DOI:
10.1039/D0RA09117K
(Paper)
RSC Adv., 2021,
11, 8239-8249
Potential therapeutic targets and molecular details of anthocyan-treated inflammatory bowel disease: a systematic bioinformatics analysis of network pharmacology†
Received
11th November 2020
, Accepted 3rd February 2021
First published on 22nd February 2021
Abstract
Anthocyans, containing anthocyanins and anthocyanidins, play a crucial role in preventing and treating inflammatory bowel disease (IBD). Most anthocyanins and their basic elements, namely anthocyanidins have been recognized for the effective treatment of IBD, but the key biomarkers of anthocyan-treated IBD remain unclear. In this study, a bioinformatics analysis based on network pharmacology was performed to demonstrate the core-targets, biological functions, and signaling pathways of most common anthocyanidins that existed in anthocyans to reveal their potential or major mechanisms. The network pharmacology of the multi-target drug molecular design with specific signal nodes was selected, which was used to analyse core targets and complete the bioinformatics analysis of core targets. The network assays indicated 44 common targeted genes, 5 of which were core targets of both six most common anthocyanidins and IBD. These 44 common targets related to major signaling mechanisms of the six most common anthocyanidins in IBD may involve following processes: promotion of intracellular metabolism and proliferation, inhibition of cell necrosis, anti-inflammation and regulation of intestinal epithelial survival mainly via pathways such as, the EGFR tyrosine kinase inhibitor resistance pathway, platelet activation, microRNAs in cancer, arachidonic acid metabolism and the cGMP-PKG signaling pathway. Thus, our findings may provide other molecular details about anthocyans in the treatment of IBD and contribute towards the use of anthocyanidins, which will be meaningful shedding light on the action mechanisms of anthocyanidins in treating IBD.
1. Introduction
Inflammatory bowel disease (IBD) is chronic and intestinal inflammation with unknown etiology and pathogenesis, which is mainly defined as Crohn's disease (CD) or ulcerative colitis (UC).1 UC is an idiopathic and chronic inflammatory disorder of the colonic mucosa, which starts in the rectum and extends to the proximity in an uninterrupted pattern through the entire colon or part of it, and bloody diarrhea, abdominal pain, and urgency are the typical presenting symptoms of UC.2 Mainly affected by the change of environmental factors and gut microbiota, CD often causes disorders of innate and adaptive immune responses, ranging from mildly active diseases such as occasional diarrhea and rectal bleeding to severely active diseases that may cause 10–20 translation of bloody bowel per day. Besides, people with CD have an increased risk of cancer, osteoporosis, anemia, nutritional deficiency, depression, infection, and thrombosis.3 However, the location of inflammation is different between CD and UC. In CD, inflammation is usually transmural, and CD can be associated with intestinal granuloma, strictures, and fistulas, while in UC, inflammation is usually limited to the mucosa, and the symptoms in CD are not typical in UC.4 In addition, IBD, including two idiopathic types of CD and UC, and atypical types of collagen colitis and refractory colitis, is characterized as chronic gastrointestinal inflammation, alternating clinical remission recurrence, and patients with severe IBD need to be treated by the colorectal resection.5 IBD has affected over 1 million individuals in the United States, while more than 2.5 million in Europe. The prevalence of IBD is highest in the Western world, affecting up to 0.5% of the total population, with a treatment cost of over 6 billion in 2004.6 There are mainly 5-aminosalicylic acid, 6-mercaptopurine, sulfasalazine, glucocorticoids, azathioprine, cyclosporine, thioguanine, methotrexate, and tacrolimus. However, long-term usage is limited due to different side effects and other adverse effects of these drugs, including, diarrhea, abdominal pain, nausea, and vomiting.7 Consequently, it is essential to highlight the exploit of new treatment and drugs for IBD to relieve the current risk and reduce costs.
It is noteworthy that many patients with IBD begin to turn to alternative medicine and turn to traditional plant-based drugs; consequently, anthocyan-rich foods are highly emphasized. Anthocyans, including anthocyanins and anthocyanidins, are a group of water-soluble plant pigments found in different parts of higher plants, mainly flowers and fruits, including leaves, stems, and roots. Depending on the pH value and the presence of chelating metal ions, they are intensely coloured in purple, red, blue, or violet.8 Notably, anthocyanins and anthocyanidins have antioxidative, antimicrobial, anti-inflammatory, and anti-mutagenic properties, which have been recognized as bioactive substances with health benefits. Anthocyanins and anthocyanidins play a crucial role in preventing and treating many chronic diseases such as metabolic disorders, cancer, eye diseases, and cardiovascular diseases, particularly IBD.9,10 Anthocyans play an anti-inflammatory role by protecting intestinal mucosal integrity, epithelial barrier function, and recovery of immune regulation, and by regulating gut microbiota, which is beneficial to IBD treatment.11,12 Moreover, anthocyanin degradation products and colonic metabolites, such as anthocyanidins, can suppress pro-inflammatory cytokine production.13 As the bioavailability of consumed anthocyanins and anthocyanidins to the plasma seems to be less than 2%, it is recognized that the anthocyans are either absorbed through the gastrointestinal tract or metabolized in the intestines or liver.8 Numerous studies have demonstrated the anti-inflammatory effects of anthocyan-rich fruits and plants (ESI Table 1†). Increasing evidence suggests anthocyans as dietary supplements, whose role in managing IBD and its associated oncogenesis is deemed to be crucial.14 Due to the existence of flavonoid ions in the anthocyanin structure and its unique electronic distribution, multiple anthocyanins are derived from natural anthocyanidins and will be hydrolyzed into anthocyanidins. There are only six anthocyanidins that have been commonly found in fruits and vegetables, namely, cyanidin, peonidin, delphinidin, pelargonidin, malvidin, and petunidin.15,16 The six most common anthocyanidins are highlighted as the basic elements of anthocyanins, and they are produced upon anthocyanin hydrolysis. Otherwise, early research has pointed out that the anthocyanidins could very well be the immediate metabolites after ingestion of anthocyanins because the h-glucosidase enzyme found in intestinal bacteria can easily hydrolyze respective anthocyanins to anthocyanidins.17,18 In addition, another research found that the anthocyanins assayed did not inhibit cell proliferation of cell lines tested at a specific concentration (200 μg ml−1), but anthocyanidins showed cell proliferation inhibitory activity.19 Therefore, herein, we summarize the anti-inflammatory activity or potential of the six most common anthocyanidins (ESI Table 2†), and their actions by inhibiting the downstream genes are shown in Fig. 1, highlighting that the downstream genes are closely connected with the anti-inflammation of the mentioned six anthocyanidins, as described in the following research.
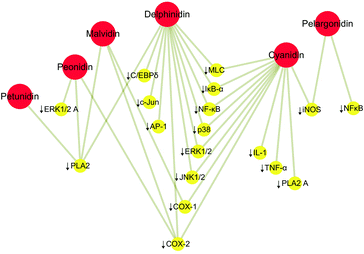 |
| | Fig. 1 Anti-inflammatory activity of the six most common anthocyanidins. ↓ represents inhibition. Red circles represent the most common anthocyanidins and yellow circles the downstream genes. | |
Cyanidin and delphinidin acted by mitigating TNFα-triggered activation of transcription factor nuclear factor kappa B (NF-κB), and downstream phosphorylation of myosin light chain (MLC), which support a particular capacity in the protection of the intestinal barrier against inflammation-induced permeabilization, in part through the inhibition of the NF-κB pathway.20 Another research pointed out that cyanidin and delphinidin supplementary in mice fed with a high-fat diet can reduce lipid deposition and inflammation in the liver, potentially related to the activation of the redox-sensitive signals IKK/NF-κB and JNK1/2, and increased expression of the PTP1B phosphatase regulated by NF-κB.21,22 In the research of anti-inflammatory activities of peonidin, treatment of JB6 P(+) cells with peonidin inhibited 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced cytochrome c oxidase assembly protein-2 (COX-2) expression, and also decreased TPA-induced neoplastic transformation and blocked TPA-induced phosphorylation of extracellular signal-regulated kinases (ERKs) in the cells to meet the anti-inflammatory effect.23 However, malvidin, peonidin, and petunidin exhibited an inhibitory effect of secretory phospholipase A (2) (sPLA2).24 It is known that sPLA2 contributes to the mediation of the inflammation, which is the main factor leading to the excessive production of arachidonic acid under inflammatory conditions.25,26 Thus, these anthocyanidins play a modulatory role, realizing their anti-inflammatory activities. Otherwise, pelargonidin accomplishes its anti-inflammatory effects by inhibiting the inducible nitric oxide synthase (iNOS) protein and mRNA expression and nitric oxide (NO) production.27 Studies have shown that compounds that inhibit the expression or activity of iNOS have potential anti-inflammatory effects.28,29
After collecting and analysing the effects of anthocyan-rich foods on IBD, the relevant anti-inflammatory activities of the six most common anthocyanidins were studied. However, the mentioned mechanism about the anthocyanidins is still unclear, and the treatment of IBD still urgently needs to be studied as the early research has pointed out the anti-inflammatory effects of anthocyanidins20–26 and also other effects on various types of cancers,17,19 making it essential to completely explore the usefulness of this natural ingredient. Thus, we aimed to find out the potential mechanisms of IBD treatment by analysing the most existing anthocyanidins. We consider that the common targets and potential signaling mechanisms of these 6 anthocyanidins for treating IBD will contribute to relevant information to IBD treatment. To explore the common targets and major signaling mechanisms of the anthocyanidins in IBD treatment, we tried to integrate bioinformatics and network pharmacology tools to predict the target genes and analyse the significant interactions between the six most common anthocyanidins and IBD. The investigative flowchart is shown in Fig. 2.
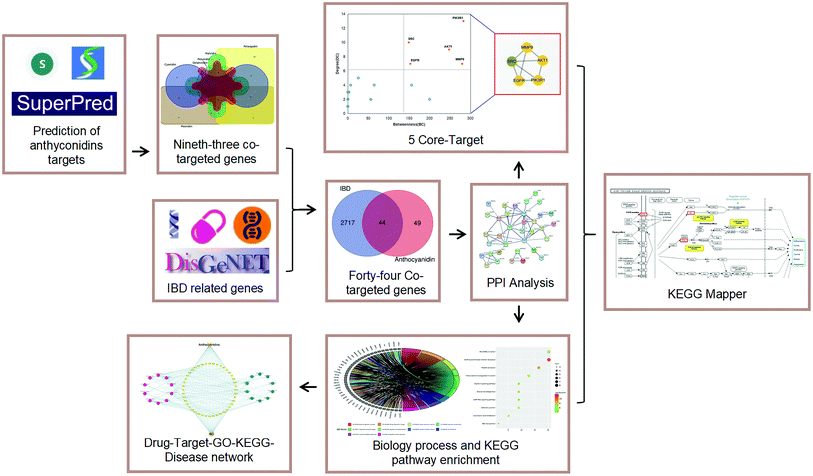 |
| | Fig. 2 Investigate flowcharts for prioritizing biotargets and signaling pathways of anthocyans in treatment of IBD. | |
2. Materials and methods
2.1 Screening potential targets of the six most common anthocyanidins and IBD
The potential targets of the most common anthocyanidins (cyanidin, delphinidin, malvidin, petunidin, pelargonidin, and peonidin) were acquired through analysis of the Traditional Chinese Medical Systems Pharmacology (TCMSP), SuperPred, and Swiss Target Prediction. TCMSP, a unique system pharmacology platform and a form of Chinese herbal medicines is used to capture the relationships between drugs, targets, and diseases. TCMSP selects the connected targets of anthocyanidins by using the SysDT formulas of Herbal Ingredients' Targets Database (HIT), and the information of TCMSP mainly comes from the Therapeutic Target Database (TTD) and PharmGKB databases.30 SuperPred is a prediction webserver for anatomical therapeutic chemical code and target prediction of compounds, which allows prognoses about the medical indication area of novel compounds and finds new leads for known targets.31 Swiss Target Prediction is an online tool since 2014. It aims to predict the most probable small molecular protein targets according to the principle of similarity, and possible targets can also be found through material structures' input on it.32 SuperPred and Swiss Target Prediction accomplish the work based on the methods, namely, 2D fragment and 3D similarity searching. Otherwise, there are authoritative and comprehensive disease analysis databases. GeneCards has been regarded as a searchable and integrative database, considered the most comprehensive genetic data database because of the comprehensive, easy-to-use information about all annotated and predicted human genes.33 The other one, Online Mendelian Inheritance in Man (OMIM), is a major repository of comprehensive and selected information on genes and genetic phenotypes and their relationships. Through this database, diseases are linked to genes related to the human genome and provide the relevant references for further research and genome tool analysis of cataloged genes.34,35 Moreover, DisGeNET, one of the largest available collections of genes and variants involved in human diseases, is homogeneously annotated with controlled vocabularies and community-driven ontologies.36 As a general platform for different research purposes, including investigating the molecular underpinnings of specific human diseases and their comorbidities, analyzing the characteristics of disease genes, generating a hypothesis on the therapeutic action and adverse effects of drugs, predicting disease genes through careful calculation, and evaluating the performance of text mining methods.36 GeneCards, OMIM, and DisGeNET could quickly find out the connected genes based on their database and special formulas, such as integrating data from expert-curated databases with information gathered through text-mining the scientific literature. GeneCards, OMIM and DisGeNET were combined to choose the gene symbol by using the keyword “Inflammatory bowel disease”. UniProt database is the most informative and widely available protein database, which can be used to obtain the targets' official genetic symbols.37 Based on the different computational formulas of all the databases, we can receive the connected targets as much as possible for a comprehensive analysis. Then, we deleted the duplicate targets of all the obtained targets. The Draw Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) was applied to show common targets of the six most common anthocyanidins. Then, using FunRich_v3.1.3, we showed the common targets of the involved anthocyanidins against IBD.
2.2 Protein–protein interaction data and screening for core targets
Based on the STRING database online platform (https://string-db.org/) (Version 11.0),38 which covers the most known human PPI information, protein–protein interaction (PPI) data of the received common targets of the involved anthocyanidins against IBD were obtained. Then, a PPI network model with species set to “Homo sapiens” was constructed by STRING, based on the different analyses of multi databases in STRING, such as, the cooccurence across genomes, co-expression, experimental/biochemical data, association in curated databases, and co-mentioned in Pubmed Abstracts, finally to receive the PPI network with selected score. The minimum required interaction score was set to “high confidence” (high confidence 0.700) and the disconnected nodes in the network were hidden. Other parameters were the default settings. After establishing the PPI network, Cytoscape (Version 3.8.1) was used to filter out the core targets by analyzing the PPI information of common targets from STRING. By calculating and analyzing the network structure and weighted reconnection between nodes, the core targets can be selected. In this work, selecting filters are based on degree values (DC) and betweenness-centrality (BC), DC represents the connection scope, the target in higher DC with the more comprehensive effect, and BC represents the number of connected targets, the targets in higher BC with more connected targets, the core targets will be selected through these filters, thus, we selected the core targets based on DC and BC through CentiScaPe 2.2 in Cytoscape_v3.8.1.
2.3 Functional processes and molecular pathway enrichment analysis of common targets
Metascape is a convenient and powerful annotation analysis tool of gene function, which can help users apply the popular bioinformatics analysis methods to batch gene and protein analysis to realize gene or protein function cognition.39 It combines functional enrichment, interactive group analysis, gene annotation, and membership search to utilize more than 40 independent knowledge bases in an integrated portal.39 In the study, the enrichment analysis of Gene Ontology in biological function (GO-BP) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were completed by Metascape using the previously identified common targets of the involved anthocyanidins and IBD. Using multiple authoritative, functional databases, we inputted the selected common targets into the website and ran the Enrichment Analysis program.
2.4 Key KEGG pathway analysis and core target position
KEGG is a database resource for understanding advanced genomic, chemical and system functions and biological systems (such as cells, organisms, and ecosystems), used to systematically analyze the function of genes to reveal the genetic and chemical blueprint of life phenomena. It is a knowledge base created manually, based on the knowledge obtained by capturing and organizing experiments in a computable form. Besides, the powerful graphic function of KEGG helps to express multiple metabolic pathways and their relationships vividly.40,41 The KEGG database (http://www.genome.ad.jp/kegg/) was used to analyze the key KEGG pathway with the lowest p-value. The core targets in the key pathway were highlighted and their anti-inflammatory effects were analyzed to accomplish a comprehensive understanding of the potential mechanisms of the anthocyanidins in treating IBD.
2.5 Assay and visualization of network relationships
Cytoscape_v3.8.1 was used to visualize the relationship between common targets of the anthocyanidins against IBD and the selected GO-BP enrichments and KEGG pathways. Moreover, the targets of the six most common anthocyanidins against IBD were studied. Enrichment of other targets of KEGG visualization is beneficial for further correlation analysis.
3. Result
3.1 Detailed data of common targets and the PPI network
TCMSP, Swiss Target Prediction and SuperPred were used to identify the potential targets of the six most common anthocyanidins. The identified targets of cyanidin, delphinidin, malvidin, pelargonidin, peonidin, and petunidin were 104, 108, 104, 114, 104, and 104, respectively. Through the Draw Venn diagram, 93 common targets of these anthocyanidins were revealed, which are shown in Fig. 3.
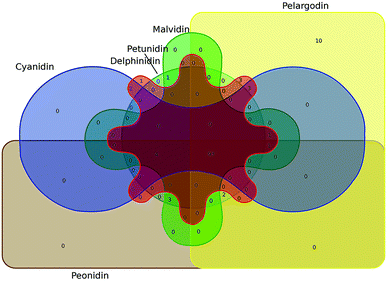 |
| | Fig. 3 Venn diagram of the common targets of the six most common anthocyanidins. | |
Following the bioinformatics analysis, 1385 known therapeutic targets for IBD were selected with a GeneCards relevance score over 12.82, a median value. Besides, 1578 and 528 known targets for IBD treatment were acquired from DisGeNET and OMIM databases, respectively. In total, 2761 therapeutic targets of IBD were collected after the duplicates were removed. The 2761 therapeutic targets of IBD were mapped onto the 93 common targets of the anthocyanidins using FunRich_v3.1.3 to construct a Venn diagram. Eventually, 44 targets were identified as the commonly targeted genes with IBD and the six most common anthocyanidins. Meanwhile, with the species set to “Homo sapiens”, the minimum required interaction score was set to “High confidence” (high confidence 0.700) and the disconnected nodes in the network were hidden. The STRING database was used to analyse 44 common targets with functionally related PPI data, as shown in Fig. 4.
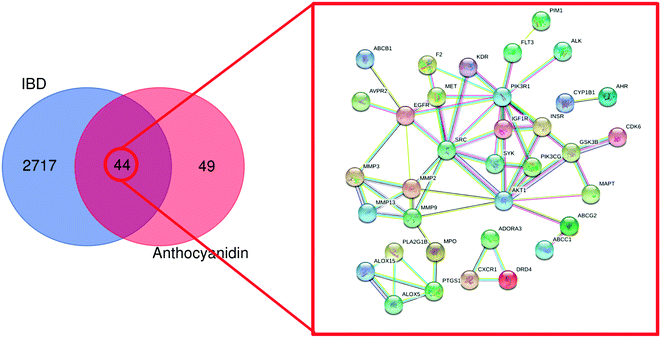 |
| | Fig. 4 PPI network of the 44 common targets. Colored nodes represent query proteins and first shell of interactors. Edges represent protein–protein associations. ABCB1: ATP-dependent translocase ABCB1; AVPR2: vasopressin V2 receptor; MMP3: stromelysin-1; MMP13: collagenase 3; ALOX15: arachidonate 15-lipoxygenase; ALOX5: arachidonate 5-lipoxygenase; PTGS1: prostaglandin G/H synthase 1; MPD: PLA2GB1: phospholipase A2; MMP9: matrix metalloproteinase-9; MMP12: matrix metalloproteinase-12; MMP2: matrix metalloproteinase-2; MMP3: matrix metalloproteinase-3; EGFR: epidermal growth factor receptor; F2: prothrombin; MET: hepatocyte growth factor receptor; SRC: proto-oncogene tyrosine–protein kinase Src; KDR: vascular endothelial growth factor receptor 2; CXCR1: C-X-C chemokine receptor type 1; DRD4: dopamine receptor D4; ADORA3: transmembrane domain-containing protein TMIGD3; PIM1: serine/threonine–protein kinase pim-1; FLT3: receptor-type tyrosine–protein kinase FLT3; PIK3R1: phosphatidylinositol 3-kinase regulatory subunit alpha; IGF1R: insulin-like growth factor 1 receptor; SYK: tyrosine–protein kinase SYK; AKT1: RAC-alpha serine/threonine–protein kinase; PIK3CG: phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform; INSR: insulin receptor; CYP1B1: cytochrome P450 1B1; AHR: aryl hydrocarbon receptor; CDK6: cyclin-dependent kinase 6; GSK3B: glycogen synthase kinase-3 beta; MAPT: microtubule-associated protein tau; ABCG2: ATP-binding cassette sub-family G member 2; ABCC1: multidrug resistance-associated protein 1. | |
3.2 Screening for core targets
The received PPI data was imported into Cytoscape_v3.8.1 to calculate the interaction network's topological parameters of anthocyanidin's targets for treatment of IBD and functionally related targets. By analyzing the network, the maximum freedom degree of the targets is 13, and the medium freedom degree of the targets is 3.314. Furthermore, the maximum betweenness–centrality value of the targets is 283, and the medium betweenness–centrality value of the targets is 48.114. The topological attributes of DC and BC were respectively presented as the node size and node color depth. The network is shown in Fig. 5. Consequently, 5 core targets with the topological attributes of BC > 149 and DC > 7 were selected, and their detailed information is listed in Table 1. These core targets were phosphatidylinositol 3-kinase regulatory subunit alpha (PIK3R1), proto-oncogene tyrosine–protein kinase Src (SRC), RAC-alpha serine/threonine–protein kinase (AKT1), epidermal growth factor receptor (EGFR), and matrix metalloproteinase-9 (MMP9).
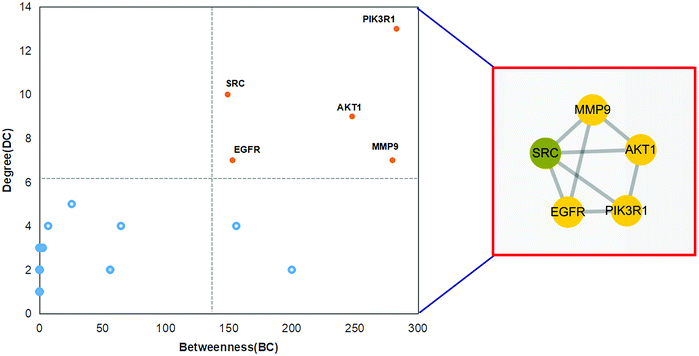 |
| | Fig. 5 Core targets of anthocyanidin in treatment of IBD. | |
Table 1 Information of the core targets
| Gene symbol |
Gene name |
Degree |
Betweenness |
Closeness centrality |
| PIK3R1 |
Phosphatidylinositol 3-kinase regulatory subunit alpha |
13 |
283.00 |
0.49 |
| SRC |
Proto-oncogene tyrosine–protein kinase Src |
10 |
149.17 |
0.50 |
| AKT1 |
RAC-alpha serine/threonine–protein kinase |
9 |
247.83 |
0.49 |
| EGFR |
Epidermal growth factor receptor |
7 |
153.00 |
0.45 |
| MMP9 |
Matrix metalloproteinase-9 |
7 |
279.83 |
0.48 |
3.3 Common targets in GO biological functions and KEGG pathway enrichments
GO and KEGG analyses of the 44 common targets were accomplished through Metascape database to obtain rich ontology clusters. GO biological functions (Table 2), and KEGG pathways (Table 3) are expressed in different forms: a bar graph, circro circles, and a bubble diagram. According to findings, GO biological functions associated with these targets mainly included the following pathways, as shown in Fig. 6: positive regulation of kinase activity, cellular response to nitrogen compounds, cellular response to reactive oxygen species, response to amyloid-beta, regulation of reactive oxygen species metabolic process, regulation of transmembrane transport, icosanoid metabolic process, cell chemotaxis, cytokine-mediated signaling pathway and regulation of bone resorption. KEGG pathway enrichment analysis was further implemented to identify and analyze the mechanism of complex regulation genes in the network, as the results indicated that the related KEGG pathways (P < 0.05) with high confidence are EGFR tyrosine kinase inhibitor resistance, platelet activation, ovarian steroidogenesis, MicroRNAs in cancer, adheren junction, transcriptional misregulation in cancer, arachidonic acid metabolism, oxytocin signaling pathway, cGMP-PKG signaling pathway, and ABC transporters, as shown in Fig. 7. In addition, all the other related GO-BP enrichments and KEGG pathways are also listed (ESI Table 3†).
Table 2 Top 10 GO biological processes of the targets
| GO-BP |
Category |
Description |
Count |
% |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10(P) 10(P) |
| GO:0033674 |
GO biological processes |
Positive regulation of kinase activity |
15 |
34.09 |
−12.995 |
| GO:1901699 |
GO biological processes |
Cellular response to nitrogen compound |
15 |
34.09 |
−12.531 |
| GO:0034614 |
GO biological processes |
Cellular response to reactive oxygen species |
10 |
22.73 |
−12.374 |
| GO:1904645 |
GO biological processes |
Response to amyloid-beta |
7 |
15.91 |
−10.918 |
| GO:2000377 |
GO biological processes |
Regulation of reactive oxygen species metabolic process |
9 |
20.45 |
−10.166 |
| GO:0034762 |
GO biological processes |
Regulation of transmembrane transport |
12 |
27.27 |
−9.576 |
| GO:0006690 |
GO biological processes |
Icosanoid metabolic process |
7 |
15.91 |
−8.738 |
| GO:0060326 |
GO biological processes |
Cell chemotaxis |
9 |
20.45 |
−8.502 |
| GO:0019221 |
GO biological processes |
Cytokine-mediated signaling pathway |
12 |
27.27 |
−7.960 |
| GO:0045124 |
GO biological processes |
Regulation of bone resorption |
5 |
11.36 |
−7.911 |
Table 3 Top 10 KEGG pathways of the targets
| KEGG |
Category |
Description |
Count |
% |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10(P) 10(P) |
| hsa01521 |
KEGG pathway |
EGFR tyrosine kinase inhibitor resistance |
8 |
18.18 |
−11.86 |
| hsa04611 |
KEGG pathway |
Platelet activation |
7 |
15.91 |
−8.64 |
| hsa04913 |
KEGG pathway |
Ovarian steroidogenesis |
5 |
11.36 |
−7.52 |
| hsa05206 |
KEGG pathway |
MicroRNAs in cancer |
8 |
18.18 |
−7.25 |
| hsa04520 |
KEGG pathway |
Adherens junction |
5 |
11.36 |
−6.71 |
| hsa05202 |
KEGG pathway |
Transcriptional misregulation in cancer |
6 |
13.64 |
−6.08 |
| hsa00590 |
KEGG pathway |
Arachidonic acid metabolism |
4 |
9.09 |
−5.32 |
| hsa04921 |
KEGG pathway |
Oxytocin signaling pathway |
5 |
11.36 |
−5.1 |
| hsa04022 |
KEGG pathway |
cGMP-PKG signaling pathway |
5 |
11.36 |
−4.96 |
| hsa02010 |
KEGG pathway |
ABC transporters |
3 |
6.82 |
−4.16 |
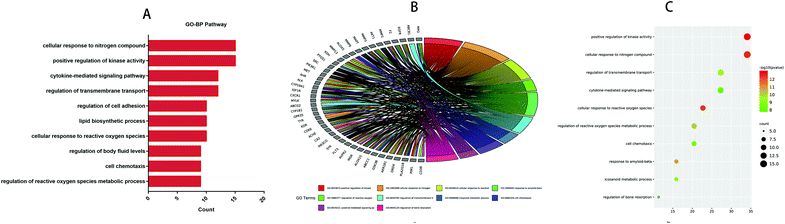 |
| | Fig. 6 Top 10 biological processes of treatment of IBD with anthocyanidins were identified. (A) Bar diagrams. (B) Circro circles. (C) Bubble diagrams. | |
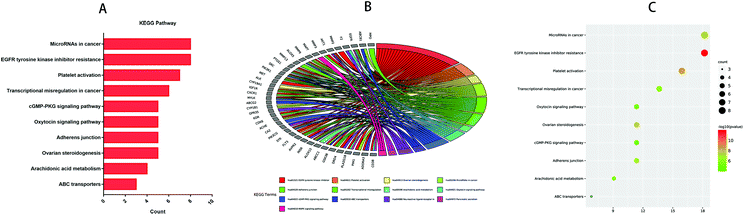 |
| | Fig. 7 Top 10 signaling pathways of treatment of IBD with anthocyanidins were identified. (A) Bar diagrams. (B) Circro circles. (C) Bubble diagrams. | |
3.4 Core targets in key KEGG pathway enrichment
Top 10 KEGG pathways were selected from the Metascape database, and these pathways are essential in cytokine storms caused by IBD. Based on the Reactome knowledge base, we analysed the most potential pathway for anthocyanidins in treatment of IBD, the EGFR tyrosine kinase inhibitor resistance pathway as shown in Fig. 8, which showed the potential targets for anthocyanidins and IBD with hit genes and p-value, simultaneously highlighting the effects of core targets based on their anti-inflammatory activities. This pathway vividly illustrates the most potential signaling mechanism of anthocyanidin-rich foods and anthocyanidins in IBD treatment.
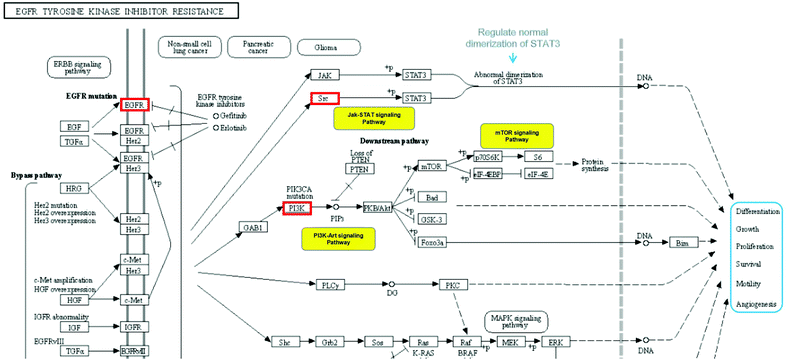 |
| | Fig. 8 Analysis of EGFR tyrosine kinase inhibitor resistance pathway. | |
3.5 Network of anthocyanidins in treatment of IBD
Based on the data of the top 10 enriched GO terms and KEGG pathways, we created a network visualization of anthocyanidins–targets–GO–KEGG–IBD using Cytoscape_v3.8.1, as shown in Fig. 9. The network shows that anthocyanidins shared 10 biological processes and 10 KEGG pathways, highlighting their combined activity against IBD targets.
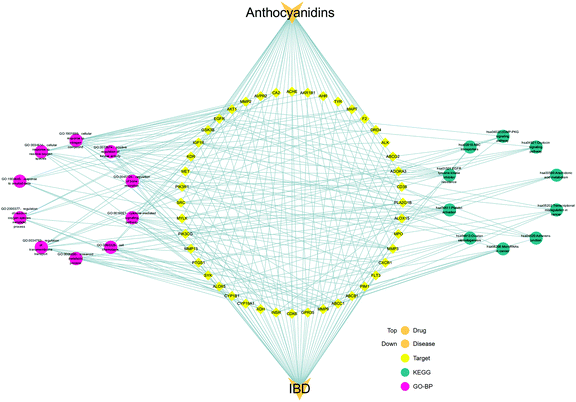 |
| | Fig. 9 Integrated visualization network of the detailed interaction of anthocyanidins–targets–GO–KEGG–IBD. | |
4. Discussion
IBD includes two main forms, namely, CD and UC, which are characterized by chronic relapsing intestinal inflammation. The incidence of IBD shows a rising trend, which has been regarded as a global medical problem.42 IBD has emerged in the USA and Europe and in newly industrialized countries in Asia and South America, and thus, has evolved into a global disease with rising prevalence.43 IBD refers to a death-causing infection and patients have a high burden of parenteral diseases. Patients with IBD have a higher risk of cancer and other organ and system complications such as osteoporosis, venous thromboembolism, and cardiovascular disease.44 Although the pathogenesis of CD and UC has been reported, the etiology of IBD is mostly unknown or no in-depth study is available, and the current treatment is not satisfactory.45–47 Therefore, there is an urgent need for effective management and treatment strategies for IBD. Based on the mentioned factors for treating IBD, people begin to shift from drug therapy to dietary regulation or targeted dietary intake to recover from IBD.48 There is a new frontier in IBD treatment that's beginning to focus, that is, anthocyans, due to the multiple functions, particularly anti-inflammatory.49 Anthocyans, including anthocyanins and anthocyanidins, are commonly present in fruits and other plant parts, and the evident pharmacological efficacy on IBD has been verified,50–53 including the connection between gut microbiota and the brain barrier. In the research talking about the influence of dietary blueberry and broccoli on cecal microbiota activity and colon morphology in a model of IBD, it was found that the number of IBD-related gut microbiota such as Enterococcus, Lactobacillus and Escherichia coli in the intestinal tract decreased significantly, and the number of organic acids such as butyric acid and propionic acid increased in the intestinal tract.52 On the one hand, the effect of anthocyan-rich food on IBD has been widely reported, and, on the other hand, the relationship between gut microbiota and neuroinflammation is also being studied. Intake of anthocyan-rich foods can slow down the activation of downstream NF-κB transcription factors induced by lipopolysaccharide in microglia during intestinal dystrophy, thereby inhibiting the various pathways associated with Alzheimer's disease.54 Similarly, anthocyanins can significantly improve the expression of pro-inflammatory cytokines (TNF-α, IL-1β and COX2) and ROS/JNK to prevent neuroinflammation and Alzheimer's pathology.55 Obviously, there is a certain relationship between anthocyan intake and gut microbiota, and changes in gut microbiota will lead to changes in anti-inflammatory factors that affect the brain barrier, such as Alzheimer's disease. Thus, the comprehensive discussion of the major potential mechanisms of anthocyans against IBD is essential for a deeper understanding of IBD treatment. In this study, we found the common pharmacological efficacy of the six most common anthocyanidins on IBD, focused on pointing out the major pharmacological mechanism and supporting other ideas for IBD treatment. Here, we used a new research route to uncover the integrative pharmacological mechanism of the six most common anthocyanidins used for treating IBD by network pharmacology and other data mining approaches. Systematic network pharmacology to identify and optimize all the core targets, functional processes, and molecular pathway enrichment analysis of the anthocyanidins used for treating IBD was employed. In our finding, 44 common targets were identified, and 5 core targets were recognized as PIK3R1, SRC, AKT1, EGFR, and MMP9.
In network pharmacology analysis, notable enriched GO biological processes included positive regulation of kinase activity, cellular response to nitrogen compound and cellular response to reactive oxygen species. According to the KEGG pathway analysis, the most potential pharmacological mechanisms of anthocyanidins in treatment of IBD includes the EGFR tyrosine kinase inhibitor resistance pathway involving 4 core targets, namely, EGFR, PIK3R1, AKT1, and SRC. EGFR is a tyrosine kinase that participates in intracellular homeostasis regulation, which is also a stimulating factor for cancerous growth. Both EGFR gene mutations and protein overexpression activate downstream pathways that are associated with cancers.56 We analyzed that the anthocyanidins may regulate the EGFR activation to induce the proliferation of epithelial cells by regulating the FOXO signaling pathway in the EGFR tyrosine kinase inhibitor resistance pathway.57 There is evidence that inhibiting the EGFR pathway by reducing the phosphorylation of eIF2α phosphorylation increases the cellular oxidative stress sensitivity.58 The other involved core targets, AKT1 and PIK3R1, AKT1, a serine/threonine kinase, are multifunctional proteins implicated in regulating cell growth, survival ratio and proliferation.59 It is noteworthy that inhibiting the PI3K/AKT/NF-κB pathway mediated by PIK3R1 to exert an anti-inflammatory effect was also be regarded as the potential mechanism of anthocyanidins to realize their effect.60 And the NF-κB pathway has been reported to be connected with the gut microbiota and brain barrier.54 Moreover, many studies have shown that SRC plays multiple roles in macrophage-mediated innate immunity, such as phagocytosis, the production of inflammatory cytokines/mediators, and the induction of cellular migration.61 Studies have suggested roles for SRC in macrophage-mediated inflammatory responses.62,63 Notably, various inflammatory diseases are closely related to macrophage activation.64 We combined and investigated the effects of anthocyanidins through the EGFR tyrosine kinase inhibitor resistance by analyzing the anti-inflammatory effects of the core pathway, and concluded that anthocyan-rich foods might treat IBD by regulating the expression of EGFR, inhibiting the PI3K-AKT signal pathway, inhibiting the abnormal dimerization of STAT3 by inactivating SRC, and pharmacologically regulating the resistance pathway of EGFR tyrosine kinase inhibitors. Furthermore, we found the other potential pharmacological mechanisms of anthocyans in treatment of IBD involved specific modulations of anti-inflammatory responses, for example, platelet activation,65 MicroRNAs in cancer,66 arachidonic acid metabolism,67 and cGMP-PKG signaling pathway.68 Based on all the results, we concluded that the anthocyans used for treating IBD may promote intracellular metabolism and proliferation, inhibition of cell necrosis, anti-inflammation, and moderate intestinal epithelial survival via the mentioned pathways, and all the results will be verified with experiments in the further research.
5. Conclusion
In summary, the bioinformatics findings we have harvested hint that the modulation of EGFR, SRC, PIK3R1, and AKT1 in the EGFR tyrosine kinase inhibitor resistance pathway may be the major pathway of the anthocyans in treatment of IBD. Otherwise, the activation of molecular pathways of platelet activation, microRNAs in cancer, arachidonic acid metabolism, and the cGMP-PKG signaling pathway should also be regarded as the potential pathways. In brief, all common targets, functional processes and molecular pathways of the six most common anthocyanidins in treatment of IBD were identified using bioinformatics and network pharmacology tools.
This study provides a systematic view of the major signaling mechanisms of anthocyans in treatment of IBD based on the analysis of the most common anthocyanidins, which may offer a new IBD treatment idea. However, these potential targets and signaling mechanisms that are predicted by bioinformatic and network pharmacology tools still need to be confirmed by further investigation. Consequently, experimental verification will be emphasized in the following research.
Author contributions
RM Huang conceived and designed the study; YQ Lin, HW Lin, and XL Li contributed to the acquisition, analysis, and interpretation of data. YQ Lin, LX Luo wrote the manuscript; LX Luo and RM Huang reviewed the paper and provided comments, and all the authors reviewed the manuscript.
Conflicts of interest
The authors declare that they have no competing interests.
Acknowledgements
This work was financially supported by the Program of Department of Natural Resources of Guangdong Province (No. GDME2018C014 and GDNRC[2020]038), the Guangzhou Planned Program in Science and Technology (No. 201803020003), Guangdong Provincial Special Fund For Modern Agriculture Industry Technology Innovation Teams (No. 2020KJ122), Key-Area Research and Development Program of Guangdong Province (No. 2020B1111030004), Basic and Applied Basic Research Program of Guangdong Province (2019A1515110201), Administration of Traditional Chinese Medicine of Guangdong Province (20201180), Science and Technology Special Project of Zhanjiang (2019A01009).
Notes and references
- K. Matsuoka and T. Kanai, Semin. Immunopathol., 2015, 37, 47–55 CrossRef CAS.
- I. Ordás, L. Eckmann, M. Talamini, D. C. Baumgart and W. J. Sandborn, Lancet, 2012, 380, 1606–1619 CrossRef.
- B. Veauthier and J. R. Hornecker, Am. Fam. Physician, 2018, 98, 661–669 Search PubMed.
- C. Abraham and J. H. Cho, N. Engl. J. Med., 2009, 361, 2066–2078 CrossRef CAS.
- M. H. Farzaei, R. Rahimi and M. Abdollahi, Curr. Pharm. Biotechnol., 2015, 16, 196–210 CAS.
- J. Miyoshi and E. B. Chang, Transl. Res., 2017, 179, 38–48 CrossRef.
- E. J. Cho, J. S. Shin, Y. S. Noh, Y. W. Cho, S. J. Hong, J. H. Park, J. Y. Lee, J. Y. Lee and K. T. Lee, J. Ethnopharmacol., 2011, 136, 428–435 CrossRef.
- R. Domitrovic, Curr. Med. Chem., 2011, 18, 4454–4469 CrossRef CAS.
- A. Sinopoli, G. Calogero and A. Bartolotta, Food Chem., 2019, 297, 124898 CrossRef CAS.
- M. Sugata, C. Y. Lin and Y. C. Shih, BioMed Res. Int., 2015, 2015, 768093 Search PubMed.
- L. Zhao, Y. Zhang, G. Liu, S. Hao, C. Wang and Y. Wang, Food Funct., 2018, 9, 2796–2808 RSC.
- J. M. Monk, W. Wu, A. L. Hutchinson, P. Pauls, L. E. Robinson and K. A. Power, J. Nutr. Biochem., 2018, 56, 215–223 CrossRef CAS.
- H. P. Amin, C. Czank, S. Raheem, Q. Zhang, N. P. Botting, A. Cassidy and C. D. Kay, Mol. Nutr. Food Res., 2015, 59, 1095–1106 CrossRef CAS.
- C. Zhang, J. M. Monk, J. T. Lu, L. Zarepoor, W. Wu, R. Liu, K. P. Pauls, G. A. Wood, L. Robinson, R. Tsao and K. A. Power, Br. J. Nutr., 2014, 111, 1549–1563 CrossRef CAS.
- Y. M. Lee, Y. Yoon, H. Yoon, H. M. Park, S. Song and K. J. Yeum, Nutrients, 2017, 9(10), 1089 CrossRef.
- A. Smeriglio, D. Barreca, E. Bellocco and D. Trombetta, Phytother. Res., 2016, 30, 1265–1286 CrossRef CAS.
- T. K. McGhie and M. C. Walton, Mol. Nutr. Food Res., 2007, 51(6), 702–713 CrossRef CAS.
- S. Meiers, M. Kemeny, U. Weyand, R. V. Gastpar, E. Angerer and D. Marko, J. Agric. Food Chem., 2001, 49, 958–962 CrossRef CAS.
- Y. Zhang, S. K. Vareed and M. G. Nair, Life Sci., 2005, 76(13), 1465–1472 CrossRef CAS.
- S. Putta, N. S. Yarla, K. E. Kumar, D. B. Lakkappa, M. A. Kamal, L. Scotti, M. T. Scotti, G. M. Ashraf, B. S. B. Rao, S. K. D, G. V. Reddy, V. V. Tarasov, S. B. Imandi and G. Aliev, Curr. Med. Chem., 2018, 25, 5347–5371 CrossRef CAS.
- E. Cremonini, A. Mastaloudis, S. N. Hester, S. V. Verstraeten, M. Anderson, S. M. Wood, A. L. Waterhouse, C. G. Fraga and P. I. Oteiza, Food Funct., 2017, 8, 2915–2923 RSC.
- E. Daveri, E. Cremonini, A. Mastaloudis, S. N. Hester, S. M. Wood, A. L. Waterhouse, M. Anderson, C. G. Fraga and P. I. Oteiza, Redox. Biol., 2018, 18, 16–24 CrossRef CAS.
- J. Y. Kwon, K. W. Lee, H. J. Hur and H. J. Lee, Ann. N. Y. Acad. Sci., 2007, 1095, 513–520 CrossRef CAS.
- A. Dreiseitel, G. Korte, P. Schreier, A. Oehme, S. Locher, G. Hajak and P. G. Sand, J. Neural Transm., 2009, 116, 1071–1077 CrossRef CAS.
- G. Y. Sun, J. Xu, M. D. Jensen and A. Simonyi, J. Lipid Res., 2004, 45, 205–213 CrossRef CAS.
- S. Yedgar, D. Lichtenberg and E. Schnitzer, Biochim. Biophys. Acta, 2000, 1488, 182–187 CrossRef CAS.
- M. Hämäläinen, R. Nieminen, P. Vuorela, M. Heinonen and E. Moilanen, Mediators Inflammation, 2007, 2007, 45673 Search PubMed.
- S. M. Evans and B. J. Whittle, Eur. J. Pharmacol., 2001, 429, 287–296 CrossRef CAS.
- J. P. Pelletier, D. Jovanovic, J. C. Fernandes, P. Manning, J. R. Connor, M. G. Currie, J. A. Di Battista and J. Martel-Pelletier, Arthritis Rheum., 1998, 41, 1275–1286 CrossRef CAS.
- J. Ru, P. Li, J. Wang, W. Zhou, B. Li, C. Huang, P. Li, Z. Guo, W. Tao, Y. Yang, X. Xu, Y. Li, Y. Wang and L. Yang, J. Cheminf., 2014, 6, 13 Search PubMed.
- J. Nickel, B. O. Gohlke, J. Erehman, P. Banerjee, W. W. Rong, A. Goede, M. Dunkel and R. Preissner, Nucleic Acids Res., 2014, 42, W26–W31 CrossRef CAS.
- A. Daina, O. Michielin and V. Zoete, Nucleic Acids Res., 2019, 47, W357–w364 CrossRef CAS.
- S. Fishilevich, R. Nudel, N. Rappaport, R. Hadar, I. Plaschkes, T. Iny Stein, N. Rosen, A. Kohn, M. Twik, M. Safran, D. Lancet and D. Cohen, Database, 2017, bax028 Search PubMed.
- J. S. Amberger, C. A. Bocchini, F. Schiettecatte, A. F. Scott and A. Hamosh, Nucleic Acids Res., 2015, 43, D789–D798 CrossRef.
- J. S. Amberger and A. Hamosh, Curr. Protoc. Bioinf., 2017, 58, 1.2.1–1.2.12 Search PubMed.
- J. Piñero, À. Bravo, N. Queralt-Rosinach, A. Gutiérrez-Sacristán, J. Deu-Pons, E. Centeno, J. García-García, F. Sanz and L. I. Furlong, Nucleic Acids Res., 2017, 45, 833–839 CrossRef.
- T. UniProt Consortium, Nucleic Acids Res., 2018, 46, 2699 CrossRef.
- C. W. Hsia, M. Y. Ho, H. A. Shui, C. B. Tsai and M. J. Tseng, Int. J. Mol. Sci., 2015, 16, 3579–3598 CrossRef CAS.
- Y. Zhou, B. Zhou, L. Pache, M. Chang, A. H. Khodabakhshi, O. Tanaseichuk, C. Benner and S. K. Chanda, Nat. Commun., 2019, 10, 1523 CrossRef.
- M. Kanehisa and S. Goto, Nucleic Acids Res., 2000, 28, 27–30 CrossRef CAS.
- M. Kanehisa, M. Furumichi, M. Tanabe, Y. Sato and K. Morishima, Nucleic Acids Res., 2017, 45, 353–361 CrossRef.
- R. J. Xavier and D. K. Podolsky, Nature, 2007, 448, 427–434 CrossRef CAS.
- G. G. Kaplan, Nat. Rev. Gastroenterol. Hepatol., 2015, 12, 720–727 CrossRef.
- C. N. Bernstein, E. I. Benchimol, A. Bitton, S. K. Murthy, G. C. Nguyen, K. Lee, J. Cooke-Lauder and G. G. Kaplan, J. Can. Assoc. Gastroenterol., 2019, 2, 73–80 CrossRef.
- Y. Z. Zhang and Y. Y. Li, World J. Gastroenterol., 2014, 20, 91–99 CrossRef.
- S. Danese and C. Fiocchi, World J. Gastroenterol., 2006, 12, 4807–4812 CrossRef CAS.
- S. Kugathasan and C. Fiocchi, Semin. Pediatr. Surg., 2007, 16, 146–153 CrossRef.
- M. H. Farzaei, F. F. El-Senduny, S. Momtaz, F. Parvizi, A. Iranpanah, D. Tewari, R. Naseri, A. H. Abdolghaffari and N. Rezaei, Expert Rev. Gastroenterol. Hepatol., 2018, 12, 1007–1024 CrossRef CAS.
- A. Scarano, E. Butelli, S. De Santis, E. Cavalcanti, L. Hill, M. De Angelis, G. Giovinazzo, M. Chieppa, C. Martin and A. Santino, Front. Nutr., 2017, 4, 75 CrossRef.
- S. Li, B. Wu, W. Fu and L. Reddivari, Int. J. Mol. Sci., 2019, 20(10), 2588 CrossRef CAS.
- W. Kalt, A. Cassidy, L. R. Howard, R. Krikorian, A. J. Stull, F. Tremblay and R. Zamora-Ros, Adv. Nutr., 2020, 11(2), 224–236 Search PubMed.
- G. Paturi, T. Mandimika, C. A. Butts, S. Zhu, N. C. Roy, W. C. McNabb and J. Ansell, Nutrition, 2012, 28(3), 324–330 CrossRef CAS.
- J. Li, T. Wu, N. Li, X. Wang, G. Chen and X. Lyu, Food Funct., 2019, 10(1), 333–343 RSC.
- H. Ma, S. L. Johnson, W. Liu, N. A. DaSilva, S. Meschwitz, J. A. Dain and N. P. Seeram, Int. J. Mol. Sci., 2018, 19(2), 461 CrossRef.
- M. S. Khan, T. Ali, M. W. Kim, M. H. Jo, J. I. Chung and M. O. Kim, Mol. Neurobiol., 2019, 56(1), 671–687 CrossRef CAS.
- L. Huang and L. Fu, Acta Pharm. Sin. B, 2015, 5, 390–401 CrossRef.
- R. Li, C. Guo, Y. Li, Z. Qin and W. Huang, Briefings Bioinf., 2020, bbaa079 CrossRef.
- M. J. Kim, W. G. Choi, K. J. Ahn, I. G. Chae, R. Yu and S. H. Back, Mol. Cells, 2020, 43, 264–275 CAS.
- S. Duggal, N. Jailkhani, M. K. Midha, N. Agrawal, K. V. S. Rao and A. Kumar, Sci. Rep., 2018, 8, 1303 CrossRef.
- B. Cai, X. Gan, J. He, W. He, Z. Qiao, B. Ma and Y. Han, Int. Immunopharmacol., 2018, 63, 198–203 CrossRef CAS.
- S. E. Byeon, Y. S. Yi, J. Oh, B. C. Yoo, S. Hong and J. Y. Cho, Mediators Inflammation, 2012, 2012, 512926 Search PubMed.
- S. A. Courtneidge, R. Dhand, D. Pilat, G. M. Twamley, M. D. Waterfield and M. F. Roussel, EMBO J., 1993, 12, 943–950 CrossRef CAS.
- Z. Korade-Mirnics and S. J. Corey, J. Leukocyte Biol., 2000, 68, 603–613 CAS.
- P. Ruytinx, P. Proost, J. Van Damme and S. Struyf, Front. Immunol., 2018, 9, 1930 CrossRef.
- D. A. Rubenstein and W. Yin, Compr. Physiol., 2018, 8, 1117–1156 Search PubMed.
- A. Amiri, M. M. Tehran, Z. Asemi, A. Shafiee, S. Hajighadimi, S. Moradizarmehri, H. R. Mirzaei and H. Mirzaei, Curr. Med. Chem., 2021, 28(2), 360–376 CrossRef CAS.
- T. Wang, X. Fu, Q. Chen, J. K. Patra, D. Wang, Z. Wang and Z. Gai, Int. J. Mol. Sci., 2019, 20(15), 3683 CrossRef CAS.
- R. Negrete, A. Hervera, S. Leánez, J. M. Martín-Campos and O. Pol, PLoS One, 2011, 6, e26688 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra09117k |
| ‡ Contributed equally. |
|
| This journal is © The Royal Society of Chemistry 2021 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article d,
Xiaoling Lie and
Riming Huang
d,
Xiaoling Lie and
Riming Huang *a
*a


![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10(P)
10(P)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10(P)
10(P)






