Open Access Article
Open Access ArticleApplying molecular networking for targeted isolation of depsipeptides†
Xiao
Lin‡
 abc,
Ling
Chai‡
d,
Hong Rui
Zhu
e,
Yongjun
Zhou
e,
Yaoyao
Shen
e,
Kai Hao
Chen
e,
Fan
Sun
e,
Bu Ming
Liu
d,
Shi Hai
Xu
*b and
Hou Wen
Lin
abc,
Ling
Chai‡
d,
Hong Rui
Zhu
e,
Yongjun
Zhou
e,
Yaoyao
Shen
e,
Kai Hao
Chen
e,
Fan
Sun
e,
Bu Ming
Liu
d,
Shi Hai
Xu
*b and
Hou Wen
Lin
 *e
*e
aInstitute of Marine Drugs, Guangxi University of Chinese Medicine, Nanning 530200, P. R. China
bCollege of Chemistry and Materials, Jinan University, Guangzhou 510632, P. R. China. E-mail: txush@jnu.edu.cn
cCollege of Pharmacy, Jinan University, Guangzhou 510632, P. R. China
dGuangxi Key Laboratory of Traditional Chinese Medicine Quality Standards, Guangxi Institute of Traditional Medical and Pharmaceutical Sciences, Nanning, 530022, P. R. China
eResearch Center for Marine Drugs, State Key Laboratory of Oncogenes and Related Genes, Department of Pharmacy, Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai 200127, P. R. China. E-mail: franklin67@126.com
First published on 13th January 2021
Abstract
LC-HRMS/MS molecular networking enabled the targeted isolation of three new neoantimycin analogs (1, 3, 5) and two known ones (2, 4) from the culture broth of Streptomyces conglobatus RJ8. After derivatization into C1-hydroxyl form compounds (6–10) respectively, the absolute structures of 1–5 were clearly determined by analyzing the hydrolyzed components from 6–10. Compounds 2 and 3 were confirmed to be a pair of epimers with different stereochemistry at C-2, and so were 4 and 5. This is the first report of the isolation and characterization of epimers of NATs. The most abundant eight compounds we obtained were subjected to a cytotoxicity assay, 1 and 6 exhibited excellent cytotoxicity with the lowest IC50 value in the picomolar range against six human carcinoma cell lines while 7 and 8 showed potent cytotoxicity against PC-9 and PC-9/GR cell lines.
Introduction
Neoantimycins (NATs) are a subfamily of antimycin-like depsipeptides characterized by a fifteen-membered tetralactone ring.1 Since NAT-A was first discovered in 1967,2 NATs have received considerable interest for their potent inhibition of tumor cells.3–6 In contrast to antimycins which induce cancer cell apoptosis by inhibiting Bcl2/Bcl-xL-related anti-apoptotic proteins7 or mitochondrial electron transport,8 the anticancer potential of NATs is related with down-regulation of oncogenic proteins GRP78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 and K-Ras mislocalization,3 and some NATs even displayed nanomolar potency for treating drug-resistant cancer.10 Prompted by NATs' therapeutic perspective, chemists have been trying to develop more NATs by biosynthetic engineering11 or chemical synthesis12 and to unravel the molecular interaction of NATs. In light of current trends, biosynthesis of NATs seems to be a more efficient approach than chemical synthesis. By rationally reprogramming and manipulating the biosynthetic pathways, NATs with enhanced biological features can be easily produced that were otherwise inaccessible using traditional synthetic methods.5,13 However, although the knowledge of NATs biosynthesis facilitates the accumulation of NAT derivatives, there are many problems to be resolved after the biosynthesis triggering more congener of NATs: how to discriminate NAT analogs from a plethora of metabolites generated through synthetic biology, and how to navigate the isolation of NAT analogs efficiently. Molecular networking (MN) is apparently an effective tool to solve the aforementioned problems.14 Based on tandem mass spectrometry profiles, the Global Natural Products Social Molecular Networking (GNPS) system organizes hundreds of thousands of MS/MS spectra and associates these spectra according to their similar mass fragmentation patterns to form visible MN maps. This approach thus provides effective and efficient ways to navigate in the metabolome of biological samples by providing key information on analogies among the detected metabolites, thereby accelerating the discovery and characterization process.15,16
9 and K-Ras mislocalization,3 and some NATs even displayed nanomolar potency for treating drug-resistant cancer.10 Prompted by NATs' therapeutic perspective, chemists have been trying to develop more NATs by biosynthetic engineering11 or chemical synthesis12 and to unravel the molecular interaction of NATs. In light of current trends, biosynthesis of NATs seems to be a more efficient approach than chemical synthesis. By rationally reprogramming and manipulating the biosynthetic pathways, NATs with enhanced biological features can be easily produced that were otherwise inaccessible using traditional synthetic methods.5,13 However, although the knowledge of NATs biosynthesis facilitates the accumulation of NAT derivatives, there are many problems to be resolved after the biosynthesis triggering more congener of NATs: how to discriminate NAT analogs from a plethora of metabolites generated through synthetic biology, and how to navigate the isolation of NAT analogs efficiently. Molecular networking (MN) is apparently an effective tool to solve the aforementioned problems.14 Based on tandem mass spectrometry profiles, the Global Natural Products Social Molecular Networking (GNPS) system organizes hundreds of thousands of MS/MS spectra and associates these spectra according to their similar mass fragmentation patterns to form visible MN maps. This approach thus provides effective and efficient ways to navigate in the metabolome of biological samples by providing key information on analogies among the detected metabolites, thereby accelerating the discovery and characterization process.15,16
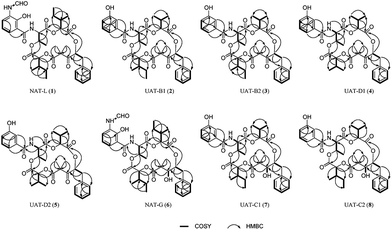
Herein, we report the application of MN led to the isolation and subsequent structure elucidation of five antimycin-like depsipeptides, neoantimycin L (1), unantimycin B1 (2), B2 (3), D1 (4), and D2 (5). 1–5 were reduced by NaBH4 to afford compounds 6–10 (Chart 1), which facilitate the absolute structural depiction of 1–5. Biological evaluation of 1–5, as well as the reduced derivatives (6–8), revealed moderate to potent cytotoxic properties against six cancer cells (human colon cancer cell line, human gastric cancer cell line, and human non-small cell lung cancer cell line as well as their respectively drug-resistant cancer cell lines).
Results and discussion
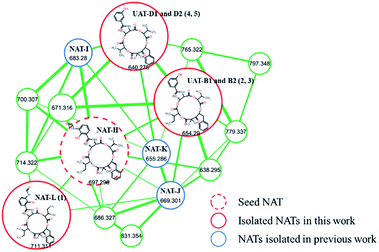
The extract of Streptomyces conglobatu RJ8 was analyzed exhaustively by UPLC-HRMS/MS, and the data were used to generate a visualized molecular networking then annotated by Cytoscape 3.6.1. A cursory search of pronated NAT-H fragment (m/z 697.296) in the global molecular network easily helped us to locate the molecular network of the NAT family, a concentrated cluster with 16 nodes (Fig. 1). The irons at m/z 697.296, m/z 683.280, m/z 669.301, and m/z 655.286 represent four reported NATs (NAT-H, I, J, and K, respectively) which were described previously.10 Ions at m/z 711.311 (1), m/z 654.290 (2 and 3), m/z 640.275 (4 and 5), and other nodes suggest that there are a multitude of NATs differing from the reported ones. Careful analysis of the MS/MS spectra of NATs family revealed that 1–5 yielded the same neutral loss pattern compared with NAT-H. In the initial fragmentation step, the NATs lose a Δm/z 332.16 Da (Δm/z 318.14 in 4 and 5) fragment, next to a loss of H2O, and then a hydroxyl acid residue, at last, a loss of CO (Fig. S1†). Besides, a mass difference of 14.02 Da between 1 and NAT-H hinted at an additional methyl or methylene group present in 1. Thereof, using the annotated molecular network (Fig. 1) as a guide, 1–5 were targeted and isolated through repetitive RP-HPLC and further structural elucidation was carried out.Neoantimycin L (1) was isolated as a pale yellow amorphous solid. Comparable HR-ESI-MS and NMR analyses (Tables 1, S1, S2 and Fig. S2†) with the reported NAT-H permitted the structural assignment of 1 (C37H46N2O12). The mass difference of 14.01 Da between 1 and NAT-H and the additional NMR signals for a methylene group CH2-21 and methyl group CH3-35, resulting in resonances at δH 1.03/δC 23.7 and δH 0.77/δC 11.1 respectively, suggested an additional methylene group present in 1. Further analysis of the 1H–1H COSY and HMBC (Fig. 2) indicated that compound 1 had two units of 2-hydroxy-3-methyl valerate, and it was a methylated congener of NAT-H.
| No. | 1 | No. | 2 | 3 | ||||
|---|---|---|---|---|---|---|---|---|
| δ C | δ H (J in Hz) | δ C | δ H (J in Hz) | δ C | δ H (J in Hz) | |||
| a Carbon resonances are not observed due to signal broadening. | ||||||||
| 1 | 202.6 | 1 | 202.4 | 202.6 | ||||
| 2 | 76.7 | 5.67, dd (7.6, 5.6) | 2 | 76.4 | 5.40, dd (10.0, 2.6) | 76.4 | 5.68, dd (7.7, 5.6) | |
| 3 | 167.0 | 3 | 168.1 | 167.2 | ||||
| 4 | 75.4 | 5.10, d (5.2) | 4 | 75.5 | 5.02, d (3.0) | 75.9 | 5.05, d (5.4) | |
| 5 | 168.0 | 5 | 168.5 | 168.2 | ||||
| 6 | 55.4 | 5.04, br s | 6 | 55.4 | 5.16, dd (9.2, 2.9) | 55.9 | 4.95, dd (8.4, 3.1) | |
| 7 | 70.8 | 5.59, m | 7 | 70.3 | 5.67, qd (6.4, 3.0) | 70.6 | 5.55, m | |
| 8 | 167.7 | 8 | 167.7 | 167.7 | ||||
| 9 | 75.6 | 4.83, d (7.6) | 9 | 75.5 | 5.21, d (8.2) | 75.6 | 4.86, d (7.7) | |
| 10 | 170.9 | 10 | 170.8 | 171.0 | ||||
| 11 | 54.1 | 11 | 55.0 | 54.2 | ||||
| 12 | 37.1 | 3.16, dd (14.2, 5.6) | 12 | 36.6 | 3.16, m | 37.1 | 3.16, dd (14.1, 7.8) | |
| 3.07, dd (14.2, 7.7) | 2.92, m | 3.06, dd (14.1, 7.8) | ||||||
| 13 | 135.4 | 13 | 136.2 | 135.4 | ||||
| 14/18 | 129.6 | 7.20, m | 14/18 | 129.2 | 7.28, overlapped | 129.6 | 7.20, m | |
| 15/17 | 128.3 | 7.30, t (7.5) | 15/17 | 128.5 | 7.33, overlapped | 128.4 | 7.28, overlapped | |
| 16 | 126.9 | 7.25, overlapped | 16 | 126.9 | 7.25, m | 127.0 | 7.25, m | |
| 19 | 36.4 | 1.79, m | 19 | 29.7 | 2.24, m | 30.0 | 2.01, m | |
| 20 | 14.2 | 0.77, overlapped | 20 | 16.1 | 0.79, d (6.9) | 16.8 | 0.69, d (6.8) | |
| 21 | 23.7 | 1.03, m | 21 | 18.3 | 0.91, d (6.9) | 17.9 | 0.83, overlapped | |
| 22 | 169.6 | 22 | 167.9 | 167.8 | ||||
| 23 | 114.8 | 23 | 135.2 | 135.0 | ||||
| 24 | N.o.a | 24 | 114.8 | 7.28, overlapped | 114.7 | 7.28, overlapped | ||
| 25 | 128.3 | 25 | 157.2 | 157.2 | ||||
| 26 | N.o.a | 8.14, m | 26 | 118.5 | 6.94, dd (8.1, 2.4) | 118.5 | 6.95, dd (7.6, 1.8) | |
| 27 | N.o.a | 6.57, br s | 27 | 129.2 | 7.28, overlapped | 129.2 | 7.28, overlapped | |
| 28 | 123.6 | 7.69, br s | 28 | 118.5 | 7.35, overlapped | 118.5 | 7.34, br d (7.8) | |
| 29 | 15.7 | 1.23, d (6.6) | 29 | 16.1 | 1.21, d (6.5) | 15.6 | 1.23, d (6.3) | |
| 30 | 35.7 | 1.88, m | 30 | 36.7 | 1.81, m | 35.7 | 1.88, qd (7.6, 3.6) | |
| 31 | 14.1 | 0.86, d (7.8) | 31 | 13.8 | 0.87, d (6.8) | 14.0 | 0.86, d (6.9) | |
| 32 | 24.0 | 1.43, m | 32 | 24.0 | 1.45, m | 24.0 | 1.43, m | |
| 1.14, m | 1.06, m | 1.13, m | ||||||
| 33 | 21.3 | 1.34, s | 33 | 21.3 | 1.41, s | 21.1 | 1.34, s | |
| 34 | 21.2 | 1.21, s | 34 | 20.7 | 1.32, s | 21.3 | 1.20, s | |
| 35 | 11.1 | 0.77 overlapped | 35 | 10.6 | 0.87, t (6.8) | 10.4 | 0.83, overlapped | |
| 36 | 10.5 | 0.84, t (7.6) | 36 | |||||
| 6-NH | 6-NH | 8.72, d (9.2) | 8.67, d (8.3) | |||||
| 24-OH | 25-OH | 9.66, s | 9.68, s | |||||
| 25-NH | 9.70 br s | |||||||
| CHO | 159.8 | 8.31, d (2.0) | ||||||
Compounds 2 and 3 were isolated as pale yellow amorphous solid. The same adduct ion and the indistinguishable peak signals in HPLC-MS chromatogram hardly made us considered that they were the same substance. The HR-ESI-MS (Table S1†) of 2 gave an adduct ion [M + H]+ at m/z 654.2924, indicating a molecular formula of C35H43NO11 (calcd for 654.2914 [M + H]+) with 15 degrees of unsaturation. After analyzing the 1H NMR, 13C NMR and HSQC spectra (Tables 1, S3 and Fig. S3†), as well as comparison with the reported spectra of unantimycin B,6 the planar structure of 2 was unambiguously assigned. The molecular formula of 3 was also elucidated as C35H43NO11 based on HR-ESI-MS (Table S1†). The 13C NMR (Tables 1, S4 and Fig. S4†) data of 3 were almost identical with those of 2, and the 1H–1H COSY and HMBC (Fig. 2) also suggested that 3 shared the same planar structure with 2, nevertheless, the 1H NMR chemical shift of the backbone between 2 and 3 showed delicate difference (e.g. δH-2 at 5.68 in 3, 5.40 in 2; δH-9 at 4.86 in 3, 5.21 in 2), hinting that 3 would be a stereoisomer of 2. Considering the structural similarities and isomeric diversities, compounds 2 and 3 were named unantimycin B1 and B2, respectively.
Compounds 4 and 5 were also a pair of epimers. The analyses of 1H NMR, 13C NMR, 1H–1H COSY, HSQC, HMBC, and ROESY spectra (Tables 2, S5, S6,†Fig. 2, S5 and S6†) in DMSO-d6 suggested that they are structurally related to 2 and 3 with the same characteristic scaffold except for a 2-hydroxyisovalerate unit tethered at C-9 (2-hydroxy-3-methyl valerate unit in both 2 and 3). 4 and 5 were named unantimycin D1 and D2, respectively.
| No | 4 | 5 | ||
|---|---|---|---|---|
| δ C | δ H (J in Hz) | δ C | δ H (J in Hz) | |
| a Values are interchangeable. | ||||
| 1 | 202.5 | 202.6 | ||
| 2 | 76.4 | 5.37, dd (10.0, 2.6) | 76.4 | 5.69, dd (7.8, 5.6) |
| 3 | 168.1 | 167.2 | ||
| 4 | 75.5 | 5.01, d (2.9) | 76.0 | 5.05, d (5.3) |
| 5 | 168.6 | 168.3 | ||
| 6 | 55.4 | 5.15, overlapped | 55.9 | 4.94, dd (8.3, 3.1) |
| 7 | 70.2 | 5.66, qd (6.4, 4.0) | 70.6 | 5.54, qd (6.4, 4.0) |
| 8 | 167.7 | 167.8 | ||
| 9 | 76.8 | 5.15, overlapped | 76.7 | 4.80, d (7.2) |
| 10 | 170.9 | 171.0 | ||
| 11 | 55.2 | 54.2 | ||
| 12 | 36.6 | 3.15, dd (14.9, 2.7) | 37.2 | 3.15, dd (14.1, 5.6) |
| 2.91, dd (14.9, 2.7) | 3.06, dd (14.0, 7.8) | |||
| 13 | 136.3 | 135.4 | ||
| 14/18 | 129.3 | 7.28, overlapped | 129.6 | 7.20, m |
| 15/17 | 128.6 | 7.34, overlapped | 128.4 | 7.28, overlapped |
| 16 | 127.0 | 7.25, m | 127.0 | 7.25, m |
| 19 | 29.7 | 2.25, m | 30.0 | 2.03, overlapped |
| 20 | 16.1 | 0.80, d (6.9) | 16.8 | 0.69, d (6.8) |
| 21 | 18.3a | 0.92, d (6.9) | 17.6a | 0.82, d (6.9) |
| 22 | 168.0 | 167.6 | ||
| 23 | 135.2 | 135.0 | ||
| 24 | 114.8 | 7.28, overlapped | 114.8 | 7.28, overlapped |
| 25 | 157.2 | 157.3 | ||
| 26 | 118.5 | 6.94, ddd (8.0, 2.5, 1.1) | 118.6 | 6.95, ddd (8.0, 2.6, 1.1) |
| 27 | 129.3 | 7.28, overlapped | 129.3 | 7.28, overlapped |
| 28 | 118.5 | 7.35, overlapped | 118.5 | 7.34, m |
| 29 | 16.2 | 1.21, d (6.4) | 15.7 | 1.23, d (6.5) |
| 30 | 30.7 | 1.99, m | 29.8 | 2.03, overlapped |
| 31 | 17.6a | 0.88, d (8.4) | 17.7a | 0.88, dd (6.8, 2.6) |
| 32 | 18.0a | 0.90, d (8.4) | 17.9a | 0.88, dd (6.8, 2.6) |
| 33 | 21.3 | 1.43, s | 21.3 | 1.34, s |
| 34 | 20.6 | 1.33, s | 21.2 | 1.21, s |
| 6-NH | 8.70, d (9.2) | 8.65, d (8.3) | ||
| 25-OH | 9.70, br s | 9.76, br s | ||
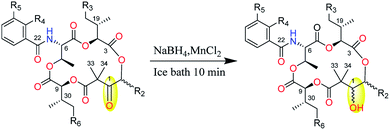
Strong curiosity leads us to further explore the stereochemical difference between compounds 2 and 3, 4 and 5. The configurational establishment of 1–5 required confirming the stereochemistry of each degraded components from 1–5. To prevent isomerization at C-2 the keto group at C-1 was reduced.17 In our previous published work, we found the ketoreductase, NatE, could specifically convert the ketone moiety of NATs to a hydroxyl group in vitro and in vivo,5 and we also successfully applied NatE to facilitate the stereochemical elucidation of two NATs with C-1 keto (NAT-J and K).10 However, this conversion didn't work for 2 and 4 when we intended to adopt the same method. More interestingly, ketone (2) and C1 hydroxyl type (8) of UAT-B were detected simultaneously in the fermentation extract of S. conglobatus RJ2 (a wide type NAT yielding strain but losing conglobatin production), which we previously supposed was attributed to the lower activity of NatE toward the substrates of NAT skeleton containing a 3-HBA moiety at our last work,6 it now seems the reason was not the lower conversion capacity but the stereoselectivity of NatE. Hence, NaBH4 was applied to reduce C1-keto of 1–5 respectively in the presence of MnC12 (Scheme 1), which was reported to give the highest selectivity for the stereoselective reduction of α-allyl β-keto esters into syn α-allyl β-hydroxy esters.18 It turned out as expected that only one main reduced product was afforded for 1–5 respectively during chemical reduction. After purification using the semi-HPLC method, five reduced products, 6–10 (Chart 1), from 1–5 correspondingly were obtained.
Compound 6 converted from 1 were confirmed to be the known NAT-G by comparison of OR and NMR (Tables 3, S7 and Fig. S7†) with those previously reported data.3 Compound 7 and 8, converted from 2 and 3 respectively, have different retention characteristics (for 7tR = 12.3 min; for 8tR = 9.0 min in 75% aqueous acetonitrile with 0.1% formic acid) on the chromatogram but share the same molecular formula as C35H45NO11 based on HR-ESI-MS (Table S1†). The spectroscopic values of 8 closely matched those reported in our previous work,6 which confirmed the stereoselectivity of NatE and explained the co-appearance of C1 hydroxyl and keto form of NAT in the broth of RJ2. After analyzing and comparing their 1H NMR, 13C NMR, HSQC and HMQC spectra (Tables 3, S8, S9, Fig. S8 and S9†) with those of 2 and 3, the structures of 7–8 were deduced to be C-1 hydroxyl form of 2–3 and allocated the names unantimycin C1 and C2 respectively. The structure of 9 and 10 converted from 4 and 5 respectively were deduced by HR-ESI-MS (Table S1†) and 1H-NMR data (ESI Fig. S10 and S11†), and named unantimycin E1 and E2 respectively.
| No. | 6 | No. | 7 | 8 | |||
|---|---|---|---|---|---|---|---|
| δ C | δ H (J in Hz) | δ C | δ H (J in Hz) | δ C | δ H (J in Hz) | ||
| a Values are interchangeable. | |||||||
| 1 | 77.8 | 3.29, d (10.5) | 1 | 75.8 | 3.48, overlapped | 77.9 | 3.29, d (10.6) |
| 2 | 71.6 | 5.45, dd (10.2, 4.6) | 2 | 76.7 | 5.42, t (7.0) | 71.8 | 5.38, dd (10.4, 4.4) |
| 3 | 167.6 | 3 | 168.4a | 168.1 | |||
| 4 | 75.7 | 5.34, d (3.7) | 4 | 76.9 | 5.17, d (4.6) | 75.4 | 5.29, d (3.4) |
| 5 | 167.6 | 5 | 168.5a | 167.8 | |||
| 6 | 55.3 | 5.14, dd (8.7, 3.3) | 6 | 55.9 | 5.15, dd (9.0, 3.1) | 55.5 | 5.05, dd (9.1, 3.4) |
| 7 | 70.7 | 5.56, m | 7 | 70.7 | 5.80, qd (6.4, 2.9) | 71.0 | 5.50, qd (6.4, 3.4) |
| 8 | 168.1 | 8 | 170.4 | 168.1 | |||
| 9 | 74.6 | 4.57, d (8.3) | 9 | 74.9 | 5.01, d (7.9) | 74.4 | 4.57, d (8.6) |
| 10 | 174.8 | 10 | 174.2 | 174.9 | |||
| 11 | 45.4 | 11 | 45.4 | 45.3 | |||
| 12 | 39.2 | 3.04, dd (14.2, 10.2) | 12 | 37.3 | 3.04, dd (13.6, 5.7) | 39.2 | 3.04, dd (14.0, 10.4) |
| 2.97, dd (14.2, 4.6) | 2.90, dd (13.7, 8.3) | 2.95, dd (14.0, 4.4) | |||||
| 13 | 137.5 | 13 | 137.4 | 137.6 | |||
| 14/18 | 129.0 | 7.22, overlapped | 14/18 | 129.3 | 7.29, overlapped | 129.1 | 7.23, overlapped |
| 15/17 | 128.3 | 7.28, m | 15/17 | 128.4 | 7.30, overlapped | 128.3 | 7.28, overlapped |
| 16 | 126.4 | 7.20, overlapped | 16 | 126.4 | 7.20, overlapped | 126.4 | 7.20, overlapped |
| 19 | 36.6 | 1.50, m | 19 | 31.5 | 1.99, overlapped | 30.3 | 1.70, m |
| 20 | 14.4 | 0.60, overlapped | 20 | 17.0 | 0.67, d (7.0) | 15.9 | 0.30, d (6.9) |
| 21 | 22.9 | 0.8, m | 21 | 18.2 | 0.67, d (7.0) | 18.4 | 0.67, d (6.9) |
| 22 | 170.0 | 22 | 167.6 | 167.8 | |||
| 23 | 114.7 | 23 | 135.1 | 135.2 | |||
| 24 | N.o.a | 24 | 114.7 | 7.29, overlapped | 114.8 | 7.28, overlapped | |
| 25 | 127.4 | 25 | 157.3 | 157.2 | |||
| 26 | N.o.a | 8.19, d (7.8) | 26 | 118.4a | 6.96, dd (8.1, 2.4) | 118.5 | 6.94, ddd (8.0, 2.5, 1.0) |
| 27 | N.o.a | 6.81, br s | 27 | 128.0 | 7.29, overlapped | 129.2 | 7.28, overlapped |
| 28 | 123.6 | 7.87, d (7.2) | 28 | 118.6a | 7.36, d (7.7) | 118.5 | 7.34, br d (7.7) |
| 29 | 15.6 | 1.21, d (6.4) | 29 | 15.9 | 1.28, d (6.4) | 15.4 | 1.20, d (6.4) |
| 30 | 35.4 | 1.85, m | 30 | 35.8 | 1.93, overlapped | 35.4 | 1.84, m |
| 31 | 14.0 | 0.87, d (7.1) | 31 | 14.7 | 0.92, d (6.9) | 13.9 | 0.86, d (6.9) |
| 32 | 24.3 | 1.50, m | 32 | 24.1 | 1.49, m | 24.2 | 1.49, m |
| 1.17, m | 1.19, m | 1.16, m | |||||
| 33 | 26.2 | 1.31, s | 33 | 23.7 | 1.10, s | 26.3 | 1.30, s |
| 34 | 22.0 | 1.25, s | 34 | 22.6 | 1.32, s | 21.9 | 1.26, s |
| 35 | 11.1 | 0.60 overlapped | 35 | 10.9 | 0.88, t (7.3) | 10.2 | 0.83, t (7.5) |
| 36 | 10.3 | 0.84, d (7.6) | 36 | ||||
| 1-OH | 4.39, d (10.4) | 1-OH | 4.85, br d (10.3) | 4.38, d (10.5) | |||
| 6-NH | 6-NH | 8.77, d (8.9) | 8.65, d (9.1) | ||||
| 24-OH | 12.7, s | 25-OH | 9.67 br, s | ||||
| 25-NH | 9.78 br s | ||||||
| CHO | 159.8 | 8.32, d (1.7) | |||||
The reduced derivatives 6–10 were used to facilitate the configurational assignment of 1–5 according to the reported methods.3,10,17 First, the relative configurations between C-1 and C-2, as well as C-9 and C-30 in each compound were determined from the HETLOC and ROESY assay. The small coupling constant constants for 3J1H–2H (∼0 Hz), 2J1C–2H (∼0 Hz) and 2J2C–1H (∼2 Hz) reasonably depicted the threo configuration between C-1 (R*) and C-2 (R*) (Fig. S12A†). On the other hand, the large 3J9H–30H coupling constant and small 3J9H–31C and 3J9H–32C coupling constants constructed two possible relative configurations of C-9 and C-30. The discrimination between these two possibilities was completed by further analyses of ROESY between the methyl signal of H-29 and that of H-31, which reasonably explained the relative configuration at C-9 and C-30 to be S* and S*, respectively (Fig. S12B†).
In the next, mosher's and Marfey's chromatographic analysis after hydrolysis of 6–10 were applied. Inferred from their planar structures, the microscale hydrolysis of 6–10 would inevitably yield the residues of isoleucic acid (Ila), 2-hydroxyisovaleric acid (Hia), 5-benzyl-4-hydroxy-3,3-dimethyldihydrofuran-2-one (Bhdo), and threonine (Thr). Hence, L-and D-Ila (prepared from L-and D-isoleucine respectively) and (2R)- and (2S)-Hia (purchased from Shanghai yuanye Bio-Technology Co., Ltd) were derivatized with the R enantiomer of Mosher's reagent respectively to yield authentic standards of the (S)-MTPA esters. (4R, 5R)-Bhdo obtained from hydrolysate of NAT-H in our previous work10 were derivatized with (R)- and (S)-MTPA-Cl to yield (S)- and (R)-MTPA esters, respectively, and (R)-MTPA ester of (4R, 5R)-Bhdo was regarded as the chromatographically equivalent surrogate for the (S)-MTPA esters of (4S, 5S)-Bhdo.3 The alkaline hydrolysates of 6–10 were respectively reacted with (R)-MTPA-Cl and subsequently dissolved in MeOH, as were the sample solution. The eleven sets of standards and samples mentioned above were subjected to C18 Mosher analysis, and the results clarified that the absolute configurations of all of the Ila moieties in 6–10 were the L-form (Fig. S13†), and the Hia residues were confirmed as S configuration (Fig. S13†). Meanwhile, Bhdo residues in 7 and 9 were determined to be 4S and 5S, 6, 8 and 10 incorporate the (4R, 5R)-Bhoa (Fig. S14†). In addition, the Thr residues in 6–10 were confirmed by advanced Marfey's analysis. L-threonine and L-allo-threonine were reacted with L- and D-FDLA (1-fluoro-2,4-dinitrophenyl-5-leucinamide), respectively. The retention times of the acid hydrolysates of 6–10 derivatized with L-FDLA were consisted with that of the L-FDLA L-threonine conjugates (m/z 412 [M–H]−), indicating the presence of L-threonine (Fig. S15†).
Therefore, the absolute configurations of 1–10 were eventually determined as shown in Chart 1: 1 as 2R, 4S, 6S, 7R, 9S, 19S, 30S; 2 and 4 as 2S, 4S, 6S, 7R, 9S, 30S; 3 and 5 as 2R, 4S, 6S, 7R, 9S, 30S; 6 as 1R, 2R, 4S, 6S, 7R, 9S, 19S, 30S; 7 and 9 as 1S, 2S, 4S, 6S, 7R, 9S, 30S; 8 and 10 as 1R, 2R, 4S, 6S, 7R, 9S, 30S.
Considering most of the NATs exhibited outstanding and broadspectrum tumor cell inhibitory activities, 1–8 were evaluated for their in vitro cytotoxicity. CCK8 bioassays were performed with the human gastric cancer cell line SGC7901, the human colon cancer cell line HCT-8, the human non-small cell lung cancer cell line PC-9, as well as their respective drug-resistant cancer cells SGC7901/DDP (resistant to cisplatin), HTC-8/T (resistant to taxol) and PC-9/GR (resistant to gefitinib). The cells were placed in 96-well plates and treated with various concentrations of the compounds for 72 h with cisplatin, taxol, and gefitinib as positive control respectively. The results (Table 4) revealed that the compounds featuring 3-formamidosalicylate unit, like 1 and 6, showed extraordinary anti-proliferative activities against all six cell lines with half-maximal inhibitory concentration (IC50) values in the range from 0.02 to 9690.0 nM. Though compounds 2–5, 7 and 8 exhibited much weaker cytotoxicity than 1 and 6, they still displayed potent and efficacious activity, especially 7 and 8 against the human non-small cell lung cancer cell lines (PC-9 and PC-9/GR) with IC50 in the range of 122.6–4228.0 nM. Besides, it may be inferred that the stereochemistry at C-1 and C-2 also plays an important role, which was reflected by the IC50 values between three pairs of diastereoisomers (2vs.3, 4vs.5, 7vs.8) against the PC-9 and PC-9/GR. Furthermore, the compounds with hydroxyl at C-1 (6–8) exhibited stronger bioactivity than their respective C-1 keto type (1–3), reconfirming the importance of the hydroxyl at C-1 position, which was demonstrated previously.10
| Compd | IC50 (nM) | ||||||
|---|---|---|---|---|---|---|---|
| SGC7901 | SGC7901/DDP | HCT-8 | HCT-8/T | PC-9 | PC-9/GR | NCM460 | |
| a Cisplatin, taxol, and gefitinib served as positive controls. | |||||||
| 1 | 1.5 ± 0.2 | 1.7 ± 0.2 | 30.3 ± 1.1 | 9690.0 ± 47.0 | 30.8 ± 1.0 | 0.5 ± 0.0 | >50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 2 | 16663.3 ± 483.3 | >20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
4091.0 ± 48.1 | 9126.3 ± 189.0 | >50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 3 | 17116.7 ± 432.9 | >20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
7866.3 ± 63.0 | 11560.0 ± 835.9 | >50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 4 | >20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
5101.0 ± 212.3 | 9614.5 ± 623.8 | >50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 5 | >20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
13821.3 ± 505.8 | 17960.1 ± 817.9 | >50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 6 | 0.02 ± 0.01 | 1.0 ± 0.1 | 11.8 ± 0.1 | 1036.7 ± 17.8 | 5.1 ± 0.1 | 779.3 ± 130.1 | >50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 7 | 9947.7 ± 353.2 | 18726.7 ± 790.3 | 18708.6 ± 376.6 | 13633.3 ± 357.2 | 122.6 ± 4.3 | 196.2 ± 2.0 | >50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| 8 | 14983.3 ± 573.9 | 17103.3 ± 285.0 | 12266.7 ± 305.3 | 19256.7 ± 249.9 | 613.6 ± 7.3 | 4228.0 ± 82.3 | >50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| cisplatina | 927.0 ± 77.6 | 5028.7 ± 198.7 | — | — | — | — | 4799.3 ± 230.9 |
| taxola | — | — | 4603.4 ± 421.6 | >20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
— | — | — |
| gefitiniba | — | — | — | — | 86.1 ± 3.0 | 8994.0 ± 105.7 | — |
Experimental
General experimental procedures
Optical rotations were measured on an Autopol VI, Serial #90079 polarimeter; UV spectra were measured in MeOH on a Persee Tu-1950 spectrophotometer. NMR spectra were recorded on a Brucker AVANCE III HD 600 MHz NMR spectrometer. Chemical shifts are reported in parts per million (δ) using the residual DMSO-d6 signals (δH 2.50; δC 39.52) as internal standards, and coupling constants (J) are reported in Hz. The high-resolution mass spectra (HR-ESI-MS) were recorded on a Thermo Scientific Q Exactive Focus instrument with a Hypersil GOLD column (100 × 2.1 mm i.d.; 1.9 μm). Preparative HPLC and semi-preparative HPLC were performed respectively on YMC-Pack Pro C18 RS (20 × 250 mm, 5 μm) column and Waters Xbridge Prep C18 (10 × 250 mm, 5 μm) column, coupled with a Waters 1525 separation module and a Waters 2998 photodiode array detector.Fermentation, extraction, and isolation
S. conglobatus RJ8 was initially grown in 50 mL TSBY medium (3% tryptone soy broth, 10.3% sucrose, 0.5% yeast extract) at 30 °C and 200 rpm to produce spore suspension. The suspension (5%) was added to the seed medium (SGCC medium) and then the production medium (SGC medium), each containing 5% glucose (autoclaved separately), 3% soybean flour (the supernatant after first autoclaving was used), 0.5% CaCO3, 0.2% (v/v) anti-foam, and 5 mg L−1 CoCl2·6H2O (omitted in the SGC medium). Fermentation was performed by inoculating 150 mL medium in a 500 mL conical flask fitted with a metal spring at 30 °C, 220 rpm for three days (for seed culture) and five days (for production culture), respectively.The production culture broth (12 L) was adjusting pH to 6 with formic acid before extracted with an equal volume of EtOAc. The EtOAc layer was concentrated under vacuum to afford an extract (8.1 g), which was then dissolved in methanol and degreased with hexane three times. The entire MeOH-soluble extract (6.6 g) was subjected to vacuum liquid chromatography (VLC) on silica gel (200–300 mesh) using a stepwise gradient elution of dichloromethane–MeOH (from 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 0
1 to 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) to yield six fractions (B1–B6). Fraction B1 (2.5 g) was subsequently passed through an ODS chromatography column eluted with a gradient of aqueous acetonitrile (from 30% to 100%) to give fourteen fractions (B1A–B1N). Fraction B1J (1.2 g) was further fractionated over a silica flash column eluted with petroleum ether (PE)–EtOAc–MeOH (from 8
1, v/v) to yield six fractions (B1–B6). Fraction B1 (2.5 g) was subsequently passed through an ODS chromatography column eluted with a gradient of aqueous acetonitrile (from 30% to 100%) to give fourteen fractions (B1A–B1N). Fraction B1J (1.2 g) was further fractionated over a silica flash column eluted with petroleum ether (PE)–EtOAc–MeOH (from 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 to 0
0 to 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0
0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v) to give ten subfractions (B1J1-J7). Subfraction B1J5 was further purified by semi-preparative HPLC (Waters Xbridge C18, 10 × 250 mm, 3 mL min−1) under isocratic conditions using 70% aqueous acetonitrile with 0.1% formic acid as the buffer to yield neoantimycin L (1) (7.9 mg, tR = 23.6 min). Subfraction B1J6 (0.7 g) was subjected to a preparative HPLC (YMC-Park C18, 20 × 250 mm, 5 μm, 8 mL min−1) and a subsequent semi-preparative HPLC (Waters Xbridge C18, 10 × 250 mm, 3 mL min−1) to afford unantimycin B1 (2) (11.3 mg, tR = 25.6 min), B2 (3) (10.1 mg, tR = 26.7 min), unantimycin D1 (4) (3.2 mg, tR = 23.2 min), and unantimycin D2 (5) (3.6 mg, tR = 24.6 min) using 65% aqueous acetonitrile with 0.1% formic acid.
1, v/v/v) to give ten subfractions (B1J1-J7). Subfraction B1J5 was further purified by semi-preparative HPLC (Waters Xbridge C18, 10 × 250 mm, 3 mL min−1) under isocratic conditions using 70% aqueous acetonitrile with 0.1% formic acid as the buffer to yield neoantimycin L (1) (7.9 mg, tR = 23.6 min). Subfraction B1J6 (0.7 g) was subjected to a preparative HPLC (YMC-Park C18, 20 × 250 mm, 5 μm, 8 mL min−1) and a subsequent semi-preparative HPLC (Waters Xbridge C18, 10 × 250 mm, 3 mL min−1) to afford unantimycin B1 (2) (11.3 mg, tR = 25.6 min), B2 (3) (10.1 mg, tR = 26.7 min), unantimycin D1 (4) (3.2 mg, tR = 23.2 min), and unantimycin D2 (5) (3.6 mg, tR = 24.6 min) using 65% aqueous acetonitrile with 0.1% formic acid.
Mass spectrometry analysis
The analysis of extract was performed on an Ultimate 3000 UHPLC system connected to a Thermo Scientific Q Exactive Focus mass spectrometer equipped with an electrospray ionization source. The spray voltage was set at 3.5 kV; the sheath gas flow rate (N2) at 40 units; the capillary temperature at 350 °C; the S lens RF level at 50; and the probe heater temperature at 300 °C. A 2 μL amount of sample solution was injected on a Hypersil GOLD column (100 × 2.1 mm i.d.; 1.9 μm) using a gradient from 30% to 100% MeCN with 0.1% formic acid in 15 min, followed by an isocratic washing step at 100% MeCN for 2 min. After the washing step, the column was re-equilibrated with 30% MeCN for 3 min prior to the next injection. The flow rate was set at 0.2 mL min−1, the column temperature at 25 °C.Molecular networking
The MeOH extract of S. conglobatus RJ8 and the fractions thereof were analyzed with UPLC-HRMS/MS. The .RAW files were converted to .mzXML format using MSConvert GUI of Proteowizzard19 and uploaded to CCMS (ccms-ftp01.ucsd.edu) A molecular network was established using the online workflow at GNPS (gnps.ucsd.edu). The data were then clustered with MS-cluster with a parent mass tolerance of 0.02 Da and an MS/MS fragment ion tolerance of 0.02 Da to create consensus spectra. Consensus spectra that contained less than two spectra were discarded. A network was then created where edges were filtered to have a cosine score above 0.7 and more than six matched peaks. Further edges between two nodes were kept in the network if and only if each of the nodes appeared in each other's respective top 10 most similar nodes. The spectra in the network were then searched against the spectral libraries of GNPS. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least six matched peaks. Results were visualized in Cytoscape 3.6.1 (www.cytoscape.org) and guided the isolation of 1–5.The conversion and purification of compounds 6–10
The reduction of the ketone moiety in 1–5 was carried out as follows. Manganese chloride (0.012 mmol) was added to a methanol (300 μL) solution of 1 (0.006 mmol) at 0 °C, NaBH4 (0.06 mmol) was slowly added under an ice bath. After stirred under ice bath for 10 min, the reaction mixture was poured into 1 M HCl and extracted with ethyl acetate (3 mL) three times. The combined organic layer was dried under a stream of N2 gas, redissolved in MeOH and purified by semi-prep HPLC (Waters Xbridge C18, 10 × 250 mm, 3 mL min−1) using 70% aqueous acetonitrile with 0.1% formic acid to afford 6 (3.5 mg, tR = 19.2 min). Analogously, 7 (3.5 mg, tR = 12.3 min in 75% aqueous acetonitrile with 0.1% formic acid), 8 (2.6 mg, tR = 9.0 min in 75% aqueous acetonitrile with 0.1% formic acid), 9 (less than 1.0 mg, tR = 10.8 min in 70% aqueous acetonitrile with 0.1% formic acid) and 10 (less than 1.0 mg, tR = 7.9 min in 75% aqueous acetonitrile with 0.1% formic acid) were converted and purified from 2–5 by the same procedure, respectively.Characterization of compounds 1–8
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 221 (4.38), 334 (3.61) nm; NMR data see Table S2 and Fig. S2;† HRESIMS m/z 711.3106 [M + H]+ (calcd for C37H47N2O12, 711.3129).
ε) 221 (4.38), 334 (3.61) nm; NMR data see Table S2 and Fig. S2;† HRESIMS m/z 711.3106 [M + H]+ (calcd for C37H47N2O12, 711.3129).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 209 (4.43), 291 (3.28) nm; NMR data see Table S3 and Fig. S3;† HRESIMS m/z 654.2924 [M + H]+ (calcd for C35H44NO11, 654.2914).
ε) 209 (4.43), 291 (3.28) nm; NMR data see Table S3 and Fig. S3;† HRESIMS m/z 654.2924 [M + H]+ (calcd for C35H44NO11, 654.2914).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 209 (4.35), 291 (3.31) nm; NMR data see Table S4 and Fig. S4;† HRESIMS m/z 654.2925 [M + H]+ (calcd for C35H44NO11, 654.2914).
ε) 209 (4.35), 291 (3.31) nm; NMR data see Table S4 and Fig. S4;† HRESIMS m/z 654.2925 [M + H]+ (calcd for C35H44NO11, 654.2914).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 209 (4.41), 291 (3.25) nm; NMR data see Table S5 and Fig. S5;† HRESIMS m/z 640.2747 [M + H]+ (calcd for C34H42NO11, 640.2758).
ε) 209 (4.41), 291 (3.25) nm; NMR data see Table S5 and Fig. S5;† HRESIMS m/z 640.2747 [M + H]+ (calcd for C34H42NO11, 640.2758).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 209 (4.42), 291 (3.27) nm; NMR data see Table S6, and Fig. S6;† HRESIMS m/z 640.2743 [M + H]+ (calcd for C34H42NO11, 640.2758).
ε) 209 (4.42), 291 (3.27) nm; NMR data see Table S6, and Fig. S6;† HRESIMS m/z 640.2743 [M + H]+ (calcd for C34H42NO11, 640.2758).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 223 (4.28), 331 (3.51) nm; NMR data see Table S7 and Fig. S7;† HRESIMS m/z 713.3262 [M + H]+ (calcd for C37H49N2O12, 713.3286).
ε) 223 (4.28), 331 (3.51) nm; NMR data see Table S7 and Fig. S7;† HRESIMS m/z 713.3262 [M + H]+ (calcd for C37H49N2O12, 713.3286).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 209 (4.29), 292 (2.91) nm; NMR data see Table S8, and Fig. S8;† HR-MS m/z 656.3069 [M + H]+ (calcd for C35H46NO11, 656.3071).
ε) 209 (4.29), 292 (2.91) nm; NMR data see Table S8, and Fig. S8;† HR-MS m/z 656.3069 [M + H]+ (calcd for C35H46NO11, 656.3071).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) 209 (4.36), 292 (3.16) nm; NMR data see Table S9, and from Fig. S9;† HR-MS m/z 656.3067 [M + H]+ (calcd for C35H46NO11, 656.3071).
ε) 209 (4.36), 292 (3.16) nm; NMR data see Table S9, and from Fig. S9;† HR-MS m/z 656.3067 [M + H]+ (calcd for C35H46NO11, 656.3071).
Preparation of MTPA-esters of 5-benzyl-4-hydroxy-3,3-dimethyldihydrofuran-2-one (Bhdo)
The (4R, 5R)-Bhdo (3.4 mg) residue was obtained from the alkali hydrolysate of NAT-H which was reported previously.9 Following protocol published by Hoye et al.,17 the Bhdo residue subsequently reacted with either (S)- or (R)- MTPA-Cl to give the corresponding (R)- or (S)- MTPA-ester, respectively.Preparation of MTPA-esters of isoleucic acid (Ila)
To a solution of L-isoleucine (200 mg) in 1 M H2SO4 (4 mL), an aqueous solution of NaNO2 (0.2 g mL−1, 4 mL) was slowly added and stirred for 2 h in an ice bath. After further stirred for 16 h at room temperature, the reaction mixture was extracted with EtOAc (4 mL × 3), The EtOAc layer was combined and washed with water (2 × 2 mL) and brine (2 × 2 mL) consecutively, then dried over anhydrous Na2SO4. The organic solvent was concentrated to afford L-Ila (colorless oil, 136 mg). Analogously, D-Ila were converted from D-isoleucine. The obtained L-Ila and D-Ila were subsequently reacted with (R)-MTPA-Cl following the protocol published by Hoye et al.20 to give the (S)-MTPA-ester of L- and D-Ila, respectively.Preparation of MTPA-esters of 2-hydroxyisovaleric acid (Hia)
(R) and (S)-2-hydroxyisovaleric acid were purchased from Shanghai yuanye Bio-Technology Co., Ltd, and reacted with (R)-MTPA-Cl following the protocol published by Hoye et al.20 to give the (S)-MTPA-ester of (2R)- and (2S)-Hia, respectively.Mosher analysis of 6–10
Compound 6–10 (100 μg) were dissolved in 3 M NaOH (100 μL) and heated to 95 °C in a sealed vial for 10 min, after which 6 M HCl (50 μL) was added to adjust pH to 2–3, then extracted with EtOAc (250 μL) three times. The combined organic layer was dried under a stream of N2 and then reacted with (R)-MTPA-Cl to produce (S)-MTPA ester derivatives. The (R)- or (S)-MTPA esters from Bhdo, the (S)-MTPA esters from Hma and Hia were adopted as reference substances to determine the absolute configuration of 6–10 using UPLC-HRMS analysis (Hypersil GOLD C18 column 100 × 2.1 mm i.d.; 1.9 μm, 0.3 mL min−1, 55% MeCN/H2O in 15 min with 0.1% formic acid).Marfey's analysis of 6–10
Compounds 6–10 (50 μg) dissolved in 300 μL of 6 M HCl were hydrolyzed at 110 °C in a sealed vial for 24 h, whereafter, the HCl was removed by evaporation under a stream of N2 gas. The hydrolysates were dissolved in 40 μL of 1 M NaHCO3 and thereafter reacted with 50 μL of Marfey's reagent (1-fluoro-2,4-dinitrophenyl-5-L-alanine amide, FDAA; 1% solution in acetone) at 40 °C for 1 h, after which, the mixture was neutralized with 1 M HCl, diluted with MeCN (800 μL) and filtered (0.45 μm PTFE) before subjected to HPLC-HRMS analysis (Hypersil GOLD C18 column 100 × 2.1 mm i.d.; 1.9 μm, 0.3 mL min−1) with linear gradient elution (30–100% solvent B in 20 min; solvent A: H2O with 0.1% formic acid, solvent B: 100% MeCN). Authentic samples of the amino acids L-Thr and L-allo-Thr were derivatized with L-FDAA and D-FDAA and analyzed by LC-HRMS under the same chromatographic condition.In Vitro cytotoxicity test
The Cell Counting Kit-8 (CCK-8) method was used for in vitro evaluation of the cytotoxicity of each obtained compound against six human cancer cell lines (SGC7901, SGC7901/DDP, HCT-8, and HCT-8/Taxinol, PC-9 and PC-9/GR) and noncancerous NCM460 colon cell line. In brief, the compounds dissolved in dimethyl sulfoxide (DMSO) were diluted with culture media to prepare serial concentrations. Each cell line was inoculated into standard 96-well, flat-bottom microplates, and incubated with DMEM (for HCT-8, PC-9/GR, and NCM460) or RPMI-1640 (for HCT-8/T, SGC7901, SGC7901/DDP, and PC-9) medium for 24 h at 37 °C in a humidified atmosphere containing 5% CO2. The adherent cells (100 μL) were then incubated with serially diluted compounds. After continuous exposure to the tested compounds at various concentrations for 72 h, 10 μL of CCK-8 solution was added and the absorbance was measured at 450 nm using a microtiter plate reader (SpectraMax 190, Molecular Devices). Cisplatin, taxol, and gefitinib (purity ≥98% Sigma, USA) were used as positive controls. All studies were performed in triplicate and the IC50 values were calculated using Prism software.Conclusions
In conclusion, a method of GNPS molecular networking for the detection and targeted isolation of antimycin-like depsipeptides is established in this study. Based on the molecular network, three new neoantimycin analogs (1, 3, 5) and two known ones (2, 4) were isolated and identified. Compounds 2 and 3 are a pair of epimer with a different configuration at C-2, and so are 4 and 5. This is the first time that the NATs with different absolute stereochemistry were isolated and confirmed. The reduced products (6–10) derived from 1–5 respectively by chemical conversion were used to complete the stereochemical assignment. In addition, the most abundant eight compounds (1–8) we obtained were evaluated in vitro assay and demonstrated a broad range of cytotoxic activities against six human cancer cell lines without inhibitory against the normal cell. It's also worth noting that 1 and 6 exhibited excellent cytotoxicity with IC50 values of 0.02–9690 nM against six human carcinoma cell lines, while 7 and 8 showed potent selective cytotoxic against PC-9 and PC-9/GR cell lines with IC50 value between 122.6 and 4228.0 nM. This work has expanded the NATs family and enrich the structure–activity relationship information about NATs.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by National Natural Science Foundation of China (No. 31670096, U1605221, 21672084, 41876145), Guangzhou Science and Technology Project (No. 201704030042), Guangxi Key Laboratory of Traditional Chinese Medicine Quality Standards Open Grant (guizhongzhongkai201603), Innovative Research Team of High-Level Local Universities in Shanghai, Construction project of Shanghai Key Laboratory of Molecular Imaging (No. 18DZ2260400), the Taishan Scholar Project from Shandong Province to H. L. and Key Program of Marine Economy Development (Six Marine Industries) Special Foundation of Department of Natural Resources of Guangdong Province (No. [2020]037).References
- J. Liu, X. Zhu, S. J. Kim and W. Zhang, Nat. Prod. Rep., 2016, 33, 1146–1165 RSC.
- G. Cassinelli, A. Grein, P. Orezzi, P. Pennella and A. Sanfilippo, Arch. Mikrobiol., 1967, 55, 358–368 CAS.
- A. A. Salim, K.-J. Cho, L. Tan, M. Quezada, E. Lacey, J. F. Hancock and R. J. Capon, Org. Lett., 2014, 16, 5036–5039 CrossRef CAS PubMed.
- C. L. Lim, T. Nogawa, A. Okano, Y. Futamura, M. Kawatani, S. Takahashi, D. Ibrahim and H. Osada, J. Antibiot., 2016, 69, 456–458 CrossRef CAS PubMed.
- Y. Zhou, X. Lin, S. R. Williams, L. Liu, Y. Shen, S.-P. Wang, F. Sun, S. Xu, H. Deng, P. F. Leadlay and H.-W. Lin, ACS Chem. Biol., 2018, 13, 2153–2160 CrossRef CAS PubMed.
- Y. Shen, F. Sun, L. Zhang, Y. Cheng, H. Zhu, S.-P. Wang, W.-H. Jiao, P. F. Leadlay, Y. Zhou and H.-W. Lin, J. Biol. Chem., 2020 DOI:10.1074/jbc.RA119.010922.
- P. S. Schwartz, M. K. Manion, C. B. Emerson, J. S. Fry, C. M. Schulz, I. R. Sweet and D. M. Hockenbery, Mol. Cancer Ther., 2007, 6, 2073–2080 CrossRef CAS PubMed.
- M. A. King and M. A. Radicchi-Mastroianni, Cytometry, 2002, 49, 106–112 CrossRef CAS PubMed.
- M. Izumikawa, J. Ueda, S. Chijiwa, M. Takagi and K. Shin-ya, J. Antibiot., 2007, 60, 640–644 CrossRef CAS PubMed.
- X. Lin, Y. Zhou, L. Liu, H. Zhu, Y. Chen, S. Wang, F. Sun, L. Chai, B. Liu, S. Xu and H.-W. Lin, Front. Chem., 2019, 7, 481 CrossRef CAS PubMed.
- T. Awakawa, T. Fujioka, L. Zhang, S. Hoshino, Z. Hu, J. Hashimoto, I. Kozone, H. Ikeda, K. Shin-Ya, W. Liu and I. Abe, Nat. Commun., 2018, 9, 3534 CrossRef PubMed.
- S. Manaviazar, P. Nockemann and K. J. Hale, Org. Lett., 2016, 18, 2902–2905 CrossRef CAS PubMed.
- W. Skyrud, J. Liu, D. Thankachan, M. Cabrera, R. F. Seipke and W. Zhang, ACS Chem. Biol., 2018, 13, 1398–1406 CrossRef CAS PubMed.
- R. Reher, M. Kuschak, N. Heycke, S. Annala, S. Kehraus, H.-F. Dai, C. E. Müller, E. Kostenis, G. M. König and M. Crüsemann, J. Nat. Prod., 2018, 81, 1628–1635 CrossRef CAS PubMed.
- M. Wang, J. J. Carver, V. V. Phelan, L. M. Sanchez, N. Garg, Y. Peng, D. D. Nguyen, J. Watrous, C. A. Kapono, T. Luzzatto-Knaan, C. Porto, A. Bouslimani, A. V. Melnik, M. J. Meehan, W.-T. Liu, M. Crüsemann, P. D. Boudreau, E. Esquenazi, M. Sandoval-Calderón, R. D. Kersten, L. A. Pace, R. A. Quinn, K. R. Duncan, C.-C. Hsu, D. J. Floros, R. G. Gavilan, K. Kleigrewe, T. Northen, R. J. Dutton, D. Parrot, E. E. Carlson, B. Aigle, C. F. Michelsen, L. Jelsbak, C. Sohlenkamp, P. Pevzner, A. Edlund, J. McLean, J. Piel, B. T. Murphy, L. Gerwick, C.-C. Liaw, Y.-L. Yang, H.-U. Humpf, M. Maansson, R. A. Keyzers, A. C. Sims, A. R. Johnson, A. M. Sidebottom, B. E. Sedio, A. Klitgaard, C. B. Larson, C. A. P. Boya, D. Torres-Mendoza, D. J. Gonzalez, D. B. Silva, L. M. Marques, D. P. Demarque, E. Pociute, E. C. O'Neill, E. Briand, E. J. N. Helfrich, E. A. Granatosky, E. Glukhov, F. Ryffel, H. Houson, H. Mohimani, J. J. Kharbush, Y. Zeng, J. A. Vorholt, K. L. Kurita, P. Charusanti, K. L. McPhail, K. F. Nielsen, L. Vuong, M. Elfeki, M. F. Traxler, N. Engene, N. Koyama, O. B. Vining, R. Baric, R. R. Silva, S. J. Mascuch, S. Tomasi, S. Jenkins, V. Macherla, T. Hoffman, V. Agarwal, P. G. Williams, J. Dai, R. Neupane, J. Gurr, A. M. C. Rodríguez, A. Lamsa, C. Zhang, K. Dorrestein, B. M. Duggan, J. Almaliti, P.-M. Allard, P. Phapale, L.-F. Nothias, T. Alexandrov, M. Litaudon, J.-L. Wolfender, J. E. Kyle, T. O. Metz, T. Peryea, D.-T. Nguyen, D. VanLeer, P. Shinn, A. Jadhav, R. Müller, K. M. Waters, W. Shi, X. Liu, L. Zhang, R. Knight, P. R. Jensen, B. Ø. Palsson, K. Pogliano, R. G. Linington, M. Gutiérrez, N. P. Lopes, W. H. Gerwick, B. S. Moore, P. C. Dorrestein and N. Bandeira, Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS PubMed.
- W. H. Gerwick, J. Nat. Prod., 2017, 80, 2583–2588 CrossRef CAS PubMed.
- Y. Umeda, K. Furihata, S. Sakuda, H. Nagasawa, K. Ishigami, H. Watanabe, W. Izumikawa, M. Takagi, T. Doi, Y. Nakao and K. Shin-ya, Org. Lett., 2007, 9, 4239–4242 CrossRef CAS PubMed.
- V. I. Boev, A. I. Moskalenko, S. L. Belopukhov and N. M. Przheval’skii, Russ. J. Org. Chem., 2015, 51, 1253–1260 CrossRef CAS.
- D. Kessner, M. Chambers, R. Burke, D. Agus and P. Mallick, Bioinformatics, 2008, 24, 2534–2536 CrossRef CAS PubMed.
- T. R. Hoye, C. S. Jeffrey and F. Shao, Nat. Protoc., 2007, 2, 2451–2458 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: HR-ESI-MS and NMR spectra of compounds 1–10. Advanced Marfey's and C18 Mosher analysis. See DOI: 10.1039/d0ra09388b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |