseminated Resistant Acinetobacter Baumannii Infection, Antimicrob. Agents Chemother., 2017, 61(10), 1–15, DOI:10.1128/AAC.00954-17.
- M. Badv, I. H. Jaffer, J. I. Weitz and T. F. Didar, An Omniphobic Lubricant-Infused Coating Produced by Chemical Vapor Deposition of Hydrophobic Organosilanes Attenuates Clotting on Catheter Surfaces, Sci. Rep., 2017, 7(1), 1–10, DOI:10.1038/s41598-017-12149-1.
- H. C. Flemming, J. Wingender, U. Szewzyk, P. Steinberg, S. A. Rice and S. Kjelleberg, Biofilms: An Emergent Form of Bacterial Life, Nat. Rev. Microbiol., 2016, 14(9), 563–575, DOI:10.1038/nrmicro.2016.94.
- F. D'Hérelle, Sur Un Microbe Invisible Antagoniste Des Bacilles Dysentériques, C. R. Acad. Sci., 1917, 165, 373–375 Search PubMed.
- F. D’Hérelle, The Bacteriophage, Its Role in Immunity, Ind. Med. Gaz., 1923, 58(9), 443–444 Search PubMed.
- C. Ferriol-González and P. Domingo-Calap, Phages for Biofilm Removal, Antibiotics, 2020, 9(5), 268, DOI:10.3390/antibiotics9050268.
- W. Ma, M. Panecka, N. Tufenkji and M. S. Rahaman, Bacteriophage-Based Strategies for Biofouling Control in Ultrafiltration: In Situ Biofouling Mitigation, Biocidal Additives and Biofilm Cleanser, J. Colloid Interface Sci., 2018, 523, 254–265, DOI:10.1016/j.jcis.2018.03.105.
- K. E. Kortright, B. K. Chan, J. L. Koff and P. E. Turner, Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria, Cell Host Microbe, 2019, 25(2), 219–232, DOI:10.1016/j.chom.2019.01.014.
- Ł. Richter, M. Janczuk-Richter, J. Niedziółka-Jönsson, J. Paczesny and R. Hołyst, Recent Advances in Bacteriophage-Based Methods for Bacteria Detection, Drug Discovery Today, 2018, 23(2), 448–455, DOI:10.1016/j.drudis.2017.11.007.
- G. P. C. Salmond and P. C. Fineran, A Century of the Phage: Past, Present and Future, Nat. Rev. Microbiol., 2015, 13(12), 777–786, DOI:10.1038/nrmicro3564.
- L. K. Harada, E. C. Silva, W. F. Campos, F. S. Del Fiol, M. Vila, K. Dąbrowska, V. N. Krylov and V. M. Balcão, Biotechnological Applications of Bacteriophages: State of the Art, Microbiol. Res., 2018, 212–213, 38–58, DOI:10.1016/j.micres.2018.04.007.
- Z. Chegini, A. Khoshbayan, M. Taati Moghadam, I. Farahani, P. Jazireian and A. Shariati, Bacteriophage Therapy against Pseudomonas Aeruginosa Biofilms: A Review, Ann. Clin. Microbiol. Antimicrob., 2020, 19(1), 45, DOI:10.1186/s12941-020-00389-5.
- F. L. Gordillo Altamirano and J. J. Barr, Phage Therapy in the Postantibiotic Era, Clin. Microbiol. Rev., 2019, 32(2), 1–25, DOI:10.1128/CMR.00066-18.
- A. S. Bhattacharjee, J. Choi, A. M. Motlagh, S. T. Mukherji and R. Goel, Bacteriophage Therapy for Membrane Biofouling in Membrane Bioreactors and Antibiotic-Resistant Bacterial Biofilms, Biotechnol. Bioeng., 2015, 112(8), 1644–1654, DOI:10.1002/bit.25574.
- S. Ayyaru, J. Choi and Y. H. Ahn, Biofouling Reduction in a MBR by the Application of a Lytic Phage on a Modified Nanocomposite Membrane, Environ. Sci.: Water Res. Technol., 2018, 4(10), 1624–1638, 10.1039/c8ew00316e.
- L. Hall-Stoodley, J. W. Costerton and P. Stoodley, Bacterial Biofilms: From the Natural Environment to Infectious Diseases, Nat. Rev. Microbiol., 2004, 2(2), 95–108, DOI:10.1038/nrmicro821.
- Z. Hosseinidoust, N. Tufenkji and T. G. M. van de Ven, Predation in Homogeneous and Heterogeneous Phage Environments Affects Virulence Determinants of Pseudomonas Aeruginosa, Appl. Environ. Microbiol., 2013, 79(9), 2862–2871, DOI:10.1128/AEM.03817-12.
- Z. Hosseinidoust, N. Tufenkji and T. G. M. van de Ven, Formation of Biofilms under Phage Predation: Considerations Concerning a Biofilm Increase, Biofouling, 2013, 29(4), 457–468, DOI:10.1080/08927014.2013.779370.
- N. Karumidze, J. A. Thomas, N. Kvatadze, M. Goderdzishvili, K. W. Hakala, S. T. Weintraub, Z. Alavidze and S. C. Hardies, Characterization of Lytic Pseudomonas Aeruginosa Bacteriophages via Biological Properties and Genomic Sequences, Appl. Microbiol. Biotechnol., 2012, 94(6), 1609–1617, DOI:10.1007/s00253-012-4119-8.
- N. Sivri, M. Jones and M. J. Allen, Pseudomonas Aeruginosa Isolated From the Marine Environments in the Istanbul Coastal Area (Turkey), Fresenius Environ. Bull., 2014, 23(12B), 1 Search PubMed.
- A. M. Holban, M. C. Chifiriuc, A. I. Cotar, C. Bleotu, A. M. Grumezescu, O. Banu and V. Lazar, Virulence Markers in Pseudomonas Aeruginosa Isolates from Hospitalacquired Infections Occurred in Patients with Underlying Cardiovascular Disease, Rom. Biotechnol. Lett., 2013, 18(6), 7243–7254 Search PubMed.
- S. Wu, G. Liu, W. Jin, P. Xiu and C. Sun, Antibiofilm and Anti-Infection of a Marine Bacterial Exopolysaccharide against Pseudomonas Aeruginosa, Front. Microbiol., 2016, 7, 1–15, DOI:10.3389/fmicb.2016.00102.
- M. R. Green, J. Sambrook, Molecular Cloning: A Laboratory Manual, 2012, vol. 1 Search PubMed.
- G. A. O'Toole, Microtiter Dish Biofilm Formation Assay, J. Visualized Exp., 2010, 47, 10–11, DOI:10.3791/2437.
- Z. Hosseinidoust, N. Tufenkji and T. G. M. van de Ven, Predation in Homogeneous and Heterogeneous Phage Environments Affects Virulence Determinants of Pseudomonas Aeruginosa, Appl. Environ. Microbiol., 2013, 79(9), 2862–2871, DOI:10.1128/AEM.03817-12.
- Z. Hosseinidoust, N. Tufenkji and T. G. M. van de Ven, Formation of Biofilms under Phage Predation: Considerations Concerning a Biofilm Increase, Biofouling, 2013, 29(4), 457–468, DOI:10.1080/08927014.2013.779370.
- Z. Hosseinidoust, T. G. M. van de Ven and N. Tufenkji, Evolution of Pseudomonas Aeruginosa Virulence as a Result of Phage Predation, Appl. Environ. Microbiol., 2013, 79(19), 6110–6116, DOI:10.1128/AEM.01421-13.
- P. Stoodley, K. Sauer, D. G. Davies and J. W. Costerton, Biofilms as Complex Differentiated Communities, Annu. Rev. Microbiol., 2002, 56(1), 187–209, DOI:10.1146/annurev.micro.56.012302.160705.
- T. Rasamiravaka, Q. Labtani, P. Duez and M. El Jaziri, The Formation of Biofilms by Pseudomonas Aeruginosa: A Review of the Natural and Synthetic Compounds Interfering with Control Mechanisms, BioMed Res. Int., 2015, 2015, 1–17, DOI:10.1155/2015/759348.
- C. J. Southey-Pillig, D. G. Davies and K. Sauer, Characterization of Temporal Protein Production in Pseudomonas Aeruginosa Biofilms, J. Bacteriol., 2005, 187(23), 8114–8126, DOI:10.1128/JB.187.23.8114-8126.2005.
- M. Simões, M. O. Pereira and M. J. Vieira, Effect of Mechanical Stress on Biofilms Challenged by Different Chemicals, Water Res., 2005, 39(20), 5142–5152, DOI:10.1016/j.watres.2005.09.028.
- X. Chen and P. S. Stewart, Biofilm Removal Caused by Chemical Treatments, Water Res., 2000, 34(17), 4229–4233, DOI:10.1016/S0043-1354(00)00187-1.
- C. Guilhen, C. Forestier and D. Balestrino, Biofilm Dispersal: Multiple Elaborate Strategies for Dissemination of Bacteria with Unique Properties, Mol. Microbiol., 2017, 105(2), 188–210, DOI:10.1111/mmi.13698.
- W. L. Simmons and K. Dybvig, Biofilms Protect Mycoplasma Pulmonis Cells from Lytic Effects of Complement and Gramicidin, Infect. Immun., 2007, 75(8), 3696–3699, DOI:10.1128/IAI.00440-07.
- A. M. Motlagh, A. S. Bhattacharjee and R. Goel, Biofilm Control with Natural and Genetically-Modified Phages, World J. Microbiol. Biotechnol., 2016, 32(4), 1–10, DOI:10.1007/s11274-016-2009-4.
- L. Hsu, P. R. Selvaganapathy, J. Brash, Q. Fang, C. Q. Xu, M. J. Deen and H. Chen, Development of a Low-Cost Hemin-Based Dissolved Oxygen Sensor with Anti-Biofouling Coating for Water Monitoring, IEEE Sens. J., 2014, 14(10), 3400–3407, DOI:10.1109/JSEN.2014.2332513.
- WHO, Annex G, Use of Disinfectants: Alcohol and Bleach, in Infection Prevention and Control of Epidemic- and Pandemic-Prone Acute Respiratory Infections in Health Care, 2014, pp. 65–66 Search PubMed.
- C. Loc-Carrillo and S. T. Abedon, Pros and Cons of Phage Therapy, Bacteriophage, 2011, 1(2), 111–114, DOI:10.4161/bact.1.2.14590.
- G. P. C. Salmond and P. C. Fineran, A Century of the Phage: Past, Present and Future, Nat. Rev. Microbiol., 2015, 13(12), 777–786, DOI:10.1038/nrmicro3564.
- J. L. Balcázar, J. Subirats and C. M. Borrego, The Role of Biofilms as Environmental Reservoirs of Antibiotic Resistance, Front. Microbiol., 2015, 6, 1–9, DOI:10.3389/fmicb.2015.01216.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra10156g |
| ‡ These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2021 |