Open Access Article
Open Access ArticleEffects of nanopatterned-surface dishes on chondrocyte growth and cell cycle progression†
Sang-Soo Han‡
a,
Myung-Ok Choab,
Kang Moo Huhb and
Sun-Woong Kang *ac
*ac
aResearch Group for Biomimetic Advanced Technology, Korea Institute of Toxicology, Daejeon, Korea. E-mail: swkang@kitox.re.kr; Tel: +82-42-610-8209
bDepartment of Polymer Science and Engineering, Chungnam National University, Daejeon, Korea
cDepartment of Human and Environmental Toxicology, University of Science and Technology, Daejeon, Korea
First published on 22nd December 2020
Abstract
Discovering and developing ideal cell culture methods is important for cell biology, drug development, and cell therapy. Recent studies have explored and demonstrated the use of nanoscale structures and patterns that influence cell behavior, such as 3D scaffolds. In this study, we analyzed the effects of nanopatterned-surface dishes using chondrocytes as model cells. Chondrocytes grown on nanopatterned dishes exhibited rounded shapes. Interestingly, chondrocytes have a lower COL10 mRNA level when cultured using nanopatterned dishes. The nanopatterned dishes induced G0-/G1-phase cell cycle arrest and reduced the rate of proliferation. Our results suggest that nanoscale structures can directly control cellular behaviors and can be used for chondrocyte cell culture without causing chondrocytes to lose their functions. These results help to elucidate cellular responses and behaviors in native-like environments, and this information can be used to improve human health.
Introduction
The conventional 2D cell culture techniques enable researchers to elucidate cellular and molecular biological processes, mechanisms of disease, and mode of drug action and to mass-produce cells for cell therapy.1 To this end, cells should be grown and maintained appropriately based on parameters such as temperature, CO2 gas concentration, cell growth medium, and presence of other nutrients. On the basis of optimization with finite-parameters for cell cultures, the efficacy of treatment modality has being monitored by changing cellular characteristics in morphology, viability, and proliferation rate within cells of the same group. However, depending on the cell type, cells lose their structural and functional characteristics during in vitro cell culture due to the differences between in vivo conditions and in vitro culture environments.2 Accordingly, when researchers aimed to conduct all researches using cell lines, it is not clear whether the loss of phenotypes comes from the culture environment or modality of target treatment. Thus to address this problem, the fully understood and normalized environmental factors for cell culture are required, and novel factors should be found to control the functions of cells during cultivation for cell expansion or maintenance.As well known, cell–cell and cell–matrix interactions play important roles in the regulation of cellular functions.3 Many factors, such as biological cues and physical cues, can affect these cellular interactions.2 Thus, the current trends of cell culture studies have focused on developing methods involving novel biological cues and on examining the influences of physical cues to control cellular functions. Indeed, numerous cellular mechanism studies have reported biological cues, including signalling and bioactive molecules in the extracellular matrix (ECM), which are easily able to create synthetic media to mimic the in vivo condition.4 Physical cues have been regulated using cell culture substrates developed with various surface charges, topographies, and mechanical properties.5 These advanced cell culture techniques enabling regulation of biological cues and physical cues are potentially better than conventional cell culture techniques for maintaining the specialized functions of different cell lines.6–8
The development of surface patterning techniques utilizing different biocompatible materials, such as inorganic non-metallic materials, metals, small molecules, and functional polymers, at the micro- and nanoscales has paved the way for the mimicking an in vivo systems about body's reactions.9–12 In recent decades, studies involving materials with regular surface topographies, such as ordered nanofibers, nanolines, nano- or microgrooves, squares and grids, have been extensively reported, and various types of topologies have been found to mimic cell microenvironments and to control cell behaviors such as cell orientation, proliferation, and ECM secretion by fibroblasts and myoblasts, osteogenic differentiation and neuronal differentiation.13–16 These studies have suggested that ideal regular topographies promoted differentiation of stem cells and secretion of ECM components and allowed rearrangement of the cell array. However, those studies were focused on stem cells,17,18 cardiomyocytes,19,20 and neural cells.21,22 In fact, no studies have evaluated the effect of nano-patterned surface on function and proliferation of primary cells that related to the loss of their phenotype during culture.
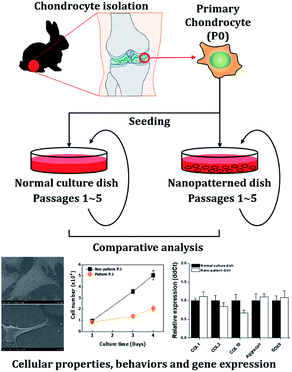
Chondrocytes are specialized cells of mesodermal origin and are the only cells found in cartilage.23 Chondrocyte culture is important for the evaluation of the therapeutic, toxicological effects of drugs on cartilaginous tissue, and cartilage tissue engineering. However, as chondrocytes in monolayer culture proliferated, the cells gradually lose their differentiated phenotype, which is indicated by the loss of synthesis of type II collagen.24 In this study, we investigated the effects of a nanopatterned surface on chondrocytes. To analyze these effects, we observed the cellular properties and behaviors, including morphology, adhesion, proliferation, viability, and cell cycle distribution of chondrocytes cultured on dishes with nanopatterned surfaces. Then, we analyzed the expression of genes related to the cell cycle and differentiation by quantitative PCR analysis (Scheme 1).
Results and discussion
Nanopatterned surface-induced changes in chondrocyte morphology
To investigate the effects of the nanopatterned dishes on chondrocyte shape and orientation at the single-cell level, SEM was performed 3 days after chondrocyte seeding. As shown in Fig. 1, the chondrocytes assumed 3D-like cell shapes on the nanopatterned dishes and were relatively small compared to the flat cells on the normal culture dishes. In the normal culture dishes, the chondrocytes were well spread, with the edges of the cells becoming very thin. On the other hand, the chondrocytes on the nanopatterned dishes were mostly parallel to one another with increased structural heights and showed adhesion to nonpore sites.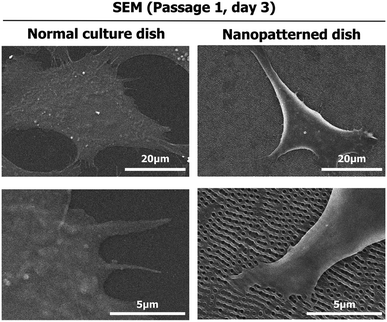 | ||
| Fig. 1 Scanning electron microscopy (SEM) images of the morphology of chondrocytes cultured on normal and nanopatterned dishes for 3 days. | ||
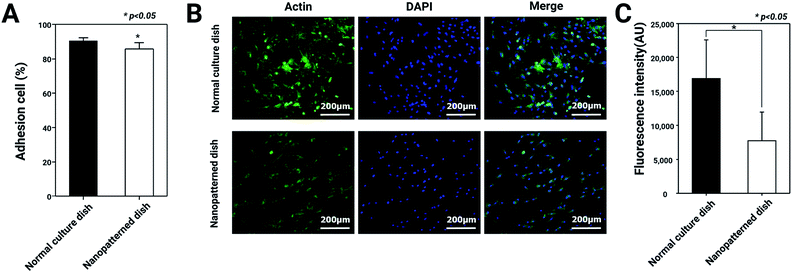
Adhesion ability and actin cytoskeletons of chondrocytes on nanopatterned dishes
The morphological changes in the chondrocytes on the nanopatterned dishes were observed using SEM. In general, cell morphology plays important roles in cell attachment, actin cytoskeleton regulation, and differentiation.25 Based on this knowledge, we hypothesized that F-actin expression and the adhesion ability of chondrocytes would decrease with culture on the nanopatterned surface. To test this hypothesis, we analyzed adhesion ability and F-actin expression in chondrocytes cultured on normal and nanopatterned dishes. As shown in Fig. 2, the percentages of adhered cells were 91.3 ± 3.1% and 86 ± 6.3% on the normal culture dishes and nanopatterned dishes, respectively. Interestingly, the chondrocyte spreading area on nanopatterned dish was 0.6 fold lower than normal culture dish (Fig. S1†), however the difference of adhesion rates between the groups was approximately 5%. In addition, in the cells cultured on the normal culture dishes, the fluorescence intensity of F-actin staining was high, and F-actin was densely arranged. On the other hand, the fluorescence intensity of F-actin staining was weaker in the cells cultured on the nanopatterned dishes than in the cells cultured on the normal culture dishes (Fig. 2C). These results suggested that the nanopatterned dishes changed the shapes of the cells to 3D structures, which decreased F-actin expression in the chondrocytes.Effects of nanopatterned dishes on chondrocyte proliferation
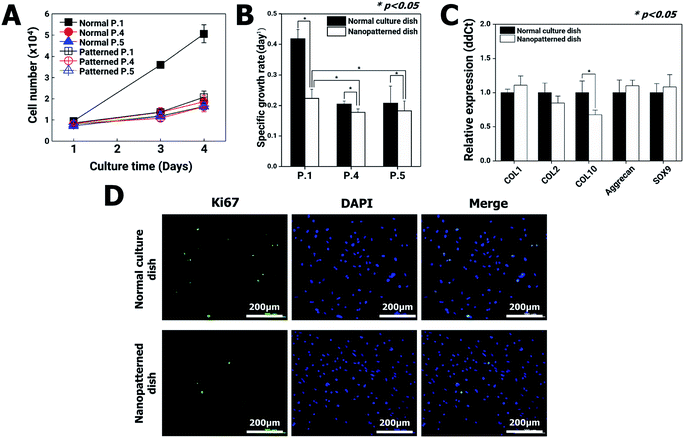
Changes in cytoskeletal dynamics that cause alterations in cellular morphology and adhesion may lead to cell death via an apoptosis-like pathway and changes in cellular processes such as growth, motility, senescence, and division.26 The nanopatterned dishes resulted in decreased F-actin protein levels in chondrocytes, which had the potential to decrease cell proliferation. To address this possibility, chondrocyte proliferation, apoptosis, senescence, gene expression, and cell viability were analyzed using the first passages of chondrocyte.On initial seeding in normal culture dishes (passage 1, P1), the chondrocytes rapidly grew, and the cell populations almost doubled each day for 4 days. After the second passage in normal culture dishes, however, the chondrocyte growth rate dramatically decreased. These results are in general agreement with those of previous reports revealing that chondrocytes gradually lose their differentiated phenotype.27 On the other hand, in the nanopatterned dishes, the growth rates of chondrocytes exhibited similar patterns following the second passage of normal culture dishes (Fig. 3A and B). To quantitatively analyze the changes in gene expression in chondrocytes cultured on nanopatterned dishes, we performed real-time qRT-PCR of several well-known chondrogenic marker genes (collagen type I (COL1), collagen type II (COL2), collagen type X (COL10), aggrecan and SOX9) using P1 chondrocytes after 4 days of culture on normal and nanopatterned dishes (Fig. 3C). The results showed that COL1, COL2, aggrecan, and SOX9 mRNA expression was not significantly different between the normal and nanopatterned dish culture groups. Interestingly, the level for COL10 mRNA in the nanopatterned dish group was 0.6-fold that in the normal dish group (n = 3, p > 0.05). Additionally, expression of Ki67, a proliferation marker, was observed in P1 chondrocytes. The Ki67 expression of the chondrocytes cultured on the nanopatterned dishes was lower than that of the chondrocytes cultured on the normal culture dishes (Fig. 3D).
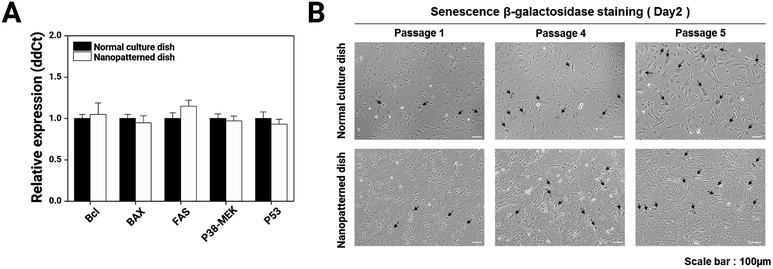
To better elucidate the effects of the nanopatterned dishes in chondrocyte culture, we performed comparative real-time qRT-PCR for many well-known apoptosis marker genes (Bcl, BAX, FAS, P38-MEK, and P53) on P1 chondrocyte cells cultured for 4 days on normal and nanopatterned dishes (Fig. 4A) and performed cellular senescence assays (Fig. 4B). The apoptosis marker genes showed similar expression levels between the normal and nanopatterned dish-cultured chondrocytes. Chondrocyte senescence also was not altered by culture on the nanopatterned dishes. These results indicated that the nanopatterned dishes affected chondrocyte proliferation.
Articular cartilage defects due to inflammatory causes, trauma, and aging are common.28,29 Due to the limited self-regeneration of cartilage after damage,30 several new procedures have recently emerged, such as implantation of cartilage tissue formed in vitro to repair widespread or severe articular cartilage damage.31–33 The successful growth of cartilage tissue in vitro requires sufficient numbers of chondrocytes34,35 with retained character. Current strategies, including micro or nanopatterned-surface culture dishes,36 3D culture techniques37–39 using collagen40 or alginate scaffolds,41 and techniques using biodegradable polymers,42 have been investigated to keep the chondrocyte function. These promising studies have suggested that ideal scaffolds enable chondrocytes to retain their functions during in vitro culture. In our study, we analyzed the effects of nanopatterned dishes with a diameter of 200 nm and a depth of 500 nm with a distance of 500 nm between pore centers on chondrocyte characteristics and behaviors. Compared with that in chondrocytes cultured on normal culture dishes, the expression of chondrogenic markers (COL1, COL2 Aggrecan, and SOX9) was similar, but the expression of COL10 was significantly lower, in chondrocytes cultured on nanopatterned dishes (Fig. 3B). COL10 is a characteristic marker of chondrocyte hypertrophy, the last stage of differentiation in the chondrogenic cell lineage.43 Therefore, the results regarding the expression of chondrogenic markers suggested that the chondrocytes cultured on the nanopatterned dishes retained their specific functions.
G0-/G1-phase arrest of chondrocytes cultured on nanopatterned dishes
On the nanopatterned dishes, chondrocytes regained round shapes like 3D-structure (Fig. 1). Cell shape is a product of the interaction of a cell with external conditions. Therefore, the change in chondrocyte shape was caused by the physical interaction with the surface of the nanopatterned dish. Chondrocyte morphology in particular directly influences specific cellular behaviors and functions and ECM production.44 However, the chondrocytes on nanopatterned dishes have shown similar to normal culture dishes about adhesion rate, cellular processes such as growth, motility, senescence, and division without cytoskeletal dynamics.In general, two types of extracellular signals, hormone/growth factor stimulation, and adhesion to the ECM are required for cells to proliferate.45,46 In addition, the destabilization of actin filaments is known to influence cell cycle progression and inhibit cell proliferation.47 Based on this knowledge, we hypothesized that the nanopatterned dishes would inhibit chondrocyte proliferation, causing cell cycle arrest. To test this hypothesis, the mRNA expression levels of key genes related to the MAPK and JAK/STAT signalling pathways and the cell cycle were investigated, and FACS cell cycle assays were performed.
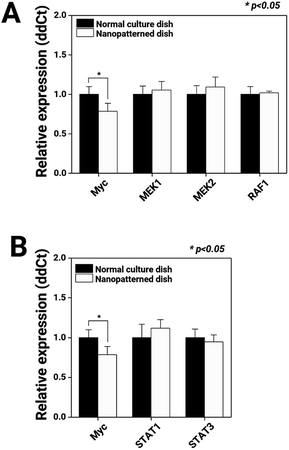
First, we observed the MAPK and JAK/STAT signalling pathways, which regulate cell proliferation, through mRNA expression analysis (Fig. 5). Compared with chondrocytes cultured on normal dishes, chondrocytes cultured on nanopatterned dishes exhibited similar expression levels of MEK1, MEK2, RAF1, STAT1, and STAT2, key genes of the MAPK and JAK/STAT signalling pathways. Interestingly, the c-Myc mRNA expression level in the nanopatterned dish group was 0.7-fold that in the normal dish group (n = 3, p > 0.05). c-Myc plays an integral role in cellular proliferation, as understood from its high levels in proliferating cells, its low levels in quiescent cells, its rapid increases during growth factor-stimulated proliferation, and its ability to stimulate S-phase entry and shorten the cell cycle.48 Additionally, the majority of these Myc-responsive targets regulate the activity of G1 cyclin-dependent kinases (CDKs), primarily CDK2, thereby promoting transport through the G1/S transition.49 Thus, in our study, the low c-Myc expression levels in the chondrocytes on the nanopatterned dishes indicated arrest in the G1 phase.
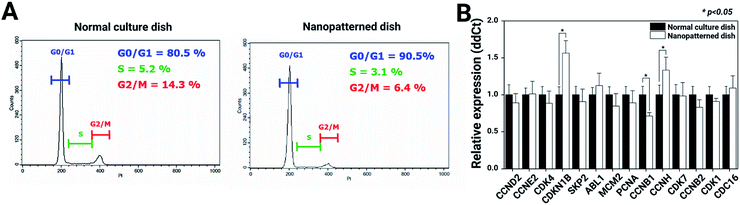
To better understand the suppressive effects on c-Myc expression, we performed a FACScan analysis of the cell cycle profile changes in chondrocytes cultured on normal vs. nanopatterned dishes (Fig. 6A). The results showed that the percentage of G0-/G1-phase cells among chondrocytes cultured on nanopatterned dishes was 90.5%; this percentage was 10% higher than that among chondrocytes cultured on normal culture dishes, while the percentages of S- and G2-/M-phase cells were 2.1% and 7.9% lower among nanopatterned dish-cultured chondrocytes than among normal dish-cultured chondrocytes. These findings indicated that the suppressive effect of the nanopatterned dishes on chondrocyte proliferation was mediated by G0/G1 cell cycle arrest. To further explore the mechanism by which the nanopatterned dishes inhibited cell proliferation and the G0/G1 transition, the real-time qRT-PCR analysis was performed to examine the mRNA expression of cell cycle-regulated genes in pure G1-, S-, G2- or M-phase chondrocytes. As shown in Fig. 6B, the mRNA expression of cyclin-dependent kinase inhibitor 1B (CDKN1B) and cyclin H (CCNH) in chondrocytes cultured on nanopatterned dishes was 1.6-fold and 1.3-fold that of chondrocytes cultured on normal dishes (n = 3, p > 0.05), respectively, and Cyclin B1 (CCNB1) mRNA expression in the nanopatterned dish group was 0.6-fold that in the normal dish group (n = 3, p > 0.05). Generally, CDKN1B expression strongly correlates with a decreased rate of cell cycle entry at the G1/S transition, and high expression of CDKN1B negatively regulates G1-phase cell cycle progression. CCNH is the catalytic subunit of the CDK-activating kinase (CAK, CCNH-CDK7) enzymatic complex. The CAK complex tightly regulates the activity of CDK1/CCNB1, which initiates entry into mitosis. CCNB1 is involved in G2-to-M-phase progression. These mRNA expression profiles suggested that the nanopatterned dishes induced G0/G1 arrest in chondrocytes.
The chondrocytes cultured on nanopatterned dishes showed lower rates of proliferation than those cultured on normal culture dishes (Fig. 3A and B) causing the cell cycle arrest. Generally, the rate of proliferation usually decreases with cell specialization, and most cells in adult animals are arrested in the G0/G1 stage of the cell cycle.50 The nanopatterned dishes induced G0-/G1-phase arrest of chondrocytes through regulation of c-Myc, CCNB1, CDKN1B, and CCNH (Fig. 5 and 6). c-Myc is a transcription factor that is involved in major biological processes, including proliferation, metabolism, cell cycle progression, apoptosis, growth, differentiation, fibrosis, and polarity establishment.48 Normally, downregulation of c-Myc causes accumulation of cells in the G0/G1 phase via reduction of cyclin E/CDK2 kinase activity and causes cells to become prone to apoptosis. However, in our study, the chondrocytes cultured on the nanopatterned dishes did not exhibit altered expression of apoptotic marker genes (Fig. 4A). In addition, analysis of the mRNA expression profiles of cell cycle-related genes showed that the chondrocytes remained at the cell cycle checkpoints of the G0/G1 and G2 phases. CCNH (upregulated) and CCNB1 (downregulated) are cyclin proteins that control the cell cycle by activating CDK enzymes. Interestingly, these cyclins showed opposing expression patterns in chondrocytes. The CCNH/CDK7 complex plays a role in CDK1/CCNB1 activation, but the expression of CCNB1 was reduced. As a result of this cyclin expression change, the chondrocytes were not allowed to progress into the G2 phase. However, many unanswered questions remain regarding the effects of the nanopatterned dishes, such as how c-Myc was downregulated; why apoptosis was not induced; how CCNB1, CDKN1B, and CCNH were regulated; and why the chondrocytes exhibited mainly G0-/G1-phase arrest.
Even though our and previous studies have shown that 3D cell culture models and nanopatterned dishes enable to mimic the in vivo condition, important challenges remain such as the need for a large number of cells for chondrocyte therapy. Ryuji et al. suggested that the combinatorial method with a 3D cell culture model and synthetic medium promoted cartilage regeneration.51 Specific proliferation medium including TGF-b1, FGF-2, and PDGF enable to increase cell number directly. This concept should provide a solution of chondrocyte culture, but a lot of research is still needed because it shows insufficient results such as incomplete cartilage formation.
A variety of nanopatterns with specific topographies, including nanopits,14 nanopillars,19 and nanogrooves,52 have been developed. Nanoscale patterning has been shown to prevent or promote cell adhesion on biomaterial implants and to direct cell growth and differentiation during the creation of in vitro-engineered tissue or the generation of in vitro tissue models. Previous nanoscale studies have used the fibroblasts and cardiomyocytes as mesoderm lineage, and have used various nanoscale topographical structures and patterns, interestingly, cellular effects similar to those in our study, including reduced proliferation and decreased cytoskeletal parameters, have been observed.14,19,52,53 Also, proliferation rate of mouse embryonic fibroblast (mEF) on nanopatterend dish was reduced (Fig. S3†). As we known, the mEF was used as feeder cells, mEF growth were fully inhibited by mitomycin-C for many embryonic stem cell (ESC) and induced pluripotent stem cells (iPSC) culture.54 Nanopatterned dish can provide the stem cell culture system with treatment of mitomycin-C. Although we need the more data to get strong insights for effects of nanoscale structure on germline or other cell lines, our and previous efforts have suggested that nanoscale structures can directly control cellular behaviors. Thus, these results can be used to develop co-culture system for various type of cells with different proliferation rate.
Conclusions
The goal of this study was to investigate the influences of nanopatterned dishes on chondrocyte functions. Chondrocytes cultured on nanopatterned dishes showed behaviors similar to those of chondrocytes cultured in 3D systems. However, the nanopatterned dishes led to decreased chondrocyte proliferation and cell cycle arrest. The biological responses of cells to nanoscale patterns and 3D structures are not currently understood. Our results help to elucidate cellular responses and behaviors in native-like environments. Furthermore, for in vitro co-culture with two or more cell types having different growth rates, growth inhibition of cells with a rapid growth rate is often required. However, current growth inhibition methods have several limitations. Chemical or ultraviolet treatments induce a reduction of the functions of cells such as cellular activity and metabolic capacity. In our study, we found nanopatterned structure can decrease growth rate without other changes in cellular functions. Thus, in stem cell studies for basic research, drug discovery, or therapeutic applications, nanopatterned dishes will be able to provide a very useful way to control cell proliferation of feeder cell lines without genotoxic chemical treatment, and this information can be used to improve human health.Experimental section
Nanopatterned surface dish
The nanopatterned 35 mm culture dishes were provided from Materials processing & Integrated Biosystems Laboratory (MIB), Pohang, Korea.55 As briefly described, the nanopatterned surface dish was composed of polystyrene (PS) and was fabricated by the procedure for the nickel nano-mold insert using the two-step anodization and the electroforming process. To enhance the hydrophilicity and the attachment of cells, the dishes were exposed to oxygen plasma (VITAI; Femto science, Dongtangiheung, Korea). The surface area of nanopatterned surface dishes is 8.8 cm2. Each pore of the nanopatterned surface dishes employed a diameter of 200 nm and a depth of 500 nm with a distance of 500 nm between pore centers (Fig. S2†).Chondrocyte isolation and culture
All animal studies were performed in compliance with the guidelines of the Institutional Animal Care and Use Committee of the Korea Institute of Toxicology (IACUC #1304-0113) and the Guidelines for the Care and Use of Laboratory Animals of the National Research Council and experiments were approved by the Animal Ethics Committee of “KIT Rabbit articular cartilage was isolated from the knee joints of New Zealand white rabbits (n = 5, Jung-Ang Lab Animal, Seoul, Korea) using a sterile scalpel. The cartilage fragments were chopped, washed in phosphate-buffered saline (PBS; pH 7.4), and digested with 0.5% collagenase type II (Sigma-Aldrich, Missouri, USA) in Dulbecco's modified Eagle's medium/nutrient mixture F-12 (DMEM/F12, Gibco, NY, USA) for 10 h. The culture medium was supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco) and a combination of the antibiotics penicillin and streptomycin (100 units per mL, Invitrogen, CA, USA). To analyze cellular behavior and gene expression, chondrocytes (1 × 104 cells per plate) were seeded in normal 35 mm culture dishes (Corning, NY, USA) and nanopatterned 35 mm culture dishes and incubated at 37 °C.Scanning electron microscopy (SEM) for cell morphology analysis
After 3 days of culture on the normal 35 mm culture dishes and nanopatterned 35 mm culture dishes, the chondrocytes were washed with 1× PBS and fixed with 2.5% glutaraldehyde/PBS for 1 h. After fixation, they were washed three times with 1× PBS for 10 min per wash. Then, the cells were dehydrated in a graded series of ethanol (50, 70, 90, and 100%) for 30 min per step and left in 100% ethanol until they were subjected to supercritical point CO2 drying. Next, the dried samples were sputter-coated with gold for SEM (JSM-6330F, MA, USA). Micrographs were taken at an accelerating voltage of 10 kV to ensure a suitable image resolution.Chondrocyte viability and adhesion assay
At different times after seeding (1, 3, and 4 days), cell samples were taken, and cell viability was measured using a Cell Counting Kit-8 (Dojindo Molecular Technologies Inc., Kumamoto, Japan) according to the manufacturer's protocol. The cells were manually counted using a haemocytometer. The specific growth rate of cells (μ) was determined using the following eqn (1)
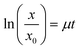 | (1) |
Immunostaining
After 3 days of incubation in vitro, chondrocytes from both groups were fixed with 3.7% paraformaldehyde in 0.1 M PBS (pH 7.4) for 15 min at ambient temperature, permeabilized with 0.5% Triton X-100 (BDH, Poole, UK) in 0.1 M PBS for 10 min and washed with 0.1 M PBS. The samples were then preincubated with 1% bovine serum albumin (BSA; Sigma) in 0.1 M PBS for 5 min to block nonspecific staining. While shielded from light, the samples were stained for filamentous actin (F-actin) with 1 U per mL Alexa Fluor 488-conjugated phalloidin (Molecular Probes, Eugene, OR, USA) in 1% BSA in 0.1 M PBS for 15 min and then washed with 0.1 M PBS for 5 min. Ki67 was labeled with an anti-Ki67 primary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 dilution, Abcam) overnight at 4 °C. The sections were then incubated with an anti-rabbit Alexa Fluor 546 secondary antibody (1
400 dilution, Abcam) overnight at 4 °C. The sections were then incubated with an anti-rabbit Alexa Fluor 546 secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution; Life Technologies) for 3 h at room temperature in a 1% BSA solution. The nuclei were stained with 1
1000 dilution; Life Technologies) for 3 h at room temperature in a 1% BSA solution. The nuclei were stained with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 DAPI (62248; Thermo Fisher Scientific) for 5 min. Finally, fluorescence was visualized using a confocal microscope, and fluorescence intensity was measured using ImageJ software (NIH, Bethesda, MD).
500 DAPI (62248; Thermo Fisher Scientific) for 5 min. Finally, fluorescence was visualized using a confocal microscope, and fluorescence intensity was measured using ImageJ software (NIH, Bethesda, MD).
Real-time qRT-PCR
For RT-qPCR, each sample consisted of a total of 5 × 105 cells in 1 mL. Total RNA was isolated in a single-step method using TRIzol reagent. cDNA synthesis was performed with 1 μg of total RNA per sample using random primers and a PrimeScript 1st Strand cDNA Synthesis Kit (Takara Biotech, Kusatsu, Shiga, Japan). Real-time PCR was performed on an ABI PRISM 7000 sequence detection system (Applied Biosystems, Foster City, CA) using specific primers (Table S1†).Cellular senescence analysis
Chondrocytes were washed twice with PBS and stained using a senescence-associated β-galactosidase (SA-β-gal) staining kit (Abcam, Cambridge, UK) according to the manufacturer's instructions. Senescent (SA-β-gal-positive) cells were counted under an inverted microscope (CKX 41SF; Olympus Corporation, Tokyo, Japan).Cell cycle analysis by FACS
The cell cycle was assessed by staining of the DNA content with propidium iodide. Briefly, primary chondrocytes were plated at a density of 2 × 104 cells per plate and incubated for 24 h. The fresh medium was applied to the culture dishes, and the cells were further incubated for 24 h. Following incubation, the cells were harvested and fixed with 70% ethanol in PBS overnight. The fixed cells were incubated with RNase A (50 μg mL−1) for 25 min before their nucleic acids were stained with propidium iodide (50 μg mL−1) for 5 min. The DNA content of 5 × 104 cells per plate was analyzed by flow cytometry (Partec GmbH, Münster, Germany), and the results are presented as histograms. The cells in distinct cell cycle phases were quantified using FloMax analysis software (Partec GmbH).Statistical analysis
The statistical significance of differences between the experimental groups was assessed via two-tailed Student's t-tests. A p-value <0.05 was considered to indicate significance.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF-2016M3A9B4919655, NRF-2019M3A9H1103331, and NRF-2020R1A2C2100794) funded by the Korean government.References
- M. J. Lerman, J. Lembong, S. Muramoto, G. Gillen and J. P. Fisher, Tissue Eng., Part B, 2018, 24, 359–372 CrossRef CAS.
- R. Edmondson, J. J. Broglie, A. F. Adcock and L. Yang, Assay Drug Dev. Technol., 2014, 12, 207–218 CrossRef CAS.
- S. Bersini, I. K. Yazdi, G. Talò, S. R. Shin, M. Moretti and A. Khademhosseini, Biotechnol. Adv., 2016, 34, 1113–1130 CrossRef CAS.
- T. Yao and Y. Asayama, Reprod. Med. Biol., 2017, 16, 99–117 CrossRef.
- M. Mirbagheri, V. Adibnia, B. R. Hughes, S. D. Waldman, X. Banquy and D. K. Hwang, Mater. Horiz., 2019, 6, 45–71 RSC.
- B. Leopold, J. Strutz, E. Weiss, J. Gindlhuber, R. Birner-Gruenberger, H. Hackl, H. M. Appel, S. Cvitic and U. Hiden, Histochem. Cell Biol., 2019, 152, 377–390 CrossRef CAS.
- M. Kapałczyńska, T. Kolenda, W. Przybyła, M. Zajączkowska, A. Teresiak, V. Filas, M. Ibbs, R. Bliźniak, Ł. Łuczewski and K. Lamperska, Arch. Med. Sci., 2018, 14, 910–919 Search PubMed.
- D. Lv, Z. Hu, L. Lu, H. Lu and X. Xu, Oncol. Lett., 2017, 14, 6999–7010 Search PubMed.
- L. Chen, C. Yan and Z. Zheng, Mater. Today, 2018, 21, 38–59 CrossRef CAS.
- H. Jeon, S. Koo, W. M. Reese, P. Loskill, C. P. Grigoropoulos and K. E. Healy, Nat. Mater., 2015, 14, 918–923 CrossRef CAS.
- K. A. Kilian, B. Bugarija, B. T. Lahn and M. Mrksich, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4872–4877 CrossRef CAS.
- H. Takahashi, M. Nakayama, K. Itoga, M. Yamato and T. Okano, Biomacromolecules, 2011, 12, 1414–1418 CrossRef CAS.
- E. N. Sevcik, J. M. Szymanski, Q. Jallerat and A. W. Feinberg, Curr. Protoc. Cell Biol., 2017, 75, 10.23.11–10.23.25 Search PubMed.
- M. J. Dalby, N. Gadegaard, M. O. Riehle, C. D. W. Wilkinson and A. S. G. Curtis, Int. J. Biochem. Cell Biol., 2004, 36, 2005–2015 CrossRef CAS.
- Y. S. Choi, L. G. Vincent, A. R. Lee, K. C. Kretchmer, S. Chirasatitsin, M. K. Dobke and A. J. Engler, Biomaterials, 2012, 33, 6943–6951 CrossRef CAS.
- F. X. Jiang, B. Yurke, B. L. Firestein and N. A. Langrana, Ann. Biomed. Eng., 2008, 36, 1565 CrossRef.
- A. Shukla, J. H. Slater, J. C. Culver, M. E. Dickinson and J. L. West, ACS Appl. Mater. Interfaces, 2016, 8, 21883–21892 CrossRef CAS.
- X. Yao, R. Peng and J. Ding, Adv. Mater., 2013, 25, 5257–5286 CrossRef CAS.
- D. H. Kim, P. Kim, I. Song, J. M. Cha, S. H. Lee, B. Kim and K. Y. Suh, Langmuir, 2006, 22, 5419–5426 CrossRef CAS.
- T. C. McDevitt, J. C. Angello, M. L. Whitney, H. Reinecke, S. D. Hauschka, C. E. Murry and P. S. Stayton, J. Biomed. Mater. Res., 2002, 60, 472–479 CrossRef CAS.
- S. Turunen, A. M. Haaparanta, R. Aänismaa and M. Kellomäki, J. Tissue Eng. Regener. Med., 2013, 7, 253–270 CrossRef CAS.
- M. R. Hynd, J. N. Turner and W. Shain, J. Biomater. Sci., Polym. Ed., 2007, 18, 1223–1244 CrossRef CAS.
- A. M. Craft, N. Ahmed, J. S. Rockel, G. S. Baht, B. A. Alman, R. A. Kandel, A. E. Grigoriadis and G. M. Keller, Development, 2013, 140, 2597–2610 CrossRef CAS.
- M. B. Goldring, K. L. Culley, E. Wondimu and M. Otero, in Kelley and Firestein's Textbook of Rheumatology, ed. G. S. Firestein, R. C. Budd, S. E. Gabriel, I. B. McInnes and J. R. O'Dell, Elsevier, 10th edn, 2017, pp. 34–59.e33, DOI:10.1016/B978-0-323-31696-5.00003-6.
- M. Bao, J. Xie, A. Piruska and W. T. S. Huck, Nat. Commun., 2017, 8, 1962 CrossRef.
- B. N. Brandhagen, C. R. Tieszen, T. M. Ulmer, M. S. Tracy, A. A. Goyeneche and C. M. Telleria, BMC Cancer, 2013, 13, 35 CrossRef CAS.
- S. W. Kang, S. P. Yoo and B. S. Kim, Biomed. Mater. Eng., 2007, 17, 269–276 CAS.
- K. Johnson, S. Zhu, M. S. Tremblay, J. N. Payette, J. Wang, L. C. Bouchez, S. Meeusen, A. Althage, C. Y. Cho, X. Wu and P. G. Schultz, Science, 2012, 336, 717–721 CrossRef CAS.
- C. Vinatier and J. Guicheux, Ann. Phys. Rehabil. Med., 2016, 59, 139–144 CrossRef CAS.
- M. Liu, X. Zeng, C. Ma, H. Yi, Z. Ali, X. Mou, S. Li, Y. Deng and N. He, Bone Res., 2017, 5, 17014 CrossRef CAS.
- C. Fan and D.-A. Wang, Eur. Polym. J., 2015, 72, 651–660 CrossRef CAS.
- J. L. Drury and D. J. Mooney, Biomaterials, 2003, 24, 4337–4351 CrossRef CAS.
- Y. Liu, G. Zhou and Y. Cao, Engineering, 2017, 3, 28–35 CrossRef CAS.
- B. J. H. Driessen, C. Logie and L. A. Vonk, Cell Biol. Toxicol., 2017, 33, 329–349 CrossRef.
- B. M. Nam, B. Y. Kim, Y. H. Jo, S. Lee, J. G. Nemeno, W. Yang, K. M. Lee, H. Kim, I. J. Jang, T. Takebe and J. I. Lee, Transplant. Proc., 2014, 46, 1145–1149 CrossRef CAS.
- J. Shaik, J. Shaikh Mohammed, M. J. McShane and D. K. Mills, J. Med. Eng., 2013, 2013, 560328 Search PubMed.
- P. Smeriglio, J. H. Lai, F. Yang and N. Bhutani, J. Visualized Exp., 2015, 104, e53085 Search PubMed.
- K. E. Benders, P. R. van Weeren, S. F. Badylak, D. B. Saris, W. J. Dhert and J. Malda, Trends Biotechnol., 2013, 31, 169–176 CrossRef CAS.
- X. Zhang, J. Zhu, F. Liu, Y. Li, A. Chandra, L. S. Levin, F. Beier, M. Enomoto-Iwamoto and L. Qin, Bone Res., 2014, 2, 14015 CrossRef CAS.
- M. B. Pabbruwe, E. Esfandiari, W. Kafienah, J. F. Tarlton and A. P. Hollander, Biomaterials, 2009, 30, 4277–4286 CrossRef CAS.
- C. C. Wang, K. C. Yang, K. H. Lin, H. C. Liu and F. H. Lin, Biomaterials, 2011, 32, 7118–7126 CrossRef CAS.
- Z. Gugala and S. Gogolewski, J. Biomed. Mater. Res., 2000, 49, 183–191 CrossRef CAS.
- A. Yamashita, S. Nishikawa and D. E. Rancourt, PLoS One, 2010, 5, e10998 CrossRef.
- M. B. Goldring, Ther. Adv. Musculoskeletal Dis., 2012, 4, 269–285 CrossRef CAS.
- S. Liu, W. R. Kam, J. Ding, M. P. Hatton and D. A. Sullivan, Invest. Ophthalmol. Visual Sci., 2013, 54, 2541–2550 CrossRef CAS.
- S. Schlie-Wolter, A. Ngezahayo and B. N. Chichkov, Exp. Cell Res., 2013, 319, 1553–1561 CrossRef CAS.
- Y. W. Heng and C. G. Koh, Int. J. Biochem. Cell Biol., 2010, 42, 1622–1633 CrossRef CAS.
- C. M. Benaud and R. B. Dickson, Oncogene, 2001, 20, 4554–4567 CrossRef CAS.
- R. C. O'Hagan, M. Ohh, G. David, I. M. de Alboran, F. W. Alt, W. G. Kaelin Jr and R. A. DePinho, Genes Dev., 2000, 14, 2185–2191 CrossRef.
- J. P. Matson and J. G. Cook, FEBS J., 2017, 284, 362–375 CrossRef CAS.
- R. Okubo, Y. Asawa, M. Watanabe, S. Nagata, M. Nio, T. Takato, A. Hikita and K. Hoshi, Regenerative therapy, 2019, 11, 306–315 CrossRef.
- W. Loesberg, J. Te Riet, F. Van Delft, P. Schön, C. Figdor, S. Speller, J. van Loon, X. Walboomers and J. Jansen, Biomaterials, 2007, 28, 3944–3951 CrossRef CAS.
- H. Sunami, I. Yokota and Y. Igarashi, Biomater. Sci., 2014, 2, 399–409 RSC.
- M. Acquarone, T. M. de Melo, F. Meireles, J. Brito-Moreira, G. Oliveira, S. T. Ferreira, N. G. Castro, F. Tovar-Moll, J.-C. Houzel and S. K. Rehen, Front. Cell. Neurosci., 2015, 9, 97 Search PubMed.
- K. J. Cha, M.-H. Na, H. W. Kim and D. S. Kim, J. Micromech. Microeng., 2014, 24, 055002 CrossRef.
- M. J. Humphries, in Extracellular Matrix Protocols, ed. C. H. Streuli, Springer, New York, NY, 2009, pp. 203–210 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra08256b |
| ‡ Present address: Applied Bioresources Research Division, Freshwater Bioresources Utilization Bureau, Nakdonggang National Institute of Biological Resource, Sangju, Korea. |
| This journal is © The Royal Society of Chemistry 2021 |