Understanding the self-ordering of amino acids into supramolecular architectures: co-assembly-based modulation of phenylalanine nanofibrils†
Prabhjot
Singh
a,
Satish K.
Pandey
b,
Aarzoo
Grover‡
c,
Rohit K.
Sharma
 c and
Nishima
Wangoo
c and
Nishima
Wangoo
 *d
*d
aCentre for Nanoscience and Nanotechnology, Panjab University, Sector 25, Chandigarh, 160014, India
bCentral Scientific Instruments Organization, Sector-30C, Chandigarh-160030, India
cDepartment of Chemistry & Centre for Advanced Studies in Chemistry, Panjab University, Sector 14, Chandigarh-160014, India
dDepartment of Applied Sciences, University Institute of Engineering and Technology (U.I.E.T.), Panjab University, Chandigarh-160014, India. E-mail: nishima@pu.ac.in
First published on 22nd December 2020
Abstract
Amino acids have emerged as promising molecular frameworks for the generation of functional materials owing to the bio-compatibility and thermodynamic stability of their self-assembled architectures. The homogeneous and heterogeneous self-assembly of all naturally occurring amino acids may demonstrate vast functional diversity owing to their common zwitterionic motif, which can serve as a library for the generation of nano/microscale supramolecular biomaterials. Furthermore, amino acids-based self-assembled amyloid structures exhibit pathological relevance, which shifts the paradigm from the self-assembly of polymeric entities to the self-assembly of simple organic molecules or metabolites. Thus, in order to understand the in-built self-ordering behaviour of amino acids, we investigated all naturally occurring amino acids (except aromatic amino acids) for their well-defined morphology and solution-phase β-sheet-like amyloid characteristics. Furthermore, this comprehensive analysis of self-assembly data led to the discovery of aliphatic chain amino acids (ACAAs; Ala, Leu, Ile and Val) as potential candidates for the modulation of phenylalanine fibril toxicity. Thus, this inclusive amino acid self-assembly analysis may pave the way for the design and development of simple organic molecules with inherited self-assembling properties for the generation of functional nanomaterials.
Introduction
The self-assembled architecture made up of small molecular motifs are recognized as the functional force of living systems.1,2 The optimum balance between intermolecular non-covalent forces to acquire structural diversity and functionality may be identified as the first step towards cellular life.3 Inspired by this, the molecular self-assembly of supramolecular entities, such as proteins, peptides, DNA and lipids, has been investigated in much detail to understand the mechanism that regulates and controls the matter flux responsible for the functional aspects of living systems.4 Specifically, self-assembled supramolecular structures of peptides and proteins have been studied in much detail due to their propensity to form amyloid aggregates.5–7Despite this, limited efforts have been made to understand the well-ordered secondary structures formed by small molecules through the available set of non-covalent forces.8,9 However, recent reports have suggested a probable link between the self-assembly of metabolites (such as amino acids and nucleotides) and amyloid-based pathological disorders.10–15 In particular, phenylalanine, tyrosine and adenine structures exhibit amyloid-like toxicity and have been further analyzed for the antibody generation. The mechanism behind many unexplained pathological disorders have been linked to generic β-sheet-type amyloid formation regardless of their molecular origin.16 In addition, proteins with α-helix structures have been reported to display amyloid formation, which extends the complexity of these pathogenic structures.17,18 The occurrence of these pathogenic structures has been linked to serious health problems, such as Alzheimer's, Parkinson's, Chronic Traumatic Encephalopathy (CTE) and phenylketonuria (PKU).19,20 The occurrence of a high phenylalanine concentration in the blood and observation of phenylalanine self-assembled fibrils in the parietal cortex brain tissue have been recognized as diagnostic tools for PKU.12,21 To date, tetrahydrobiopterin with a restricted phenylalanine diet has been used as a treatment for patients suffering from PKU.22 Further, branched chain amino acids have been clinically adopted as a dietary therapy approach, which is believed to inhibit phenylalanine transport across the blood–brain barrier.23 However, the mechanistic of branched chain amino acids to lower the adverse health effects in PKU is still unclear.24
In this regard, various attempts have been made to understand the self-assembly mechanism of phenylalanine so that it can be used to devise an effective strategy to inhibit its amyloid formation for different motifs, mainly by inhibiting the π–π stacking interaction, which is generic in amyloids containing an aromatic residue.25–31 In our earlier report, we investigated the role of the zwitterionic part in the self-assembly of aromatic amino acids using dielectric modulation of the solvent system.32 Observed shifts in morphological patterns were suggested to be correlated with the electrostatic interactions between zwitterionic ends in varying dielectric solvents. Further, aromatic and non-aromatic moieties immobilized over gold nanoparticles were demonstrated to have a modulating effect on the self-assembly of aromatic amino acids.33 Successively, we reported the formation of a phenylalanine dimer assembly nanostructure as the basic building block of amyloids forming nanofibrils, which have been suggested to be responsible for the nerve cell damage in patients suffering from PKU.34 Therefore, looking at the role of self-assembled amino acids in various pathological disorders and understanding the molecular level self-assembly mechanism of amino acid motifs, herein, we report the self-assembly of all naturally occurring amino acids (except for aromatic amino acids, which was reported in a previous report) in a variety of nano to microscale organizational habits.32 It is pertinent to note that we report novel self-assembled structures for all the amino acids (except aromatic, cysteine and methionine) for the first time in this paper. This investigation of the self-assembled pattern generation for all amino acids was divided into five categories based on their hydration potential as well as hydrophobicity index. Specifically, the self-assembled architectures for glycine (Gly), alanine (Ala), leucine (Leu), isoleucine (Ile), methionine (Met), valine (Val) (fern-like crystallite), proline (Pro) (rod crystallite) and cysteine (Cys) (globular floral) were reported in category I; threonine (Thr) (spear-like crystallite) and serine (Ser) (capsule structure) in category II; asparagine (Asn) and glutamine (Gln) (dendritic floral) in category III; aspartic acid (Asp) and glutamic acid (Glu) (membrane-like structure) in category IV; lysine (Lys) (microrod crystallite), and arginine (Arg) (nanorod crystallite) and histidine (His) (fibril) in category V were investigated under an optimum set of conditions using optical and field emission scanning electron microscopy (FE-SEM). The observed self-assembled architectures in these categories indicated the occurrence of an inherited structure–morphology relationship for amino acids that may pave the way for the development of the first of its kind of systematic nanofabrication tool using simple organic molecules. Furthermore, all amino acids were investigated for the amyloid characteristics using an optimized aqueous medium thioflavin T (ThT) assay.
The morphological assessment of all the self-assembled amino acids and the presence of a common zwitterionic motif in monomers inspired us to investigate the role of the co-assembly (in binary mixture) of various amino acids to inhibit phenylalanine fibril formation. Remarkably, aliphatic chain amino acids (ACAAs; Ala, Leu, Ileu and Val) demonstrated a peculiar hindrance to the phenylalanine fibril formation. Furthermore, solution-phase ThT assay for the co-assembled system indicated there was an inhibition of the β-sheet-like structure formation as displayed by phenylalanine. Further, validation of the inhibitory effect exhibited by the co-assembly of ACAAs to reduce phenylalanine cytotoxicity was carried out by using the MTT assay. These intriguing results clearly confirmed the inhibitory role played by ACAAs in regulating the amyloid formation of phenylalanine. Thus, understanding amino acid self-assembly based on microscopic analysis led to the first-of-its-kind molecular structure–morphology pattern relationship. Furthermore, comprehensive analysis of amino acid self-assembly depicted the role of ACAAs in the disruption of phenylalanine fibril formation, which is notable evidence supporting the potential of dietary therapy to work against phenylketonuria.35
Materials and methods
Materials
All the α-amino acids were purchased from Sigma-Aldrich and used as such. Milli-Q water (resistivity ∼18.2 MΩ) used for all the self-assembly experiments. The methanol used for the self-assembly experiments was of analytical grade with >99% purity.Optical microscopy
Optical images of the amino acid self-assembled structures were captured using a light microscope with an in-built camera attachment by depositing 10 μL of the sample solution on a microscopic glass slide.FE-SEM analysis
FE-SEM was used to investigate the morphology of the self-assembled structures. In general, a 10 μL solution of amino acid solution at 1 mg mL−1 concentration was evaporated slowly on the smooth surface of a Ag film pasted on the FE-SEM step. Pt coating was applied on the samples to make them conductive, followed by FE-SEM analysis on a Hitachi, SU8010 electron microscope, operating at 10–15 kV.Confocal laser scanning microscopy
The fluorescence emission signal in the ThT binding assay as well as the intrinsic emission of the self-assembled structures were captured using the Zeiss LSM 510 confocal laser scanning microscope. In general, the samples were prepared by air drying 10 μL of each sample solution on a microscopic glass slide.Fluorescence spectroscopy
The fluorescence spectra were recorded on an Agilent Technologies Cary Bundle instrument. For ThT optimization, various concentrations varying from 0.0001 to 1.0000 mg mL−1 were prepared in an aqueous medium and the fluorescence spectra recorded at an excitation wavelength of 370 nm. For ThT assay, ageing of the amino acid solutions (concentration 80 mM) were done by placing them in a closed vessel and leaving undisturbed at room temperature for 15 days, followed by the addition of optimized ThT concentration (2.5 μM) for the binding assay. The solution-phase ThT binding assay for all the amino acids was performed using optimized ThT as the final concentration in the sample solutions.Cell viability assay
The cell cytotoxicity of the amino acid samples was assessed against HeLa cells via a 3-(4,5)-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. Briefly, HeLa cells were seeded (2 × 104 cells per well) in triplicate in a 96-well culture plate containing RPMI-1640 (supplemented with 5% fetal bovine serum) and incubated for 24 h. Cells were treated with amino acid self-assembled structures at different concentrations for the cytotoxicity assay. After the required duration, the medium was aspirated and 100 μL fresh medium was added to each plate. Thereafter, the cells were treated with 20 μL of MTT solution (2.5 mg mL−1 in PBS) and again left to incubate for 4 h at 37 °C. The supernatant was separated and the resulting formazan crystals were dissolved in 100 μL DMSO. The samples were measured for their absorbance at 570 nm with a microplate reader. The comparative viability of cells was calculated by the O.D. 570 ratio of the treated and untreated cells.Result and discussion
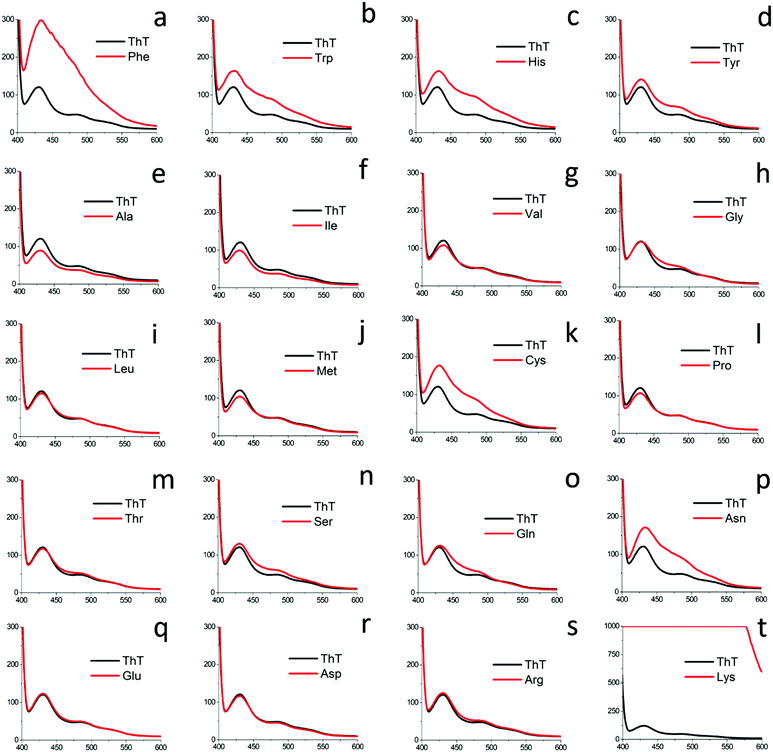
The amyloid-like self-assembled fibril morphology of aromatic amino acids inspired us to perform a comprehensive investigation of the patterned structures for all naturally occurring amino acids. Moreover, the occurrence of a common zwitterionic motif in all the natural amino acids and its indispensable role in determining the self-assembly of amino acids was an added motivation behind this comprehensive investigation.36,37 In this regard, we analyzed the self-assembly of all naturally occurring amino acids (except aromatic amino acids) by dividing them into five categories depending on their hydration potential as well as hydrophobicity index.38–40Category I: self-assembly of glycine and amino acids with a neutral side chain (Gly, Ala, Val, Leu, Ile, Cys, Met)
Considering the properties of amino acids, such as their hydration potential as well as hydrophobicity index, allowed us to devise a strategy to organize all naturally occurring amino acids (except aromatic amino acids) into five categories with a gradual change of properties from a low hydration potential and high hydrophobicity index (category I) to high hydration potential and low hydrophobicity index (category V) [ESI,† Table S1].Based on this rationale, the self-assembly of glycine (Gly), alanine (Ala), leucine (Leu), isoleucine (Ile), valine (Val), proline (Pro), methionine (Met) and cysteine (Cys) was investigated in category I amino acids [Fig. 1(a–h)]. For this purpose, amino acid solutions were prepared in a water and methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) [MW 1.1] solvent system and their self-assembled patterns were observed using optical microscopy and FE-SEM. Remarkably, glycine, ACAAs (Ala, Leu, Ile, Val) and methionine (Met) displayed the formation of exceptionally similar self-assembled patterns forming micro-crystallite structures arranged in fern-like architectures in both water and [MW 1.1] solvent as evident by the optical microscopy and FE-SEM results [Fig. 1(a–f) and ESI,† Fig. S1–S6].
1) [MW 1.1] solvent system and their self-assembled patterns were observed using optical microscopy and FE-SEM. Remarkably, glycine, ACAAs (Ala, Leu, Ile, Val) and methionine (Met) displayed the formation of exceptionally similar self-assembled patterns forming micro-crystallite structures arranged in fern-like architectures in both water and [MW 1.1] solvent as evident by the optical microscopy and FE-SEM results [Fig. 1(a–f) and ESI,† Fig. S1–S6].
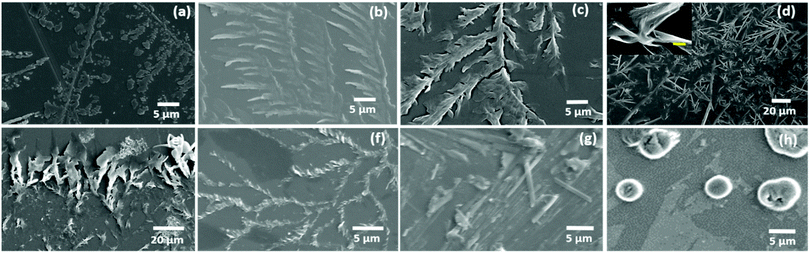 | ||
| Fig. 1 FE-SEM images of Gly (a), Ala (b), Leu (c), Ile (d), Val (e), Met (f), Pro (g) displaying the formation of microcrystals arranged in fern-like architectures, and Cys (h) displaying the formation of globular structures [ESI,† Table S2 for specific set of conditions]. | ||
The formation of a fern-like crystallite for glycine may be solely due to the inter-zwitterionic interactions. In a recent report, the formation of similar needle-like micro crystals for glycine displayed a high piezoelectric coefficient of magnitude equivalent to barium titanate or lead zirconate titanate.41
The occurrence of common micro-crystallite structures demonstrated the ability of zwitterionic interactions to play a dominant role in the molecular recognition in the self-assembly of category I amino acids as depicted by the tightly packed inter-zwitterionic interactions in the single-crystal structures of these amino acids.36 Remarkably, isoleucine, one of the lowest soluble (34 mg mL−1) aliphatic amino acid in water, exhibited restricted crystalline growth to form a nano-sized curved sheet-like morphology approximately 100 nm wide in the MW 1.1 solvent system [Fig. 1d inset, scale bar 400 nm]. The nanodimensional order observed in the case of the isoleucine self-assembled structures suggested that van der Waals interactions between the aliphatic side chain may also play a decisive role in controlling the crystallinity along with the zwitterionic dipolar hydrogen bonds interaction.
The self-assembly of proline did not display any ordered self-assembled architecture at 1 mg mL−1 concentration, which may be due to its high solubility (1620 mg mL−1) in water [ESI,† Fig. S7(a)]. Thus, proline self-assembly was generated using concentration and solvent modulations (5 mg mL−1 and 80% methanol in water), which led to the formation of micro-rod-type crystallite structures [Fig. 1(g) and ESI,† Fig. S7(b, c)]. Interestingly, the self-assembly of cysteine exhibited globular structures in water and MW 1.1 [Fig. 1(h) and ESI,† Fig. S8]. In addition to globular structures, the optical microscopic images for cysteine also displayed an elongated rod-like morphology, which may be detected as fibril-like structures in recent work exhibiting amyloid-like toxicity.42 The interesting morphological shift in the case of cysteine from the ACAAs self-assembled structures may be due to weak S–S interactions as depicted by theoretical studies on the intermolecular interactions of chalcogens.43 Thus, not only the ionic interactions of category I amino acids, but also the van der Waals intermolecular interactions exhibited by the side chain also play a vital role in determining the molecular recognition responsible for the formation of ordered assembly structures.
Category II: self-assembly of amino acids with an hydroxyl side chain (Thr, Ser)
Threonine and serine have greater hydration potentials (−4.88 and −5.06, respectively) as well as hydrophobicity index values (0.7 and 0.8, respectively) than category I amino acids [ESI,† Table S1]. The self-assemblies of threonine and serine were investigated in category II using a water and MW 1.1 solvent system [ESI,† Fig. S9 and S10]. Interestingly, threonine displayed an aligned spear-like structure of identical sizes, as evident through the FE-SEM micrographs [Fig. 2(a) and ESI,† Fig. S9]. The morphological shift from a fern-like structure (category I) to aligned spear-like structure for threonine could be attributed to the introduction of a hydroxyl group in the amino acid side chain. Furthermore, the hydroxyl group in the Thr side chain greatly enhanced the solubility of amino acids from ∼35 mg mL−1 (Ile) to ∼97 mg mL−1 (Thr), which further depicted the ability of threonine to exhibit polar interactions, which may have provided directionality to the self-assembled micro-crystallite structures as visualized by the FE-SEM images. On the other hand, self-assembly analysis of freshly prepared serine at 1 mg mL−1 exhibited the layer formation in FE-SEM micrograph [ESI,† Fig. S10(a)]. The observed deposited layer may be due to extensive solute–solvent interactions owing to the high solubility (425 mg mL−1) of serine in water. Thus, in order to eliminate the solvent and to visualize the structures, self-assembly was performed at 50 °C with an incubation period of 24 h. Interestingly, serine was observed to form orderly aligned capsule-shaped structures both in water and in the MW 1.1 solvent system [Fig. 2(b) and ESI,† Fig. S10 (b, c)]. The hierarchy of the architectural shift from micro-crystalline (Category I) to orderly spear-like crystals (Thr) (Category II) to formation of a capsule-like morphology (Ser) points towards the inherent structure–morphology relationship for self-assembled amino acids.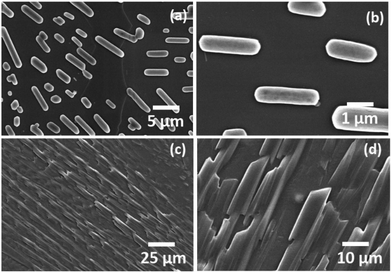 | ||
| Fig. 2 FESEM images depicting the self-assembled architectures of Ser (a and b) and Thr (c and d) using water as the solvent. | ||
Category III: self-assembly of amino acids with an amide linkage in the side chain (Gln, Asn)
The amide bond is the most abundant chemical linkage in biological systems and provides rigidity and directionality to the protein and peptide secondary structures.Consequently, the self-assembly analysis of asparagine (hydration potential, −9.68 and hydrophobicity index −3.5) and glutamine (hydration potential, −9.38 and hydrophobicity index, −3.5) were performed in category III using water and MW 1.1 as the solvent system. The self-assembled structures of both glutamine and asparagine displayed the formation of floral dendritic assemblies with a stacked fibrillary network showing a drastic morphological shift from category I and category II amino acids [Fig. 3(a, b) and ESI,† Fig. S11, S12]. This morphological shift may be correlated to the increase in side chain polar interactions as evident from the hydration potential data [ESI,† Table S1].
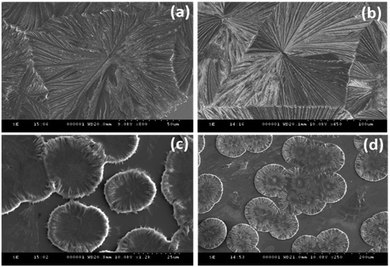 | ||
| Fig. 3 FE-SEM images depicting the self-assembled architectures of Gln (a), Asn (b), Glu (c) and Asp (d). | ||
Category IV: self-assembly of amino acids with a carboxylic group in the side chain (Glu, Asp)
The self-assembly analysis was continued for amino acids with carboxylic acid in the side chain using water and MW 1.1 solvent system. The self-assembly analysis of glutamic acid and aspartic acid was performed in various solvent systems [ESI,† Fig. S13(a–c)] and S14(a–c)]. The observed micrographs revealed the formation of membrane-like structures for glutamic acid in water and 0.1 M HCl whereas aspartic acid displayed similar membrane-like architectures in 0.1 M aqueous ammonia solution. The outer layer of membrane-like morphology for glutamic acid and aspartic acid structures displayed inter-structural diffusion as depicted by FE-SEM images which indicated the soft and permeable nature of these higher order assemblies [Fig. 3(c and d)]. The formation of completely different self-assembled architectures for category IV may be attributed to the higher value of hydration potential (Glu = −10.20; Asp = −10.95) and hydrophobicity index (Glu = −3.5; Asp = −3.5) as compared to category I to III amino acids which indicated increase in polarity of side chain. Despite higher hydration potential, category IV amino acids exhibited low solubility in water (Asp ∼ 5 mg mL−1 and Glu ∼ 8 mg mL−1) that may be due to decreased solute–solvent interactions. Therefore, the formation of such well-ordered membrane-like architectures may be due to optimum balance between inter-side-chain interactions depicted by higher value of hydration potential and low water solubility in water.At this stage, a conceptual hypothesis can be introduced based on the intriguing results, which may give a primitive and first-of-its-kind description of the structural–morphological relationship for amino acid self-assembly. The gradual shift in the self-assembled morphology from category I (crystallite) to category IV (soft membrane) amino acids with the increase in hydration potential value strengthens the possibility of performing an impeccable molecular design linked to the fabrication of nano/microscale architectures [Fig. 4].
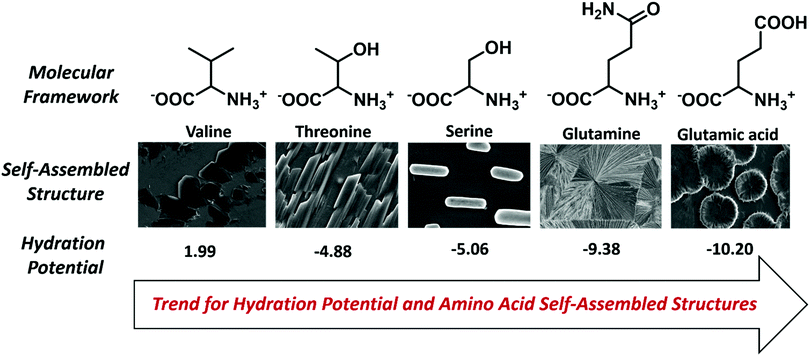 | ||
| Fig. 4 Pictorial description of the hydration potential trend and structural–morphological shift for amino acid self-assembled architectures. | ||
The self-assembly analysis of amino acids in aqueous media with varying degrees of dielectric constant indicated that zwitterionic dipolar interactions play a dominating role in imparting ionic crystallinity to supramolecular structures. The high order morphology generated by these ionic interactions may be termed as hard crystallinity, which seldom is affected by slight changes in the side chain van der Waals interactions as displayed by category I amino acids. On the other hand, an increase in the contribution of inter-side-chain polar interactions between amino acid monomers may shift the morphology from hard crystallinity to soft crystallinity as depicted by the morphological trends from category II to category IV amino acids.
Category V: self-assembly of amino acids with a cationic moiety in the side chain (Lys, His and Arg)
The amino acids with a cationic side chain (hydration potential for Lys, His and Arg are −9.52, −10.27 and −19.92, respectively) exhibited very high solubility in water. This behaviour may be responsible for the suppression of the intermolecular recognition and thereby may impose a restriction on the formation of ordered assembly structures [ESI,† Fig. S15].Therefore, pH modulation was done for self-assembly generation of the cationic amino acids. The self-assembly of lysine was done using 0.1 M NaOH, and it displayed elongated assemblies, which were also visualized as exhibiting an aligned perpendicular growth over the surface [Fig. 5(a and b)].
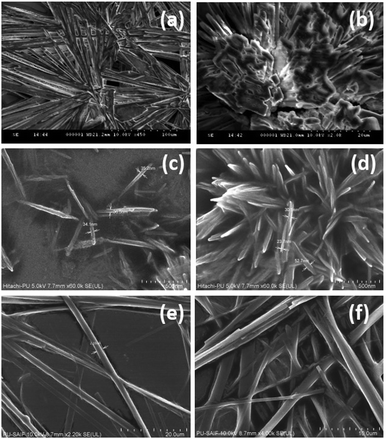 | ||
| Fig. 5 FE-SEM images depicting the self-assembled architectures of Lys (a and b), Arg (c and d) and His (e and f). | ||
Further, arginine displayed nanoscale rod-like crystallinity, which grew to form snowy flake-type structures in 0.1 M aqueous NaOH solution [Fig. 5(c and d)]. The solubility of histidine is higher than the other aromatic amino acids, which may be one of the reasons behind the inability of histidine to show π-stacking-based fibril formation. In this regard, the self-assembly of histidine was performed in basic medium (pH 8 using NaOH) using a water and methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solvent system [Fig. 5(e and f)]. Remarkably, histidine demonstrated the formation of a long-range fibril morphology in the basic medium. Thus, in-depth analysis of the structure–morphological relationship under varying conditions led us to modulate the histidine self-assembled architectures from dendritic rod structures to fibrils.32
1) solvent system [Fig. 5(e and f)]. Remarkably, histidine demonstrated the formation of a long-range fibril morphology in the basic medium. Thus, in-depth analysis of the structure–morphological relationship under varying conditions led us to modulate the histidine self-assembled architectures from dendritic rod structures to fibrils.32
Thus, all the naturally occurring amino acids displayed the formation of highly ordered self-assembled architectures under optimum conditions, which represents the uniqueness of their molecular framework. The structure–morphology relationship of amino acids can be explored to fabricate controlled but diverse nano or microscale materials using synthetic organic chemistry.
Investigation into the amyloid characteristics exhibited by all naturally occurring amino acids
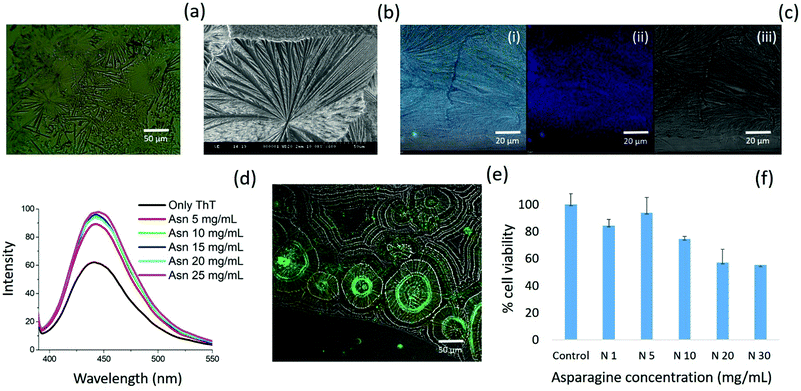
Further, asparagine exhibited a concentration-dependent enhancement in β-sheet formation in aqueous medium as depicted by the ThT assay [Fig. 7(d)]. The specific binding of ThT with asparagine self-assembled patterns was further confirmed by CLSM analysis [Fig. 7(e)]. Finally, the cytotoxicity analysis of the asparagine self-assembled structures was validated by MTT assay, which showed up to 50% cell death at a saturated concentration (30 mg mL−1) of asparagine in water [Fig. 7(f)]. Thus, the comprehensive self-assembly analysis of all naturally occurring amino acids provided valuable information regarding the asparagine amyloid-like behaviour that may be one of the missing links in understanding prion-based pathological disorders.
Application towards the anti-amyloid approach against phenylketonuria
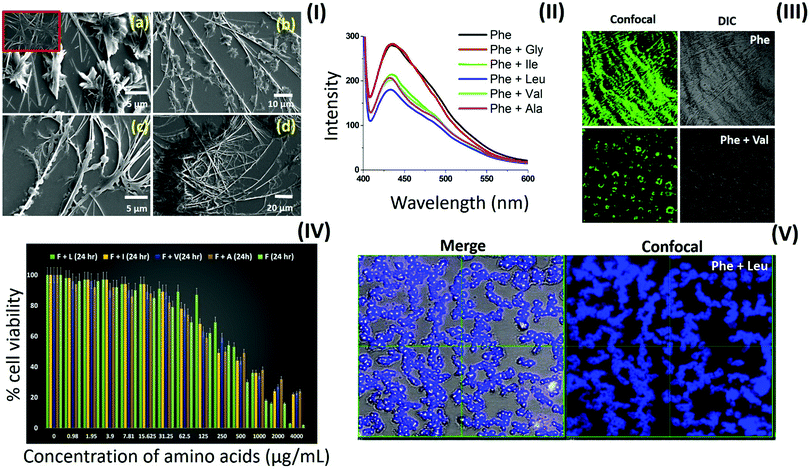
Our inclusive analysis of amino acid self-assembled structures in water and MW solvent system provided valuable insights into the role of zwitterionic and side chain intermolecular interactions in determining the structural–morphological relationship. This understanding may have wide implications in various fields, such as for the development of therapeutics for metabolite amyloids, bioinspired materials design and the generation of multi-component ordered assemblies. The category I amino acids displayed the formation of a fern-like crystallite structure, which may have been driven mainly via electrostatic inter-zwitterionic interactions. Recently, efforts have been made to explore co-crystallization to synergistically expand the structural complexity of amino acid-based supramolecular structures.8 Thus, an amino acid co-assembly approach based on the new understanding of amino acid interactions via a common zwitterionic motif can be utilized to alter the toxicity profile of phenylalanine fibrils. To test this hypothesis, first, the deposited phase co-assembly of phenylalanine with all the amino acids was investigated by optical microscopy [ESI,† Fig. S18–S36]. The phenylalanine fibril pattern was observed to undergo a modification upon co-assembly with ACAAs (Ala, Leu, Ile and Val). Furthermore, this morphological modulation of phenylalanine fibrils was confirmed by electron microscopy [Fig. 8(I)]. Remarkably, the FE-SEM images displayed a transverse growth of microstructures on the longitudinal fibril axis. This peculiar modification of phenylalanine fibrils by the co-assembly of aliphatic amino acid may be due to intermolecular heterogeneous interactions between phenylalanine and ACAAs through the zwitterionic ends.Furthermore, the modification of phenylalanine fibrils by ACAAs led us to investigate their anti-amyloid action as a modal treatment for PKU. For this purpose, the ThT assay was performed for phenylalanine co-assembled structures with ACAAs at an 80 mM concentration using only phenylalanine as a control [Fig. 8(II)]. The results showed that ThT had a suppressed intensity in the case of the co-assembled states compared to the controlled phenylalanine solution, demonstrating the ability of ACAAs to inhibit the phenylalanine β-sheet-type fibril formation in aqueous media. Furthermore, the deposited phase ThT assay of phenylalanine and valine demonstrated a complete morphological shift from fibril to disc-like aggregates [Fig. 8(III)]. The deposited phase ThT binding assay for the leucine and phenylalanine co-assembled state also demonstrated a decrease in fluorescence intensity, suggesting lower β-sheet characteristics in the co-assembled state [ESI,† Fig. S37]. The modulatory effect the ACAAs displayed on phenylalanine fibrils was correlated with the decrease in cytotoxicity in the co-assembled state when performing in vitro cytotoxicity analysis. The results of the MTT assay displayed a considerable decrease in cell toxicity for the phenylalanine:ACAAs co-assembled systems as compared to the phenylalanine-only self-assembled structures [Fig. 8(IV)]. Recently, single amino acid-based ordered assemblies have displayed unique photoluminescent characteristics, which have suggested to be due to a quantum level clustering of hydrogen bond interactions.34,50–52 In this context, phenylalanine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) leucine (1
leucine (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) co-assembly revealed a complete morphology shift for phenylalanine fibrils to form spherical structures with unique intrinsic fluorescence emission as visualized by CLSM [Fig. 8(V)]. Thus, morphological analysis, ThT assay and cytotoxicity analysis confirmed the ability of the ACAAs to modify the phenylalanine fibril assembly in such a way that may be utilized for the treatment of PKU. Furthermore, the peculiar modification of the amyloidic characteristics of co-assembled architectures comprising unique intrinsic emission may have potential biotechnological applications. These intriguing results may have wide implications in the design of a preventive strategy against phenylalanine fibril toxicity assembly as well as for the development of multi-component supramolecular structures with improved physical properties.
1) co-assembly revealed a complete morphology shift for phenylalanine fibrils to form spherical structures with unique intrinsic fluorescence emission as visualized by CLSM [Fig. 8(V)]. Thus, morphological analysis, ThT assay and cytotoxicity analysis confirmed the ability of the ACAAs to modify the phenylalanine fibril assembly in such a way that may be utilized for the treatment of PKU. Furthermore, the peculiar modification of the amyloidic characteristics of co-assembled architectures comprising unique intrinsic emission may have potential biotechnological applications. These intriguing results may have wide implications in the design of a preventive strategy against phenylalanine fibril toxicity assembly as well as for the development of multi-component supramolecular structures with improved physical properties.
Conclusions
The self-assembling analysis for all naturally occurring amino acids in comparable physical conditions have provided much needed understanding about the mode of amino acid molecular recognition that may lead to tailor made design for the nano/microscale self-assembled architectures using synthetic molecules. In particular, Category I amino acids including Ala, Leu, Ileu, Val, and Met demonstrated fern-like crystallite assembly structure which may be owing to the dominating zwitterionic bridging interactions. The self-assembly of serine and threonine (with hydroxyl group in side chain) in Category II displayed the formation of aligned capsule and spear-like structures. Further, the amino acids with amide (Category III; Gln, Asn) and acidic side chain (Category IV; Glu, Asp) demonstrated the formation of well-ordered floral dendritic and membrane-like structures, respectively. Interestingly, pH modification of amino acid with cationic side chain (Category V; Histidine, Lysine and Arginine) demonstrated the formation of well-defined fibrils for histidine and high aspect ratio structures for lysine and arginine. The investigation of solution-phase β-sheet-like structures using optimized ThT assay demonstrated the amyloid-like etiology for phenylalanine, tyrosine, tryptophan, histidine and cysteine which correlates to the earlier reports. Further, asparagine displayed the formation of β-sheet-like structure for the first time. The asparagine amyloid characteristics was validated using histological, morphological and cytotoxicity assay which further extended the generic amyloid paradigm to the amino acid metabolites. Finally, the understanding of the role of the zwitterionic motif to form bridging interactions and their direct effect on the supramolecular morphology stimulated the heterogeneous co-assembly approach that led to the discovery of ACAAs (Ala, Leu, Ileu, and Val) as anti-amyloid agents for phenylalanine toxic fibrils. The formation of well-defined co-assembled structures also delineated the role of bridging zwitterionic interactions in stabilizing the amino acid supramolecular architectures.Amino acids as natural building blocks have emerged as a unique molecular library with inherited functional diversity that may be used for the generation of self-assembled functional materials capable of transforming healthcare and energy systems. The ability of heterogeneous molecular recognition displayed by amino acids may be utilized to tackle the metabolic amyloid aggregation using controlled co-assembly techniques. Furthermore, the in-depth understanding of well-ordered fabrication displayed by amino acids may be vital for the generation of non-amyloidic nanomaterials for biomedical applications. The amino acid self-assembly data may also serve as a guiding tool for the design and synthesis of organic molecules with inherited self-assembling properties. Thus, the present study may pave the way for the controlled fabrication of a new class of nano/microscale biomaterials for diverse biomedical applications.
CRediT authorship contribution statement
Prabhjot Singh: Writing – original draft, Investigation, Methodology, Conceptualization and Experimentation. Satish K. Pandey: Biological experimentation, Investigation. Aarzoo Grover: Experimentation. Rohit K. Sharma: Manuscript editing and Methodology. Nishima Wangoo: Supervision, Conceptualization, Manuscript writing and review, Overall supervision, Research support.Data availability
The data required to reproduce these findings can be made available on request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Science Engineering and Research Board (SERB) of India grant no. ECR/2017/000874 and DST-PURSE grant awarded to NW. PS thanks University Grant Commission (UGC), India, for his senior research fellowship.References
- J. W. Szostak, D. P. Bartel and P. L. Luisi, Synthesizing life, Nature, 2001, 409, 387–390 CrossRef CAS.
- G. M. Whitesides and M. Boncheva, Beyond molecules: Self-assembly of mesoscopic and macroscopic components, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 4769–4774 CrossRef CAS.
- P.-A. Monnard and D. W. Deamer, Membrane self-assembly processes: Steps toward the first cellular life, Anat. Rec., 2002, 268, 196–207 CrossRef CAS.
- B. A. Grzybowski and W. T. S. Huck, The nanotechnology of life-inspired systems, Nat. Nanotechnol., 2016, 11, 585–592 CrossRef CAS.
- F. Chiti and C. M. Dobson, Amyloid formation by globular proteins under native conditions, Nat. Chem. Biol., 2009, 5, 15–22 CrossRef CAS.
- C. Soto and S. Pritzkow, Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases, Nat. Neurosci., 2018, 21, 1332–1340 CrossRef CAS.
- E. Gazit, Self Assembly of Short Aromatic Peptides into Amyloid Fibrils and Related Nanostructures, Prion, 2007, 1, 32–35 CrossRef.
- S. Bera, S. Mondal, Y. Tang, G. Jacoby, E. Arad, T. Guterman, R. Jelinek, R. Beck, G. Wei and E. Gazit, Deciphering the Rules for Amino Acid Co-Assembly Based on Interlayer Distances, ACS Nano, 2019, 13, 1703–1712 CrossRef CAS.
- N. Amdursky and M. M. Stevens, Circular Dichroism of Amino Acids: Following the Structural Formation of Phenylalanine, ChemPhysChem, 2015, 16, 2768–2774 CrossRef CAS.
- S. Shaham-Niv, L. Adler-Abramovich, L. Schnaider and E. Gazit, Extension of the generic amyloid hypothesis to nonproteinaceous metabolite assemblies, Sci. Adv., 2015, 1, e1500137 CrossRef.
- S. Shaham-Niv, P. Rehak, L. Vuković, L. Adler-Abramovich, P. Král and E. Gazit, Formation of Apoptosis-Inducing Amyloid Fibrils by Tryptophan, Isr. J. Chem., 2017, 57, 729–737 CrossRef CAS.
- L. Adler-Abramovich, L. Vaks, O. Carny, D. Trudler, A. Magno, A. Caflisch, D. Frenkel and E. Gazit, Phenylalanine assembly into toxic fibrils suggests amyloid etiology in phenylketonuria, Nat. Chem. Biol., 2012, 8, 701–706 CrossRef CAS.
- D. Zaguri, T. Kreiser, S. Shaham-Niv and E. Gazit, Antibodies towards Tyrosine Amyloid-Like Fibrils Allow Toxicity Modulation and Cellular Imaging of the Assemblies, Molecules, 2018, 23(6), 1273, DOI:10.3390/molecules23061273.
- E. Gazit, Metabolite amyloids: a new paradigm for inborn error of metabolism disorders, J. Inherited Metab. Dis., 2016, 39, 483–488 CrossRef CAS.
- D. Laor, D. Sade, S. Shaham-Niv, D. Zaguri, M. Gartner, V. Basavalingappa, A. Raveh, E. Pichinuk, H. Engel, K. Iwasaki, T. Yamamoto, H. Noothalapati and E. Gazit, Fibril formation and therapeutic targeting of amyloid-like structures in a yeast model of adenine accumulation, Nat. Commun., 2019, 10, 62 CrossRef CAS.
- S. Auer, F. Meersman, C. M. Dobson and M. Vendruscolo, A generic mechanism of emergence of amyloid protofilaments from disordered oligomeric aggregates, PLoS Comput. Biol., 2008, 4, e1000222 CrossRef.
- K. Stroobants, J. R. Kumita, N. J. Harris, D. Y. Chirgadze, C. M. Dobson, P. J. Booth and M. Vendruscolo, Amyloid-like Fibrils from an α-Helical Transmembrane Protein, Biochemistry, 2017, 56, 3225–3233 CrossRef CAS.
- E. Tayeb-Fligelman, O. Tabachnikov, A. Moshe, O. Goldshmidt-Tran, M. R. Sawaya, N. Coquelle, J.-P. Colletier and M. Landau, The cytotoxic Staphylococcus aureus PSMα3 reveals a cross-α amyloid-like fibril, Science, 2017, 355, 831–833 CrossRef.
- G. Scott, A. F. Ramlackhansingh, P. Edison, P. Hellyer, J. Cole, M. Veronese, R. Leech, R. J. Greenwood, F. E. Turkheimer, S. M. Gentleman, R. A. Heckemann, P. M. Matthews, D. J. Brooks and D. J. Sharp, Amyloid pathology and axonal injury after brain trauma, Neurology, 2016, 86, 821–828 CrossRef CAS.
- V. E. Johnson, W. Stewart and D. H. Smith, Traumatic brain injury and amyloid-β pathology: a link to Alzheimer's disease?, Nat. Rev. Neurosci., 2010, 11, 361–370 CrossRef CAS.
- N. Blau, F. J. van Spronsen and H. L. Levy, Phenylketonuria, Lancet, 2010, 376, 1417–1427 CrossRef CAS.
- K. Michals-Matalon, G. Bhatia, F. Guttler, S. K. Tyring and R. Matalon, Response of Phenylketonuria to Tetrahydrobiopterin, J. Nutr., 2007, 137, 1564S–1567S CrossRef CAS.
- T. D. Douglas, A. M. Nucci, A. M. Berry, S. T. Henes and R. H. Singh, Large neutral amino acid status in association with P:T ratio and diet in adult and pediatric patients with phenylketonuria, JIMD Rep., 2019, 50, 50–59 CrossRef.
- K. Chu, Phenylketonuria: Research and development of nutrition, AIP Conf. Proc., 2020, 2208, 20042 CrossRef CAS.
- V. Singh, R. K. Rai, A. Arora, N. Sinha and A. K. Thakur, Therapeutic implication of L-phenylalanine aggregation mechanism and its modulation by D-phenylalanine in phenylketonuria, Sci. Rep., 2014, 4, 3875 CrossRef.
- A. De Luigi, A. Mariani, M. De Paola, A. Re Depaolini, L. Colombo, L. Russo, V. Rondelli, P. Brocca, L. Adler-Abramovich, E. Gazit, E. Del Favero, L. Cantù and M. Salmona, Doxycycline hinders phenylalanine fibril assemblies revealing a potential novel therapeutic approach in phenylketonuria, Sci. Rep., 2015, 5, 15902 CrossRef CAS.
- T. D. Do, W. M. Kincannon and M. T. Bowers, Phenylalanine Oligomers and Fibrils: The Mechanism of Assembly and the Importance of Tetramers and Counterions, J. Am. Chem. Soc., 2015, 137, 10080–10083 CrossRef CAS.
- H. W. German, S. Uyaver and U. H. E. Hansmann, Self-Assembly of Phenylalanine-Based Molecules, J. Phys. Chem. A, 2015, 119, 1609–1615 CrossRef CAS.
- E. Gazit, A possible role for π-stacking in the self-assembly of amyloid fibrils, FASEB J., 2002, 16, 77–83 CrossRef CAS.
- S. Shaham-Niv, P. Rehak, D. Zaguri, A. Levin, L. Adler-Abramovich, L. Vuković, P. Král and E. Gazit, Differential inhibition of metabolite amyloid formation by generic fibrillation-modifying polyphenols, Commun. Chem., 2018, 1, 25 CrossRef.
- D. Banik, R. Dutta, P. Banerjee, S. Kundu and N. Sarkar, Inhibition of Fibrillar Assemblies of L-Phenylalanine by Crown Ethers: A Potential Approach toward Phenylketonuria, J. Phys. Chem. B, 2016, 120, 7662–7670 CrossRef CAS.
- P. Singh, S. K. Brar, M. Bajaj, N. Narang, V. S. Mithu, O. P. Katare, N. Wangoo and R. K. Sharma, Self-assembly of aromatic α-amino acids into amyloid inspired nano/micro scaled architects, Mater. Sci. Eng., C, 2017, 72, 590–600 CrossRef CAS.
- P. Singh, N. Narang, R. K. Sharma and N. Wangoo, Interplay of Self-Assembling Aromatic Amino Acids and Functionalized Gold Nanoparticles Generating Supramolecular Structures, ACS Appl. Bio Mater., 2020, 3, 6196–6203 CrossRef CAS.
- P. Singh, N. Wangoo and R. K. Sharma, Phenylalanine dimer assembly structure as the basic building block of an amyloid like photoluminescent nanofibril network, Soft Matter, 2020, 16, 4105–4109 RSC.
- H. K. Berry, M. K. Bofinger, M. M. Hunt, P. J. Phillips and M. B. Guilfoile, Reduction of cerebrospinal fluid phenylalanine after oral administration of valine, isoleucine, and leucine, Pediatr. Res., 1982, 16, 751–755 CrossRef CAS.
- C. H. Görbitz, Crystal structures of amino acids: from bond lengths in glycine to metal complexes and high-pressure polymorphs, Crystallogr. Rev., 2015, 21, 160–212 CrossRef.
- S. Bera, S. Mondal, S. Rencus-Lazar and E. Gazit, Organization of Amino Acids into Layered Supramolecular Secondary Structures, Acc. Chem. Res., 2018, 51, 2187–2197 CrossRef CAS.
- J. Kyte and R. F. Doolittle, A simple method for displaying the hydropathic character of a protein, J. Mol. Biol., 1982, 157, 105–132 CrossRef CAS.
- J. Popot and C. De, Vitry, Integral Membrane Protein, Encycl, Cancer, 2011, 1884 Search PubMed.
- R. Wolfenden, L. Andersson, P. M. Cullis and C. C. B. Southgate, Affinities of Amino Acid Side Chains for Solvent Water, Biochemistry, 1981, 20, 849–855 CrossRef CAS.
- S. Guerin, A. Stapleton, D. Chovan, R. Mouras, M. Gleeson, C. McKeown, M. R. Noor, C. Silien, F. M. F. Rhen, A. L. Kholkin, N. Liu, T. Soulimane, S. A. M. Tofail and D. Thompson, Control of piezoelectricity in amino acids by supramolecular packing, Nat. Mater., 2018, 17, 180–186 CrossRef CAS.
- N. Gour, C. Kanth, P. B. Koshti, V. Kshtriya, D. Shah, S. Patel, R. Agrawal-Rajput and M. K. Pandey, Amyloid-like Structures Formed by Single Amino Acid Self-Assemblies of Cysteine and Methionine, ACS Chem. Neurosci., 2019, 10, 1230–1239 CrossRef CAS.
- C. Bleiholder, D. B. Werz, H. Köppel and R. Gleiter, Theoretical Investigations on Chalcogen−Chalcogen Interactions:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) What Makes These Nonbonded Interactions Bonding?, J. Am. Chem. Soc., 2006, 128, 2666–2674 CrossRef CAS.
What Makes These Nonbonded Interactions Bonding?, J. Am. Chem. Soc., 2006, 128, 2666–2674 CrossRef CAS. - M. R. H. Krebs, E. H. C. Bromley and A. M. Donald, The binding of thioflavin-T to amyloid fibrils: localisation and implications, J. Struct. Biol., 2005, 149, 30–37 CrossRef CAS.
- M. Biancalana and S. Koide, Molecular mechanism of Thioflavin-T binding to amyloid fibrils, Biochim. Biophys. Acta, Proteins Proteomics, 2010, 1804, 1405–1412 CrossRef CAS.
- A. I. Sulatskaya, A. V. Lavysh, A. A. Maskevich, I. M. Kuznetsova and K. K. Turoverov, Thioflavin T fluoresces as excimer in highly concentrated aqueous solutions and as monomer being incorporated in amyloid fibrils, Sci. Rep., 2017, 7, 2146 CrossRef.
- B. H. Toyama and J. S. Weissman, Amyloid structure: conformational diversity and consequences, Annu. Rev. Biochem., 2011, 80, 557–585 CrossRef CAS.
- P. Banerjee, A. Pyne and N. Sarkar, Understanding the Self-Assembling Behavior of Biological Building Block Molecules: A Spectroscopic and Microscopic Approach, J. Phys. Chem. B, 2020, 124, 2065–2080 CrossRef CAS.
- Y. Zhang, V. H. Man, C. Roland and C. Sagui, Amyloid Properties of Asparagine and Glutamine in Prion-like Proteins, ACS Chem. Neurosci., 2016, 7, 576–587 CrossRef CAS.
- D. Pinotsi, A. K. Buell, C. M. Dobson, G. S. Kaminski Schierle and C. F. Kaminski, A Label-Free, Quantitative Assay of Amyloid Fibril Growth Based on Intrinsic Fluorescence, ChemBioChem, 2013, 14, 846–850 CrossRef CAS.
- S. Shaham-Niv, Z. A. Arnon, D. Sade, A. Lichtenstein, E. A. Shirshin, S. Kolusheva and E. Gazit, Intrinsic Fluorescence of Metabolite Amyloids Allows Label-Free Monitoring of Their Formation and Dynamics in Live Cells, Angew. Chem., Int. Ed., 2018, 57, 12444–12447 CrossRef CAS.
- X. Chen, W. Luo, H. Ma, Q. Peng, W. Z. Yuan and Y. Zhang, Prevalent intrinsic emission from nonaromatic amino acids and poly(amino acids), Sci. China: Chem., 2018, 61, 351–359 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0qm00784f |
| ‡ Department of Chemistry, University of Vermont, Burlington, Vermont-05405, USA. |
| This journal is © the Partner Organisations 2021 |