Open Access Article
Open Access ArticleOzonation enhancement of low cost double-stranded DNA binding dye based fluorescence measurement of total bacterial load in water†
Jiwon Choia,
Beelee Chua *b and
Ahjeong Son
*b and
Ahjeong Son *a
*a
aDepartment of Environmental Science and Engineering, Ewha Womans University, 52 Ewhayeodae-gil, Seodaemun-gu, Seoul 03760, Republic of Korea. E-mail: ason@ewha.ac.kr; ahjeong.son@gmail.com; Fax: +82-2-3277-3275; Tel: +82-2-3277-3339
bSchool of Electrical Engineering, Korea University, 145 Anam-ro, Seongbuk-gu, Seoul 02841, Republic of Korea. E-mail: bchua@korea.ac.kr; chuabeelee@gmail.com; Tel: +82-2-3290-4639
First published on 21st January 2021
Abstract
We demonstrated the feasibility of using ozonation to enhance the performance of dsDNA binding dye SYBR Green I in the fluorescence measurement of total bacterial load in water. Unlike its membrane permeable but expensive equivalent such as SYTO82 dye, SYBR Green I is many times cheaper but membrane impermeable. Ozonation allowed SYBR Green I dye to permeate the membrane and bind with the dsDNA within by first breaching it. Using E. coli K12 bacteria at serial dilution ratios from 1/1 (980 CFU mL−1) to 1/200, we achieved corresponding quantification from 618.7 ± 9.4 to 68.0 ± 1.9 RFU (100 to 11.00% normalized RFU). In comparison, plate counting and optical density measurement were only able to quantify up till a serial dilution ratio of 1/50 (40 CFU mL−1 and 0.0421, respectively). Most importantly with ozonation, the sensitivity of SYBR Green I dye based fluorescence measurement was improved by ∼140 to 210% as compared to that without ozonation. Given its low electrical power consumption, lab-on-chip compatibility and reagent-less nature, ozonation is highly compatible with portable fluorimeters to realize low-cost monitoring of total bacterial load in water.
1. Introduction
Since the first microscopic observation of bacteria in 1676 by the Dutch scientist Antonie van Leeuwenhoek, methods for bacterial observation and detection have evolved significantly. Among the conventional and well-known methods are microscopy, plate counting, optical density measurement, and quantitative polymerase chain reaction (qPCR).1–5 As of today, some of these methods are still largely revered as the gold standard for bacterial detection and quantification. In the last couple of decades, parallel advances in biosensors have also seek to supplant these gold standard methods with equivalent performance at lower cost and hence better community accessibility. These biosensors employ synthesized nanomaterials (such as DNA–quantum dot complexes or aptamer) to capture the target bacterial gene.6–8 They also occasionally require additional reagents (such as glucose oxidase).9,10 Quantitative detection is achieved via a variety of methods such as colorimetry,11–13 fluorescence measurement14–16 or electrochemical means (coulometry and amperometry).17–19 These biosensors are also often target selective and are useful for distinguishing pathogenic strains from non-pathogenic strains. Leveraging on the advances in microfluidics,20–24 these biosensors also promise unprecedented community access to sensitive and selective bacterial detection. In other words, bacterial outbreaks should have belonged to history.Interestingly, bacterial outbreaks are still no less common today. For example in 2020, E. coli and salmonella outbreaks via clover sprouts and onions affected multiple states in the United States.25 In the same year, over 200 kindergarten teachers and students in Korea were involved in the E. coli food poisoning incident.26 This insuinates that the advances in bacterial detection technologies have not been translated into tangible benefits in the community. Further rumination will suggest a re-visit of the bacterial detection technologies paradigm and its translation to community protection. The preponderance of platitudes on the need for high selectivity and hence nanomaterial synthesis might have even ironically de-railed the translational efforts. Perhaps all we need is a veritable easy and rapid screening method that quantifies total bacterial load, in particular Gram negative bacteria such as Escherichia coli and Salmonella, to avert an outbreak. For example, upon the rapid detection of a threshold bacterial load in a sample, health advisory can be issued immediately while the sample is further analyzed with other methods. Also note that existing adenosine triphosphate (ATP) based testing meters are not able to detect Gram negative bacteria effectively.27
A cogent approach would be to leverage on three decades of development in intercalating fluorescence dye based epifluorescence microscopy and flow cytometry for total bacterial, viral as well as protozoans load detection and enumeration.28–32 In this study, we will attempt to contribute to the cost effectiveness, sensitivity and portability of existing techniques by employing ozonation to enhance cell membrane permeability. In this way, low cost dsDNA (double stranded DNA) binding dyes such as SYBR Green I could be employed with improved sensitivity. Note that SYBR Green I dye is seven fold less costly than its membrane permeable equivalent such as SYTO82 dye.33
Ozonation was selected as the preferred method of breaching the cell membrane because it is reagent-less, lab-on-chip integrable and has low electrical power consumption.34,35 There are other cell membrane breaching methods such as the use of ethanol, salts, enzymes, sonication, strong lasers and atomization.36–40 Unfortunately they will either need reagent reservoirs, high electrical power or transportation of the sample beyond the vial. Therefore they are less ideal than ozonation for the eventual integration with fluorescence measurement based portable analyzers.41–43 However ozone is also known to damage dsDNA such as inducing dsDNA backbone cleavages via hydroxyl radicals production.44 Note that SYBR Green I dye binds with the dsDNA via minor groove binding, electrostatic interactions and intercalation.45,46 Ozonation induced damage could compromise the binding of SYBR Green I to the dsDNA. Therefore it is necessary that we also demonstrate the feasibility and compatibility of ozonation enhancement. In particular, it will be important to shield the dsDNA from most of the extracellular ozonation by terminating the ozonation prior to whole cell lysis.
In order to examine the feasibility and compatibility of ozonation enhancement with SYBR Green I dye based fluorescence measurement of total bacterial load in water, we used the Gram negative Escherichia coli (E. coli) K12 as a target bacterium at varying concentrations. As shown in Fig. 1a, this method employed in this study consists of three steps. Step 1: ozonation of sample. Step 2: addition of a low cost dsDNA binding fluorescence dye (SYBR Green I). Step 3: fluorescence measurement. As shown in Fig. 1b, the ozonation first breaches the bacterial cell wall membrane. This enables the SYBR Green I dye to enter the cytosol and binds with the dsDNA. In this way, the total fluorescence is indicative of the total bacterial load.
 | ||
| Fig. 1 Ozonation enhanced dsDNA binding dye based fluorescence measurement of total bacterial load (a) step-by-step procedure (b) principle of operation. | ||
Using different ozonation duration, the output of the ozonation device was first characterized using an ozone meter. Since ozonation would breach the cell wall membrane and inactivate the cell, the effectiveness of the ozonation device was first investigated via plate counting. Its effectiveness on a given sample was inversely proportional to the number of colony forming units (CFU). After the addition of the SYBR Green I dye into the ozonated sample, the opportune amount of ozonation and entry of the SYBR Green I dye into the cytosol and its subsequent binding with the dsDNA was also verified via confocal microscopy. The results were compared with non-ozonated sample. The detection range of ozonation enhanced SYBR Green I dye based fluorescence measurement was examined at 8 different concentrations (corresponds to ∼0 to 1000 CFU mL−1). The improvement to the fluorescence measurement sensitivity as a result of ozonation was also examined. In order to highlight its efficacy, its results were also compared with that of plate counting as well as optical density measurement (another rapid screening method).
2. Materials and methods
2.1 Design and operation of the electrical discharge apparatus
As shown in Fig. 2a, the custom assembled ozonation device primarily consisted of an ionizer, a high voltage DC–DC converter and an air pump. Both high voltage DC–DC converter (spark arc ignition coil module, 3–6 VDC input, > 10 kVDC output, cost ∼ USD 2) and air pump (JQB032-3A, 3 VDC, TCS Electrical Co. Ltd, China, cost ∼ USD 3) are available commercially. The ionizer is constructed using a hollow brass cylinder (length = 40 mm, ID = 6.5 mm) and a stainless steel hypodermic needle (length = 15 mm, OD = 0.7 mm). The stainless steel hypodermic needle functions as the pin cathode and the hollow brass cylinder functions as a the cylinder anode. The inter-electrode spacing is ∼3 mm. They are connected to the negative and positive polarity output of high voltage DC–DC converter respectively. A silicone tubing (HelixMark Medical LLC, USA, OD = 2.5 mm, ID = 1.5 mm) channels the air from the air pump into the ionizer. The ozone-laden air from the ionizer exits via the acrylic conduit (ID = 2 mm). The overall dimensions of the device is 20 × 35 × 160 mm. It is powered by two AA size batteries. Fig. 2b shows the photo of the actual device. | ||
| Fig. 2 Custom assembled ozonation device (a) schematic (b) photo. Experimental setup for (c) mass flow (d) ozone measurement. (e) calculation model for ozone production rate. | ||
Prior to operation, the end of the acrylic conduit is first submerged into the sample. The battery switch is turned on and it simultaneously powered the high voltage DC–DC converter and the air pump. As a result, a high voltage is stressed between the pin cathode and cylinder anode. This created a partial electrical discharge (corona discharge) and an electrical plasma that is confined to the tip of the pin cathode. Ozone is generated within the electrical plasma and is driven through the acrylic conduit by the air pump. As the ozone bubbled into the sample, it breached the cell wall membrane of the bacteria by either direct molecular ozone reaction or via the formation of hydroxyl radical. Both ozone and hydroxyl radical are high reactive and capable of causing damage to the outer layer of protein surrounding nucleic acid and the fatty acid double layer. This allows the fluorescence dye to enter into the cell and binds with the dsDNA within.
2.2 Characterization of custom assembled ozonation device
As shown in Fig. 2c, the flow rate of the custom assembled ozonation device was first characterized via a mass flow meter (Model 4043, TSI Inc, MN, USA) at an operation voltage of ∼2.5 V (400 mA). The ozone production rate was also characterized by connecting the acrylic conduit to one end of a test cylinder (length = 40 mm, ID = 12 mm). The other end of the test cylinder was connected to an ozone meter (Model A-22, EcoSensors, NM, USA). An exhaust orifice (diameter = 2 mm) facilitated the displacement of the air within the test cylinder by the output from the ozonation device (Fig. 2d). The measurements by the ozone meter were recorded over an interval of 10 min and were repeated three times. As shown in Fig. 2e, the ozonation production rate could be estimated by assuming instantaneous mixing of ozone in the test cylinder. This means at a given instance, the ozone concentration detected by the ozone sensor (Csensor) is given by:
 | (1) |
Therefore, the rate of change of ozone concentration detected by the ozone meter is given by:
 | (2a) |
Since ozone concentration Cdevice is in turn given by the ozone mass per unit volume or Δmdevice/ΔVdevice, we can re-write eqn (2a) as follows:
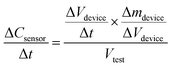 | (2b) |
Re-arranging eqn (2b) gives us the ozone production rate Δmdevice/Δt in terms of the rate of ozone concentration change with respect to time as measured by the ozone meter.
 | (3) |
2.3 Target bacteria culture
E. coli K12 in stationary state with OD600 ∼ 1.5 and ∼ 0.2 were cultured prior to the respective experiments. Difco™ LB Broth, Miller (Luria-Bertani, BD Biosciences, San Jose, CA, USA) was used to culture E. coli K12. As per manufacturer's recommendation, 15 g of LB broth was mixed with 500 mL of distilled water. The media and glassware were autoclaved (Biofree, Seoul, Korea) for 1 h at 120 °C. E. coli K12 was added to the sterilized media at a ratio of 100 to 1 prior to culturing in a shaking incubator (Wise Cube, Daihan Scientific, Gangwondo, Korea) for 24 h at 37 °C.2.4 Effects of different ozonation duration
The effects of different ozonation duration on the bacterial cell viability (hence membrane integrity) were characterized via plate counting method. The E. coli K12 culture (OD600 ∼ 1.5) was first serially diluted with PBS buffer (0.1 M, pH 7.4). The experiment was conducted with the E. coli K12 culture at 10−6 serial dilution. Two mL of each diluted bacterial culture was added to a 15 mL falcon tube and it was subject to ozonation by the custom assembled device for 1, 2, 5, and 10 min. After ozonation, 100 μL of each sample was plated on LB agar plate. The acrylic conduit of the custom assembled ozonation device was rinsed with autoclaved deionized water after each use. The agar plates were incubated in an oven at 37 °C for 24 h. Each plate was subsequently divided into 4 parts to facilitate counting.2.5 Confocal microscope visualization
Confocal microscopy was employed to visualize the bacterial cell wall membrane breach by the opportune amount of ozonation as well as the subsequent entry and dsDNA binding by SYBR Green I dye. The visualization was conducted with E. coli K12 culture (OD600 ∼ 1.5) at 10−6 serial dilution. It was washed twice using PBS buffer (0.1 M, pH 7.4) prior to 10 min of ozonation. Both ozonated and non-ozonated samples were then observed by confocal microscope (Super Resolution Confocal Microscope, Leica Microsystems, Wetzlar, Germany). At 100× magnification, the scan speed was 400 Hz and the immersion medium was HCS PL APO 100×/1.40 oil. At 20× magnification, the scan speed was 400 Hz and the immersion medium was HC PL APO CS2 20x/0.75 water.2.6 Quantification using ozonation enhanced SYBR Green I dye
As mentioned earlier, SYBR Green I nucleic acid gel stain (SYBR Green, Invitrogen, Eugene, OR, USA) was employed as the DNA intercalating dye. SYBR Green I (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×) was diluted to 10× with Tris–HCl buffer (0.02 M of Tris-base, pH 8.0, Sigma-Aldrich, Saint Louis, MO, USA) containing 0.005% of sodium dodecyl sulfate (CAS 151-21-3, Sigma-Aldrich). The experiment was performed using serially diluted E. coli K12 culture (1/1, 1/2, 1/5, 1/10, 1/20, 1/50, 1/100, and 1/200) of stationary growth state (OD600 ∼ 0.2). Each sample was washed twice using PBS buffer (0.1 M, pH 7.4) and resuspended in 50 μL PBS buffer. Subsequently each sample was ozonated for 10 min. The ozonated sample (50 μL) and SYBR Green I dye 10× (50 μL) were mixed at ambient temperature and transferred into 96 well plate. The fluorescence of sample was measured three times by M2 Spectramax spectrofluorometer (Molecular Devices LLC, San Jose, CA, USA) at λex = 254 nm and λem = 525 nm. The emission spectra from 450 to 700 nm at λex = 254 nm was also measured. The serially diluted culture was also subjected to plate counting and optical density measurement (at 600 nm) for comparison.
000×) was diluted to 10× with Tris–HCl buffer (0.02 M of Tris-base, pH 8.0, Sigma-Aldrich, Saint Louis, MO, USA) containing 0.005% of sodium dodecyl sulfate (CAS 151-21-3, Sigma-Aldrich). The experiment was performed using serially diluted E. coli K12 culture (1/1, 1/2, 1/5, 1/10, 1/20, 1/50, 1/100, and 1/200) of stationary growth state (OD600 ∼ 0.2). Each sample was washed twice using PBS buffer (0.1 M, pH 7.4) and resuspended in 50 μL PBS buffer. Subsequently each sample was ozonated for 10 min. The ozonated sample (50 μL) and SYBR Green I dye 10× (50 μL) were mixed at ambient temperature and transferred into 96 well plate. The fluorescence of sample was measured three times by M2 Spectramax spectrofluorometer (Molecular Devices LLC, San Jose, CA, USA) at λex = 254 nm and λem = 525 nm. The emission spectra from 450 to 700 nm at λex = 254 nm was also measured. The serially diluted culture was also subjected to plate counting and optical density measurement (at 600 nm) for comparison.
3. Results and discussion
3.1 Characterization of custom assembled ozonation device
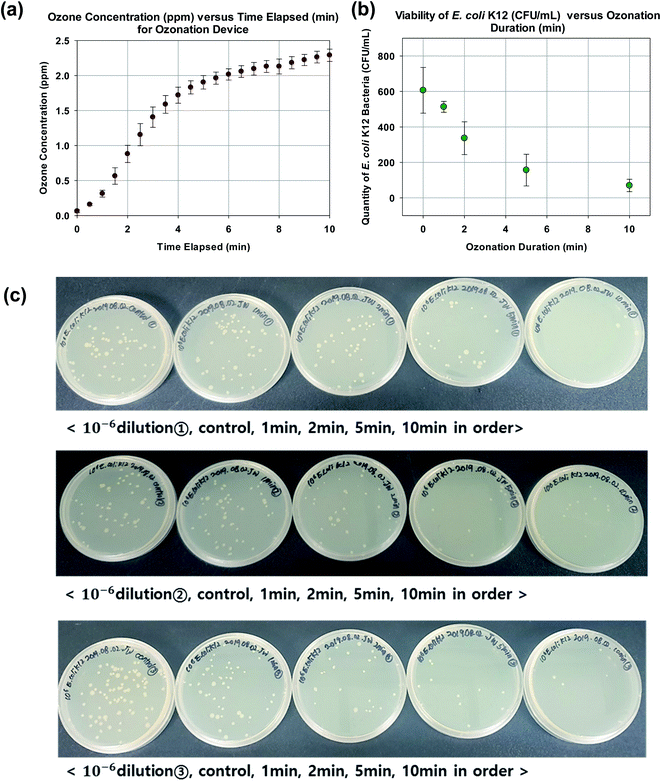
As shown in Fig. 3a, the ozone sensor detected a rapid increase of ozone to 1.406 ± 0.145 ppm in 3 min. This is followed by a more gradual increase to 1.904 ± 0.096 ppm at 5 min and finally to 2.290 ± 0.087 ppm at 10 min. In order to facilitate the estimation of the ozone production rate in accordance to eqn (3), the rate of change in ozone concentration with respect to time as measured by the ozone sensor is given by the gradient of the regression line as 0.221 ppm min−1. The test volume Vtest is ∼ 4.52 × 10−3 L. Since 1 ppm of ozone is equivalent to 2140 mg L−1 in air, the ozone production rate of the custom assembled ozonation device is estimated to be ∼2.14 mg min−1 or 128 mg h−1 (at a flow rate of 0.6 L min−1).3.2 Effects of different ozonation duration
As expected, the increment in ozonation duration corresponded to a decrement in viable E. coli K12 bacteria (Fig. 3b and c). The ozonation durations of 0, 1, 2, 5, and 10 min corresponded to a plate count of 607 ± 129, 513 ± 31, 337 ± 92, 157 ± 90, and 70 ± 35 CFU mL−1 respectively (10−6 dilution). Specifically an ozonation duration of 10 min resulted in ∼90% reduction in viability. In other words, the cell membrane of ∼90% of the bacteria was breached and ready for SYBR Green I dye to enter. Therefore an ozonation duration of 10 min was selected as a nominal setting for subsequent experiments.3.3 Confocal microscope visualization
As shown Fig. 4a and b, the SYBR Green I dye penetrated the bacterial cells and bind with the dsDNA within. In addition, the SYBR Green I dye penetrated a larger number of ozonated bacterial cells (Fig. 4d) as compared to non-ozonated ones (Fig. 4c). In particular, the ozonated sample had a fluorescent intensity of 6.63 μm−2, whereas that of non-ozonated sample was 2.06 μm−2. The ratio of the number of fluorescent bacteria cells per total bacterial cells in the fixed area was 79.3 ± 10.1% and 49.5 ± 14.4% for ozonated and non-ozonated samples, respectively (Fig. 4e). Note that the penetration of SYBR Green I dye into non-ozonated bacterial cells were higher than expected. This could be attributed to the fragility of laboratory cultured bacteria. Nevertheless, the results from both ozonated and non-ozonated samples are significantly different (t-test, P-value = 0.00099). More importantly, the ozonated bacterial cells were intact after ozonation. This suggests that it is possible that the dsDNA within was conceivably shielded from the extracellular ozonation and the damage was minimized.3.4 Quantification of E. coli K12 bacteria using ozonation enhanced SYBR Green I dye
The emission spectra of SYBR Green I dye in ozonated samples at different serial dilution ratios is shown in Fig. 5a. At the emission wavelength of 525 nm, the fluorescence decreased as the sample became more diluted. For serial dilution ratios of 1/1, 1/2, 1/5, 1/10, 1/20, 1/50, 1/100 and 1/200, the fluorescence measurements (10 min ozonation) yielded ∼618.8 ± 9.4, 446.5 ± 23.1, 281.2 ± 4.9, 181.7 ± 5.0, 128.3 ± 4.3, 88.4 ± 1.1, 75.4 ± 5.0 and 68.0 ± 1.9 RFU at λem = 525 nm. In order to gain further insight of the results' significance, the relation between serial dilution ratios, quantity of bacteria via plate counting, and optical density measurement is presented in Fig. 5b and Table 1. For serial dilution ratios of 1/1, 1/2, 1/5, 1/10, 1/20, 1/50, 1/100 and 1/200, plate counting yielded 980, 500, 230, 110, 40, 10, 0 and 0 CFU mL−1, whereas optical density measurement yielded 0.2062, 0.1248, 0.0741, 0.0563, 0.0475, 0.0421, 0.0502 and 0.0382. Note that even though plate counting and optical density measurement were not performed in triplicate, they correlated very well (y = 81.34 + 0.40x, R2 = 0.99).| Serial dilution ratio | Plate counting (CFU mL−1) | Optical density measurement (OD600) | SYBR Green I dye fluorescence, ozonation 10 min (RFU) |
|---|---|---|---|
| 1/1 | 980 | 0.2062 | 618.7 ± 9.4 |
| 1/2 | 500 | 0.1248 | 446.5 ± 23.2 |
| 1/5 | 230 | 0.0741 | 281.2 ± 4.9 |
| 1/10 | 110 | 0.0563 | 181.7 ± 5.0 |
| 1/20 | 40 | 0.0475 | 128.3 ± 4.3 |
| 1/50 | 10 | 0.0421 | 88.4 ± 1.1 |
| 1/100 | 0 | 0.0502 | 75.4 ± 5.0 |
| 1/200 | 0 | 0.0382 | 68.0 ± 1.9 |
As shown in Fig. 5c, the proposed method (ozonation 10 min) was able to quantify E. coli K12 bacteria over a concentration range of 0 to 980 CFU mL−1 (y = 102.77 + 0.57x, R2 = 0.96). Interestingly, the fluorescence of the non-ozonated samples appeared to be also indicative of the E. coli K12 bacterial concentration. This is consistent with the confocal microscopy observation in Fig. 4a and e where the bacterial membrane was already breached prior to ozonation. As mentioned earlier, this was expected because the bacteria was laboratory cultured and tended to be more fragile than its environmental counterparts. Therefore, the results with non-ozonated samples do not invalidate the improvement of sensitivity acquired as a result of ozonation.
In order to further appreciate the improvement of sensitivity by ozonation, the fluorescence measurement is first background subtracted and then divided by the respective bacterial quanitity in CFU mL−1. This gives the sensitivity of the SYBR Green I based fluorescence measurement in terms of RFU/CFU mL−1 as shown in Fig. 5d. It is apparent that with ozonation, the sensitivity of the fluorescence measurement was improved for the entire range of bacterial quantity from 0 to 980 CFU mL−1. The improvement by ozonation is further elucidated by examining the ratio of ozonated to non-ozonated sample. As shown in Fig. 5e, for bacterial quantity of 10 CFU mL−1, ozonation enhancement was at 213% (sensitivity improved by 113%). As expected, this enhancement decreased as the bacterial quantity increased. Nonetheless for bacterial quantity of 980 CFU mL−1, the ozonation enhancement was at 140% (sensitivity improved by 40%).
3.5 Comparison of ozonation enhanced SYBR Green I dye based fluorescence measurement with plate counting and optical density measurement
As shown in Fig. 6a to c, the quantification results of plate counting, optical density measurement and SYBR Green I dye based fluorescence measurement were plotted in accordance to the serial dilution ratios. Plate counting was able to quantify E. coli K12 bacteria from serial dilution ratios of 1/1 to 1/100 (980 to 0 CFU mL−1, Fig. 6a). Similarly, optical density measurement was able to quantify at serial dilution ratios from 1/1 to 1/50 (corresponding absorbance from 0.2062 to 0.0421, Fig. 6b). However at serial dilution ratio of 1/100, the absorbance instead increased to 0.0502. On the other hand, the ozonation enhanced SYBR Green I dye based fluorescence measurement was able to quantify E. coli K12 bacteria for the entire range of serial dilution ratios from 1/1 to 1/200 (618 ± 9.4 to 68.0 ± 1.9 RFU, Fig. 6c).In order to facilitate a more intuitive comparison between these three methods, the respective quantification units are normalized as shown in Fig. 6d and Table 2. The normalized quantification of plate counting and optical density measurement ranged from 100 to 1.02% and 100 to 20.42% respectively for serial dilution ratios from 1/1 to 1/50. The normalized quantification of ozonation enhanced SYBR Green I dye ranged from 100 to 11.00% for serial dilution ratios from 1/1 to 1/200. In other words, the ozonation enhanced SYBR Green I dye was able to quantify more diluted samples than plate counting and optical density measurement. More importantly, it demonstrated the feasibility and compatibility of ozonation as a means of enhancing the performance of low cost dsDNA binding dye in fluorescence measurement.
| Serial dilution ratio | Serial dilution ratio (decimal) | Normalized plate counting (%) | Normalized optical density measurement (%) | Normalized SYBR Green I dye fluorescence, ozonation 10 min (%) |
|---|---|---|---|---|
| 1/1 | 1 | 100 | 100 | 100 |
| 1/2 | 0.5 | 51.02 | 60.52 | 72.16 |
| 1/5 | 0.2 | 23.47 | 35.94 | 45.46 |
| 1/10 | 0.1 | 11.22 | 27.30 | 29.37 |
| 1/20 | 0.05 | 4.08 | 23.04 | 20.75 |
| 1/50 | 0.02 | 1.02 | 20.42 | 14.28 |
| 1/100 | 0.01 | 0 | 24.35 | 12.19 |
| 1/200 | 0.005 | 0 | 18.53 | 11.00 |
3.6 Potential, limitations and future work
In contrary to the prevalent suggestion that ozonation is incompatible with dsDNA–dye binding based bacterial monitoring methods, we have shown that it is possible to employ ozonation and still yield a significant increase in fluorescence measurement. As mentioned earlier, the increase in fluorescence measurement ranged from 40 to over 100% more than that without ozonation. This means the dsDNA damage due to ozonation did not critically undermine the method via the impediment of SYBR Green I dye binding to the dsDNA. It is important to note that the ozonation was extracellular and the dsDNA within should be mostly shielded by the bacterial cell membrane. Subsequently the SYBR Green I dye would enter via the breach to bind with the dsDNA within. This was evident from the fluorescence within whole cells as shown in Fig. 4a to d. This also suggested that the extracellular (in solution) and intracellular (within the breached bacterial cell) concentration of SYBR Green I dye should be different and it would have implication on the dominant binding mode (either minor groove binding or intercalation). Therefore in the future work, it will be useful to elucidate the amount of intracellular ozone penetration and hence the potential damage to dsDNA within. It will also be useful to determine the intracellular dye to base pair ratio and establish the dominant binding mode.As mentioned earlier, the other portable total bacterial monitoring device commonly used by food safety professionals is the adenosine triphosphate (ATP) based testing meter and it is unable to detect Gram negative bacteria effectively.27 Therefore the presented compatibility between ozonation and dsDNA binding fluorescence dye method will lay the foundation for a low cost total bacterial monitoring device that can detect Gram negative bacteria effectively.
4. Conclusion
We have demonstrated that the use of ozonation for 10 min could significantly improve the performance of the low cost membrane impermeable dsDNA binding dye SYBR Green I in the fluorescence measurement of E. coli K12 bacteria. Ozonation has been shown to increase the permeability of cell membrane without effectively damaging the dsDNA and preventing subsequent binding. The dye binding with dsDNA after ozonation was first visually verified via confocal microscopy. Subsequently we were able to quantify E. coli K12 bacteria samples at different serial dilution ratios ranging from 1/1 (980 CFU mL−1) down to 1/200 (0 CFU mL−1) with corresponding fluorescence of 618.7 ± 9.4 and 68.0 ± 1.9 RFU respectively. In comparison, the quantification by both plate counting and optical density measurement were only satisfactory down to a serial dilution ratio of 1/100. Most importantly, ozonation was able to increase the sensitivity of the fluorescence measurement by at least ∼140% (980 CFU mL−1) and as high as ∼210% (10 CFU mL−1). In other words, ozonation has enabled low cost membrane impermeable dye based fluorescence measurement to outperform plate counting and optical density measurement. Since ozonation is reagent-less, lab-on-chip compatible and has low electrical power consumption, it is readily compatible with existing portable fluorimeters.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the National Research Foundation of Korea (NRF-2019R1A2C2084233 and NRF-2019R1F1A1058562).References
- J. R. Porter and A. Van Leeuwenhoek, Bacteriol. Rev., 1976, 40, 260–269 CrossRef CAS.
- A. Trusov, R. Bumgarner, R. Valijev, R. Chestnova, S. Talevski, C. Vragoterova and E. S. Neeley, Int. J. Tuberc. Lung Dis., 2009, 13, 836–841 Search PubMed.
- S. D. Brugger, C. Baumberger, M. Jost, W. Jenni, U. Brugger and K. Mühlemann, PLoS One, 2012, 7, e33695 CrossRef CAS.
- C. Begot, I. Desnier, J. D. Daudin, J. C. Labadie and A. Lebert, J. Microbiol. Methods, 1996, 25, 225–232 CrossRef.
- C. Jian, P. Luukkonen, H. Yki-Järvinen, A. Salonen and K. Korpela, PLoS One, 2020, 15, e0227285 CrossRef CAS.
- G.-Y. Kim, X. Wang, H. Ahn and A. Son, Environ. Sci. Technol., 2011, 45, 8873–8880 CrossRef CAS.
- X. Wang, M. R. Liles and A. Son, Soil Biol. Biochem., 2013, 58, 9–15 CrossRef CAS.
- Y.-W. Zhao, H.-X. Wang, G.-C. Jia and Z. Li, Sensors, 2018, 18, 2518 CrossRef.
- J. Sun, J. Huang, Y. Li, J. Lv and X. Ding, Talanta, 2019, 197, 304–309 CrossRef CAS.
- M. Yu, H. Wang, F. Fu, L. Li, J. Li, G. Li, Y. Song, M. T. Swihart and E. Song, Anal. Chem., 2017, 89, 4085–4090 CrossRef CAS.
- S. Qiu, Z. Lin, Y. Zhou, D. Wang, L. Yuan, Y. Wei, T. Dai, L. Luo and G. Chen, Analyst, 2015, 140, 1149–1154 RSC.
- C. Wang, X. Gao, S. Wang and Y. Liu, Anal. Bioanal. Chem., 2020, 412, 611–620 CrossRef CAS.
- T. N. Le, T. D. Tran and M. I. Kim, Nanomaterials, 2020, 10, 92 CrossRef CAS.
- E.-H. Lee, B. Chua and A. Son, Environ. Sci. Technol., 2018, 52, 1375–1385 CrossRef CAS.
- K. A. Mitchell, B. Chua and A. Son, Biosens. Bioelectron., 2014, 54, 229–236 CrossRef CAS.
- I. V. Martynenko, D. Kusić, F. Weigert and S. Stafford, et al., Anal. Chem., 2019, 91, 12661–12669 CrossRef CAS.
- G. A. Zelada-Guillén, S. V. Bhosale, J. Riu and F. X. Rius, Anal. Chem., 2020, 82, 9254–9260 CrossRef.
- E. Cesewski and B. N. Johnson, Biosens. Bioelectron., 2020, 159, 112214 CrossRef CAS.
- L. Sepunaru, K. Tschulik, C. Batchelor-McAuley, R. Gavish and R. G. Compton, Biomater. Sci., 2015, 3, 816–820 RSC.
- D. Zhang, H. Bi, B. Liu and L. Qiao, Anal. Chem., 2018, 90, 5512–5520 CrossRef CAS.
- R. Narang, S. Mohammadi, M. M. Ashani, H. Sadabadi, H. Hejazi, M. H. Zarifi and A. Sanati-Nezhad, Sci. Rep., 2018, 8, 15807 CrossRef.
- H. Li, P. Torab, K. E. Mach, C. Surrette, M. R. England, D. W. Craft, N. J. Thomas, J. C. Liao, C. Puleo and P. K. Wong, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 10270–10279 CrossRef CAS.
- B. C. Dhar and N. Y. Lee, BioChip J., 2018, 12, 173–183 CrossRef CAS.
- C. Wang, F. Madiyar, C. Yu and J. Li, J. Biol. Eng., 2017, 11, 9 CrossRef.
- C. Morales, Salmonella Cases Linked to Onions Increase to Nearly 900, The New York Times, 9th Aug 2020 Search PubMed.
- A. Kim, E. coli outbreak among preschoolers growing, The Korea Herald, 25th Jun. 2020 Search PubMed.
- D. E. Turner, E. K. Daugherity, C. Altier and K. J. Maurer, J. Am. Assoc. Lab. Anim. Sci., 2010, 49, 190–195 CAS.
- F. Hammes, M. Berney, Y. Wang, M. Vital, O. Köster and T. Egli, Water Res., 2008, 42, 269–277 CrossRef CAS.
- E. Siebel, Y. Wang, T. Egli and F. Hammes, Drinking Water Eng. Sci., 2008, 1, 1–6 CrossRef.
- T. Falcioni, S. Papa and J. M. Gasol, Appl. Environ. Microbiol., 2008, 74, 1767–1779 CrossRef CAS.
- A. Patel, R. T. Noble, J. A. Steele, M. S. Schwalbach, I. Hewson and J. A. Fuhrman, Nat. Protoc., 2007, 2, 269–276 CrossRef CAS.
- R. T. Noble and J. A. Fuhrman, Aquat. Microb. Ecol., 1998, 14, 113–118 CrossRef.
- H. Gudnason, M. Dufva, D. D. Bang and A. Wolff, Nucleic Acids Res., 2007, 35, e127 CrossRef.
- E.-H. Lee, H.-J. Lim, A. Son and B. Chua, Analyst, 2015, 140, 7776–7783 RSC.
- E.-H. Lee, B. Chua and A. Son, Sens. Actuators, B, 2015, 216, 17–23 CrossRef CAS.
- L. O. Ingram, J. Bacteriol., 1981, 146, 331–336 CrossRef CAS.
- T. C. Marentis, B. Kusler, G. G. Yaralioglu, S. Liu, E. O. Hæggström and B. T. Khuri-Yakub, Ultrasound Med. Biol., 2005, 31, 1265–1277 CrossRef.
- E. Marti and J. L. Balcázar, Appl. Environ. Microbiol., 2013, 79, 1743–1745 CrossRef CAS.
- H.-H. Lai, P. A. Quinto-Su, C. E. Sims, M. Bachman, G. P. Li, V. Venugopalan and N. L. Allbritton, J. R. Soc., Interface, 2008, 5(SUPPL.2), S113–S121 CAS.
- H.-J. Lim, E.-H. Lee, Y. Yoon, B. Chua and A. Son, J. Appl. Microbiol., 2016, 120, 379–387 CrossRef CAS.
- H.-J. Lim, B. Chua and A. Son, Biosens. Bioelectron., 2017, 94, 10–18 CrossRef CAS.
- D. Kim, H.-J. Lim, Y.-G. Ahn, B. Chua and A. Son, Talanta, 2020, 219, 121216 CrossRef CAS.
- H.-J. Lim, A.-R. Kim, M.-Y. Yoon, Y. Yoo, B. Chua and A. Son, Biosens. Bioelectron., 2018, 121, 1–9 CrossRef CAS.
- K. Ito, S. Inoue, Y. Hiraku and S. Kawanishi, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2005, 585, 60–70 CrossRef CAS.
- H. Zipper, H. Brunner, J. Bernhagen and F. Vitzthum, Nucleic Acids Res., 2004, 32, e103 CrossRef.
- A. I. Dragan, R. Pavlovic, J. B. McGivney, J. R. Casas-Finet, E. S. Bishop, R. J. Strouse, M. A. Schenerman and C. D. Geddes, J. Fluoresc., 2018, 22, 1189–1199 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra08742d |
| This journal is © The Royal Society of Chemistry 2021 |