Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A versatile and straightforward process to turn plastics into antibacterial materials†
Slim
Hadiouch
a,
Marc
Maresca
b,
Didier
Gigmes
a,
Guilherme
Machado
 c,
Aurelien
Maurel-Pantel
c,
Aurelien
Maurel-Pantel
 c,
Sabrina
Frik
c,
Sabrina
Frik
 d,
Johanna
Saunier
d,
Ariane
Deniset-Besseau
e,
Najet
Yagoubi
d,
Lukas
Michalek
f,
Christopher
Barner-Kowollik
d,
Johanna
Saunier
d,
Ariane
Deniset-Besseau
e,
Najet
Yagoubi
d,
Lukas
Michalek
f,
Christopher
Barner-Kowollik
 f,
Yohann
Guillaneuf
f,
Yohann
Guillaneuf
 a and
Catherine
Lefay
a and
Catherine
Lefay
 *a
*a
aAix-Marseille Univ., CNRS, Institut de Chimie Radicalaire, UMR 7273, F-13397 Marseille, France. E-mail: catherine.lefay@univ-amu.fr
bAix Marseille Univ., CNRS, Centrale Marseille, iSm2, 13397 Marseille, France
cAix Marseille Univ, CNRS, Centrale Marseille, LMA, Marseille, France
dUniversité Paris Saclay, UFR de pharmacie, Matériaux et Santé, 92290 Chatenay Malabry, France
eInstitut de Chimie Physique (ICP), CNRS UMR 8000, Univ. of Paris-Sud, Université Paris-Saclay, Orsay, France
fCentre of Materials Science, School of Chemistry and Physics, Queensland University of Technology (QUT), 2 George Street, Brisbane, QLD 4000, Australia
First published on 23rd November 2021
Abstract
We demonstrate that antibacterial activity can be conferred to common plastic materials using amphiphilic cationic methacrylate-based block copolymers, specifically quaternized poly(butyl methacrylate)-b-poly(N,N-dimethyl-aminoethyl methacrylate) (PBMA-b-PDMAEMA) with 64 mol% of DMAEMA and Mn close to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1. With 0.5–2 wt% of these copolymers simply dispersed in the given matrix by extrusion, the modified materials prove to be active against E. coli, S. aureus, Listeria monocytogenes and enterohemorrhagic E. coli without toxicity against two cell lines, HaCaT and L929 fibroblasts, while keeping the mechanical properties of the materials intact. In addition, the study of the mechanism of action shows that the antibacterial materials target the bacterial membrane, which is expected to avoid antibacterial resistance. Our protocol is a cost-effective solution to generate antibacterial materials for several applications, including packaging or medical devices, without modification of the production process.
000 g mol−1. With 0.5–2 wt% of these copolymers simply dispersed in the given matrix by extrusion, the modified materials prove to be active against E. coli, S. aureus, Listeria monocytogenes and enterohemorrhagic E. coli without toxicity against two cell lines, HaCaT and L929 fibroblasts, while keeping the mechanical properties of the materials intact. In addition, the study of the mechanism of action shows that the antibacterial materials target the bacterial membrane, which is expected to avoid antibacterial resistance. Our protocol is a cost-effective solution to generate antibacterial materials for several applications, including packaging or medical devices, without modification of the production process.
Introduction
Infection remains the third leading cause of death in developed countries and the second worldwide. Indeed, the continuing decline of effectiveness of existing antibiotic options due to the ability of microbes to develop and disseminate mechanisms of resistance against multiple classes of traditional antimicrobials raise international healthcare concerns as they are associated with increased morbidity and mortality and also higher hospital costs.1–3 Antimicrobial surfaces and antimicrobial materials are thus becoming a very important class of products that could contribute to mitigate the problem. Among them, antimicrobial plastics have received considerable attention for numerous applications, including appliances, filters, packaging, nonwoven fabrics and textiles, and sanitary materials.4 To confer antibacterial functionality to plastics, surface modification based on an array of methodologies has been exploited, such as surface casting, immersion, surface finishing, plasma treatment and deposition.5,6 Although these methods afford plastic products with effective antibacterial properties, the processing methods are complex and are associated with high costs. Moreover, the antibacterial properties will be weakened if the antibacterial coating is damaged.To obtain antibacterial functions, blending is a simpler method. Many antibacterial compounds can be employed and include either inorganic species (such as copper nanoparticles, silver nanoparticles and silver-doped glass) or organic entities (for example quaternary ammonium salts, N-haloamines, natural biocides such as thymol or carvacrol and classic antibiotics).4,7,8 However, the sometimes low antimicrobial efficiency and leaching of antimicrobial components along with the poor durability of their antimicrobial properties limit their application.9 To prepare long-lasting or non-leaching antibacterial composites, polymer-based compounds were used as additives to impart antibacterial properties to pristine materials.10–13 Such an approach is usually carried out either with a small library of known antibacterial polymers such as poly(hexamethylene guanidine hydrochloride) (PHMG), chitosan or poly(tert-butylamino methacrylate).14,15 In these cases, because of the incompatibility of mixing the polymers with each other, a significant amount of the additive has to be inserted into the matrix, potentially altering the material properties (mechanical properties, chemical and thermal resistance, etc.) of the plastics.
More complex polymers that feature a backbone similar to the matrix and are decorated with antibacterial moieties or antibacterial polymers18–24 have been developed to enhance the compatibilization of the additive to the plastics. Such approaches are efficient, yet specific macromolecular antibacterial architectures have to be designed for each matrix. Recently, we developed cationic amphiphilic poly(butyl methacrylate)-b-poly(N,N-dimethyl-aminoethyl methacrylate) (PBMA-b-PDMAEMA) diblock copolymers based on a methacrylate backbone that are potent antibacterial copolymers in solution as well as promising additives to specific polymer matrices (polystyrene (PS) and poly(methyl methacrylate) (PMMA)).16,17N,N-Dimethyl-aminoethyl methacrylate (DMAEMA) was chosen as the hydrophilic monomer and butyl methacrylate (BMA) as the hydrophobic monomer. A structure–activity analysis was subsequently performed, and we selected diblock copolymers with a molar composition of 64 mol% DMAEMA and obtained permanent cationic charges after quaternization of the amine groups by methyl iodide (MeI) to obtain the highest antibacterial activity (i.e., low MIC values) along with the lowest toxicity. In addition, a number-average molar mass (Mn as measured by SEC/DMF) of close to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1 was targeted. Such large number average molecular weights, Mn, compared to those typically employed in antibacterial copolymers, were expected to ensure a good dispersion of the copolymer in common industrial organic polymer matrices, while avoiding any leaching of the active copolymer and thus a loss of activity over time.
000 g mol−1 was targeted. Such large number average molecular weights, Mn, compared to those typically employed in antibacterial copolymers, were expected to ensure a good dispersion of the copolymer in common industrial organic polymer matrices, while avoiding any leaching of the active copolymer and thus a loss of activity over time.
Herein, we demonstrate that quaternized PBMA-b-PDMAEMA copolymers can be efficiently used as additives to confer antibacterial properties to four model industrial organic polymer matrices: poly(ethylene terephthalate glycol) (PETG), polylactide (PLA) and both high density and low density polyethylenes (HDPE and LDPE respectively). Such matrices were chosen since they have a large range of applications, especially for packaging, and are produced on a very large scale. The dispersion of the copolymer is performed by simply co-extruding a low amount of the antibacterial copolymer (≤2 wt%) within the given matrix without impacting the innocuity of the material and its inherent mechanical properties (Fig. 1). Such an approach constitutes a simple, efficient and versatile solution to limit bacterial infections.
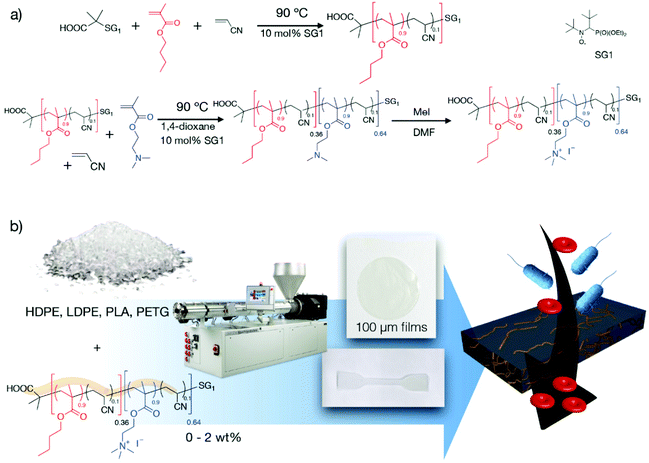 | ||
Fig. 1 (a) Synthesis of the antibacterial diblock copolymer B(A10%)Me as previously described16,17 (PBMA-b-PDMAEMA, FDMAEMA = 0.64, Mn = 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 g mol−1, quaternization with MeI). (b) Preparation of non-leaching antibacterial materials via the blending of maximum 2 wt% of the B(A10%)Me antibacterial diblock copolymer with various organic matrices by extrusion. 400 g mol−1, quaternization with MeI). (b) Preparation of non-leaching antibacterial materials via the blending of maximum 2 wt% of the B(A10%)Me antibacterial diblock copolymer with various organic matrices by extrusion. | ||
Experimental section
Materials
Commercial organic matrices, i.e. PLA (Natureworks), PETG (Skygreen S2008), HDPE (Dow Chemical KT 10000) and LDPE (Total LA 0710), were used. All the reagents and solvents were used without further purification. The synthesis of the B(A10%)Me copolymer is described in the ESI† following our previous study. Human normal skin cells HaCaT were obtained from Creative Bioarray (Shirley, NY 11967, USA) and mouse fibroblastic lung cells—L929 (C3H/An)—were purchased from Sigma.Elaboration of antibacterial organic materials
The extrusion step was performed with a Thermo Scientific HAAKE MiniLab II mini-extruder. The pellets were dried at 90 °C under vacuum for 4 h before extrusion. Once dry, 5 g of pellets were manually loaded into the extruder. For mechanical testing, tensile specimens (type 5A) were prepared by injection moulding using a Thermo Scientific HAAKE Minijet II injector. The polymer melt at the exit of the extruder was collected in the nozzle heated to 200 °C of the injector and then injected with a pressure of 500 bar in a test specimen mold (tensile bar ISO527-2-5A) heated at 60 °C. Bioassays were performed on circular shaped films of 3 cm diameter and 100 μm thickness. Upon exiting the extruder, the recovered rushes were cooled under air and then cut into small pellets, and finally shaped using a Specac hydraulic press equipped with heating platens and a thin film kit. The pellets were pressed between two aluminium sheets heated at 170 °C and under 2 tons of pressure for 10 s.Antibacterial tests
The bactericidal activity of matrices was evaluated as previously described.16 The bacteria used in the study were either Gram positive (Staphylococcus aureus, ATCC CRM-6538P) or Gram negative (Escherichia coli, ATCC 8739) and were routinely grown on lysogeny broth (LB) agar plates. The bactericidal activity of the different matrices was evaluated according to an adapted ISO 22196 procedure. Briefly, pieces of matrices (squares of 4 × 4 mm) were sterilely prepared from extruded films and were placed at the bottom of the wells of sterile polypropylene 96-well microplates (Greiner BioOne). Overnight bacterial suspensions (3 ml of LB broth) of E. coli or S. aureus were prepared from single colonies isolated on LB agar plates. Tubes were incubated overnight at 37 °C under agitation (200 rpm). The next day, bacterial suspensions (optical density OD > 1.0) were diluted 1/100 in 3 ml of fresh culture media and incubated at 37 °C and 200 rpm until the bacteria reached log phase growth (OD around 0.6). The bacteria were then diluted in sterile phosphate buffered saline (PBS) to reach a cell density of around 105 bacteria per ml. 10 μl of these suspensions (corresponding to ∼1000 bacteria) were added onto the surface of matrices. After 60 min of incubation at 37 °C, the bacteria were collected by adding 90 μl of sterile PBS into the wells containing the matrices and repeatedly pipetted up and down. The bacteria were then serially diluted (1 in 10 dilution) in sterile PBS before plating 10 μl of each bacterial dilution onto LB agar plates. After overnight incubation at 37 °C, the plates were observed, and colonies were counted allowing the determination of the bacterial killing. Experiments were conducted in independent triplicate (n = 3). A similar protocol was used to evaluate the bactericidal activity of the matrices against food-borne bacterial pathogens, using Listeria monocytogenes (ATCC 15313) and enterohemorrhagic E. coli EHEC (ATCC K88). Finally, the bactericidal activity of the matrices was evaluated using the brine of commercial mozzarella naturally containing mixed bacterial community. Commercial mozzarella was bought from a food supermarket (Mozzarella Galbani, from Carrefour supermarket). The brine was collected sterilely and 10 μl of brine was used to evaluate the bactericidal effect of the matrices as explained above.Extractables and leachables
To identify and quantify the extractables, we used a dissolution/precipitation process. 6 g of the material were dissolved by refluxing in 50 mL of hot toluene under stirring. After complete dissolution, the polymer was precipitated by slowly pouring 100 mL of methanol under stirring. The precipitated polymers were removed from the additive solution by filtration on filter paper. The solution that contained the extractables was evaporated using a rotavapor at 60 °C under vacuum. To identify and quantify the leachables, 6 g of the material were immersed in 15 mL of ethanol for 72 h at 37 °C in glass vials. The material was then removed and ethanol that contained the leachables for these conditions of storage was evaporated using a rotavapor at 60 °C under vacuum.Before injection in the HPLC system, the dry residue containing the extractables or leachables was dissolved in a mixture of 0.5 mL of tetrahydrofuran (HPLC grade Prolabo) and 0.5 mL of acetonitrile (HPLC grade Prolabo) and the solution was filtered on a 0.2 μm filter. 20 μL of the solution were injected on a Lichrospher 100 RP 18 (5 microns) using a 100% acetonitrile phase; the apparatus was a HPLC Ultimate 3000 series (Dionex) equipped with an automatic injector, a gradient pump and a UV detector with a diode array (190–400 nm). Characterization and possible identification of the compounds were performed by using their retention time and their UV spectrum and by comparing them to those of references.
Results and discussion
Antibacterial activity
Synthetic antimicrobial copolymers are inspired by antimicrobial peptides, i.e. synthetic amphiphilic cationic copolymers.25 Since the pioneering work of DeGrado and colleagues,11 studies covering synthetic antimicrobial copolymers experienced an exponential growth with more than 1500 publications per year. These constructs have many advantages such as simple manufacturing on a large scale. Another advantage of these synthetic copolymers compared to peptides is that a large library of monomers can be used, as well as diverse architectures (generally random or block copolymers), lengths (Mn between 1000 to 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) and compositions (various hydrophilic/hydrophobic balances).26–29 This chemical variety allows identifying optimum antimicrobial systems that feature the best compromise between high antimicrobial activity and low toxicity.25 Several families of polymers have been reported in the literature, including polyacrylate and polymethacrylate derivatives, poly(oxa)norbornene, polymaleimides, polyionenes and polycarbonates.25,30 Herein we prepare an antibacterial copolymer constituted of a methacrylate backbone16 due to the exothermic mixing enthalpy of PMMA derivatives with a wide range of commercially relevant plastics (e.g., poly(vinyl chloride) PVC, poly(styrene-co-acrylonitrile) PSAN, poly(vinylidene fluoride) PVDF, epoxy matrices, etc.), allowing for their facile incorporation into different organic matrices.31 The diblock copolymer was synthesized via the NMP technique32 using acrylonitrile ACN as the comonomer33 to ensure good control of the polymerization reaction (PBMA-b-PDMAEMA, FDMAEMA = 0.64, Mn = 18
000 g mol−1) and compositions (various hydrophilic/hydrophobic balances).26–29 This chemical variety allows identifying optimum antimicrobial systems that feature the best compromise between high antimicrobial activity and low toxicity.25 Several families of polymers have been reported in the literature, including polyacrylate and polymethacrylate derivatives, poly(oxa)norbornene, polymaleimides, polyionenes and polycarbonates.25,30 Herein we prepare an antibacterial copolymer constituted of a methacrylate backbone16 due to the exothermic mixing enthalpy of PMMA derivatives with a wide range of commercially relevant plastics (e.g., poly(vinyl chloride) PVC, poly(styrene-co-acrylonitrile) PSAN, poly(vinylidene fluoride) PVDF, epoxy matrices, etc.), allowing for their facile incorporation into different organic matrices.31 The diblock copolymer was synthesized via the NMP technique32 using acrylonitrile ACN as the comonomer33 to ensure good control of the polymerization reaction (PBMA-b-PDMAEMA, FDMAEMA = 0.64, Mn = 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 g mol−1). ACN was used to lower the average main equilibrium constant, K, between the macroradical and macroalkoxyamine. This equilibrium is shifted towards active species in the case of methacrylate derivatives, impeding the obtainment of good control/livingness of the polymerization. The addition of only 10% of some comonomers restored the correct equilibrium, enabling living and controlled polymerization.33
400 g mol−1). ACN was used to lower the average main equilibrium constant, K, between the macroradical and macroalkoxyamine. This equilibrium is shifted towards active species in the case of methacrylate derivatives, impeding the obtainment of good control/livingness of the polymerization. The addition of only 10% of some comonomers restored the correct equilibrium, enabling living and controlled polymerization.33
DMAEMA was selected as the hydrophilic monomer and BMA as the hydrophobic monomer. The copolymer was quaternized using methyl iodide prior to use (Fig. 1a).16 To be consistent with our previous work,16,17 this copolymer will be denoted B(A10%)Me herein. Specifically, “B”, “A” and “Me” refer respectively to the block copolymer, acrylonitrile and methyl iodide. To prepare the antibacterial materials, pellets of commercial grades of various polymer matrices (LDPE, HDPE, PETG, PLA) and the antibacterial diblock copolymer B(A10%)Me are blended during the co-extrusion using a micro-extruder that allows working on a gram scale (Fig. 1b). The experimental conditions of the extrusions are given in the ESI (ESI – Table S1†) and are very close to the recommendations of the matrix suppliers. The strips that were obtained directly from the micro-extruders were hot pressed to obtain films of 3 cm diameter and 100 micron thickness (refer to section I.4 of the ESI†). Also, it was possible to directly employ – after micro-extrusion – a mini-injection moulding machine to prepare samples for mechanical testing (tensile bar ISO527-2-5A). We used a ratio of 0, 0.5 and 2 wt% B(A10%)Me copolymer in the organic matrices to ensure that the antibacterial agent is only an additive, expecting that the mechanical properties of the matrices will not be altered.
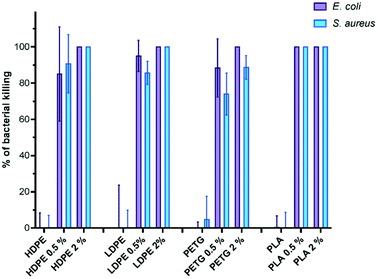
The antibacterial activity of the prepared solid materials was subsequently investigated using S. aureus and E. coli as model Gram-positive and Gram-negative bacteria respectively, following an ISO-22196 adapted procedure.16 Indeed, the size of the sample is supposed to be 5 × 5 cm in this procedure and thus requiring a high amount of the additive and film to be prepared. To screen the various matrices and concentration of the additives more efficiently, the surface that was used in our study was decreased to 4 × 4 mm, enabling the use of 96-well plates. The tests were performed in triplicate with an exposure time of 1 h (Fig. 2). It has been reported that synthetic antibacterial copolymers are very efficient, displaying biocide action after a few minutes.34 The results shown in Fig. 2 correspond to the percentage of bacteria that was killed compared to the negative control.
In all cases, the pristine matrices did not show any antibacterial activity. The incorporation of 2 wt% of B(A10%)Me proved to be sufficient to confer high antibacterial activity to commercial PETG, LDPE, HDPE and PLA matrices. The bacterial killing ranged from 88% (for PETG) to 100% (for HDPE, LDPE and PLA) on S. aureus and was 100% with all matrices for E. coli.
At 0.5 wt% of incorporation of B(A10%)Me, bactericidal activities were lower for HDPE, LDPE and PETG matrices (bacterial killing ranging from 85 to 95% for E. coli and from 74 to 90% for S. aureus). Importantly, among the various matrices, only the additivated PLA matrix remained fully active with no viable bacteria (either S. aureus or E. coli) observed after one hour of contact with only 0.5 wt% of incorporation of the B(A10%)Me copolymer. These tests confirm the antibacterial activity of these films, but more studies are required to investigate their antibiofilm behaviour as well as their effectiveness against aging.
Mechanism of action
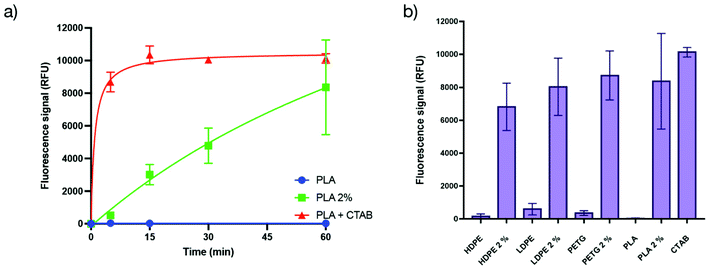
The mechanism of action of such antibacterial methacrylate-based copolymers has been studied in the literature and is hypothesized to be mainly a membrane destabilization mechanism leading to the lysis of the bacterial membrane.35 Thus we were interested in investigating the mechanism of action of the herein presented antibacterial copolymer embedded into a polymer matrix. A propidium iodide assay was used to establish if the bactericidal activity of the matrices is related to membranolytic effects.36 Indeed, such compounds are known to enter cells and form a fluorescent complex with DNA only after membrane damage and are therefore good probes to monitor membrane integrity.36–38 Incubation of E. coli as model bacteria and propidium iodide onto PLA – that displayed very good antibacterial properties – was monitored by fluorescence (the details of the test are described in the ESI†).Cetyltrimethylammonium bromide (CTAB) was used as a positive control (added in a solution on a non-additivated PLA surface) since this compound is known to lyse efficiently all the bacteria present in the medium and would represent the upper limit for the fluorescence value. The experiments performed using the additivated PLA present an increase in the fluorescence value with time, reaching the upper limit within the error margin of the positive control (Fig. 3a), confirming that the antibacterial activity is based on the permeabilization of the bacterial membrane. The comparison of the increase in the fluorescence intensity between the positive control and the additivated PLA shows that the process of permeabilization of the copolymer embedded into the matrix is slower than the liquid biocide reference, yet remains rapid (∼90% permeabilization in less than 1 h) compared to the activity of leachable biocides that generally requires hours to be efficient. The other matrices (HDPE, LDPE and PETG) were also tested and showed similar behaviours within the error margin (Fig. 3b).
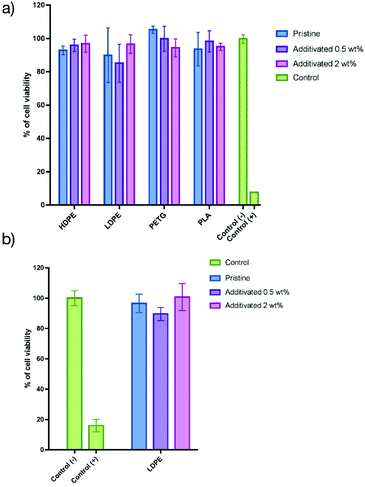
Cytotoxicity study
Once the antibacterial activity had been demonstrated, we proceeded to assess the in vitro innocuity of such materials using human normal keratinocytes as a model (HaCaT cells). Such kinds of organic matrices could be utilized for applications in which the main contact is with the skin. The cells were incubated for 48 h on the films and the cell viability was determined using the resazurin assay (the details of the tests are described in the ESI†). The results are presented in Fig. 4a. The cell viability on all pristine polymer organic matrices was close to 100% compared to the positive control.When 0.5 wt% of the additive was embedded into the matrices, the cell viability was identical to that of the pristine matrices, demonstrating that there is no impact on the additive at such low concentrations. When the amount of the additive was increased from 0.5 to 2 wt%, the HaCaT cell viability (Fig. 4a) was still close to 100%, showing that the additive does not impact the innocuity of all the materials. In order to assess if these results are dependent on the cell line, similar experiments were performed using L929 fibroblasts (Fig. 4b) using LDPE films. In that case, the cell viability is also not dependent on the concentration of the additive, showing that this approach confers antibacterial activity without altering the cell viability for all the matrices.
The main advantage of blending the antibacterial copolymer with the matrix is related to the non-leaching of the active antibacterial additive. The co-extrusion ensures a long-lasting efficiency compared to leaching approaches (either antibiotics/biocides or inorganic salts), avoiding the increase of antibiotic resistance due to the dispersion of antibiotics in the environment. Besides the action mechanism of the antibacterial copolymer involving alteration of the bacterial membrane, the eventual leaching of such materials in the environment will not induce bacterial resistance. The non-leaching feature was assessed by using LDPE as the model polymer and ethanol as the solvent.
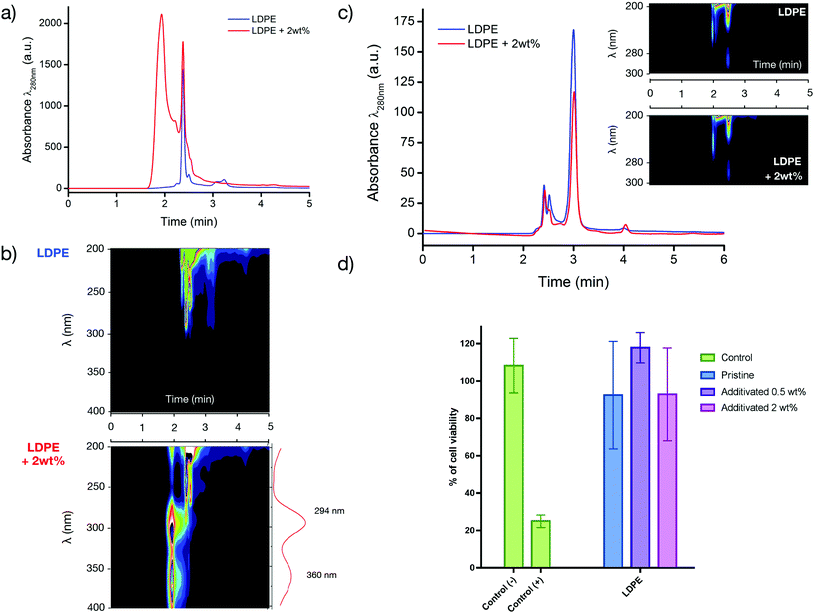
First, we determined the extractables, i.e., all the molecules that will be able to diffuse and migrate outside the polymer matrix and that will be extracted using aggressive solvent conditions. To identify and quantify the extractables, a dissolution/precipitation process was conducted using toluene under reflux. After complete dissolution, the matrix was precipitated by slowly pouring 100 mL of methanol under stirring. The filtrate was subsequently analysed by HPLC (Fig. 5a). A peak at 1.5–2 min was observed that was in good agreement with the B(A10%)Me copolymer (refer to Fig. S3 in the ESI†). Unlike extractables that require the dissolution of the matrix, leachables refer to the compounds that are effectively leached inside a media under conditions that are generally milder and closer to normal conditions of use. In our case, the films were immersed in 15 mL of ethanol for 72 h at 37 °C in glass vials.
The material was subsequently removed and the supernatant ethanol that contained the leachables (for these conditions of storage) was analysed (Fig. 5c).
Analysis of the leachables in ethanol by HPLC showed similar chromatograms for the LDPE with and without the antibacterial additive (Fig. 5c): the main leachable in this case was BHT, a phenolic antioxidant that was originally present in the pristine matrix. To confirm that the antibacterial copolymer and/or degradation side-products do not leach out and do not impact the biocompatibility of the matrix, the strips was also immersed in a culture medium for 48 h at 37 °C and this medium was then later used for incubating L929 fibroblasts. The results presented in Fig. 5d showed no decrease in cell viability, invariant to the amount of the antibacterial copolymer used as an additive (Fig. 5d). The absence of cytotoxicity of the additivated matrix is of prime importance to envision a biomedical application of the prepared antibacterial materials.
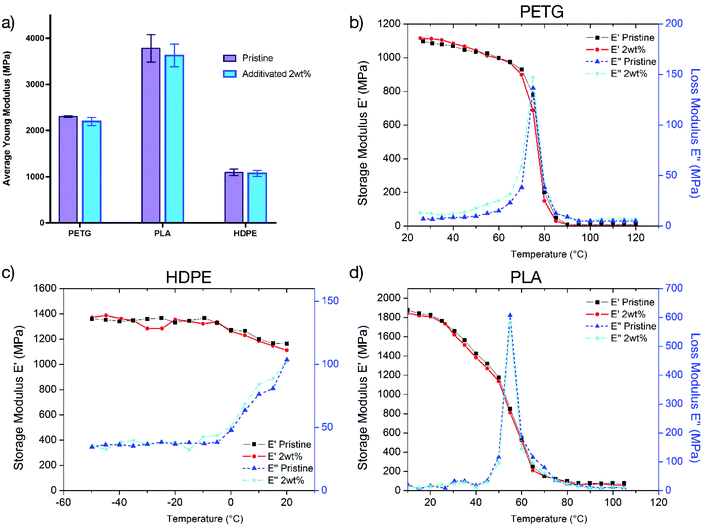
Mechanical properties
With applications ranging from biomaterials to packaging, the mechanical properties of the materials are of paramount importance. The modification of the materials to confer antibacterial activity should ideally not affect their mechanical properties. In our approach, the use of low amounts (0.5–2 wt%) of the antibacterial copolymer as an additive is not expected to alter the bulk properties of the materials. For confirmation, tensile tests were performed on PETG, PLA and HDPE samples to determine Young's modulus. The samples of both pristine and additivated matrices (2 wt%) were prepared by injection moulding.We assessed the materials with the higher amount of the antibacterial copolymer to determine the maximum modification of the mechanical properties. The comparison of these values (Fig. 6a, details in the ESI†) demonstrates that the incorporation of the copolymer in the matrix does not change the mechanical properties of the pristine material.
To confirm the conservation of the mechanical properties, dynamic mechanical analysis (DMA) tests were performed on PETG, PLA and HDPE samples to determine the viscoelastic properties over a range of temperatures.
The comparison of storage (E′) and loss moduli (E′′) curves as a function of temperature shown in Fig. 6b–d demonstrates that the materials have the same viscoelastic properties over a large range of temperature. In conclusion, the addition of the copolymer into the polymer matrix does not change the mechanical properties of the materials over a large range of temperatures and strain levels.
Surface characterization
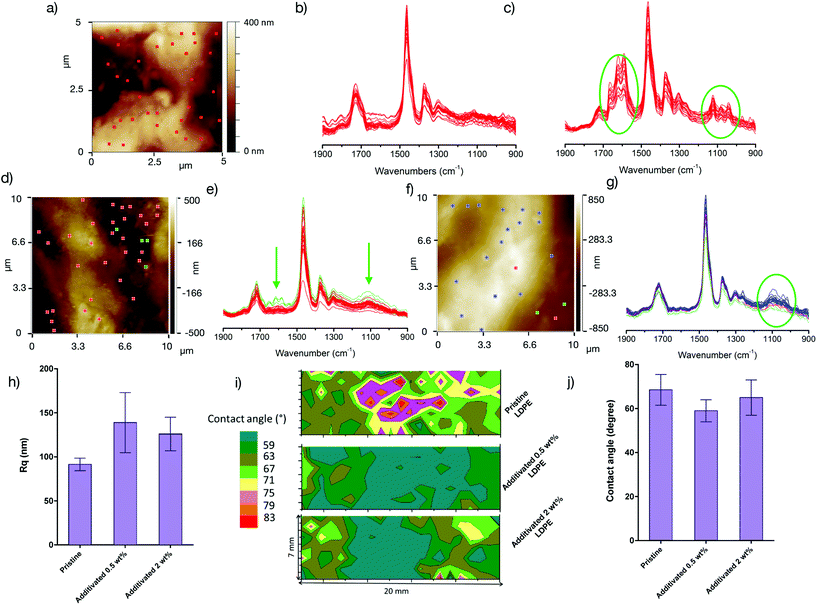
According to the antibacterial studies, the use of a macromolecular antibacterial additive that is not leached out in the medium confers good activity by membrane permeabilization even with a small amount of the copolymer. The localization of the copolymer in the materials is thus an intriguing question. Due to the blending in the micro-extruder, one would expect a homogeneous dispersion of the amphiphilic copolymer into the polymer matrix, but the partial or complete migration of the copolymer to the surface cannot be excluded. To answer this question, we performed time-of-flight secondary ion mass spectrometry (ToF-SIMS) analyses on PETG, LDPE and PLA materials containing 2 wt% of copolymers (see the ESI† – section VI for details). Using ToF-SIMS, the specific mass response of the copolymer was detected in the background of the matrix. Even if all the copolymer migrated towards the surface of the materials, the low amount of the additive was still difficult to detect. As shown in Fig. S2,† there is no increase in the intensity in the ToF-SIMS spectrum of the organic matrix with the copolymer at m/z values corresponding to peaks in the copolymer spectrum, for any of the three materials. These results showed that it was not possible to detect the antibacterial additive on the surface samples even when using the sensitive ToF-SIMS analysis. These indirect results prove that whatever the matrix is, the copolymer does not migrate to the surface and likely stays well dispersed in the material.To obtain further insights, we focused on the matrix that differs the most from the copolymer in terms of the structure (LDPE with an apolar totally carbon-based backbone) to maximize the chance to detect the copolymer. The surface of the LDPE films was characterized by atomic force microscopy (AFM) in the tapping mode and nanoscale infrared spectroscopy (nano-IR), allowing the acquisition of local Fourier transform infrared spectroscopy (FTIR) spectra of the sample with a high lateral resolution (all the techniques are described in detail in the ESI†). The pristine LDPE film has a heterogeneous and rough (Fig. 7a) surface as shown by the local FTIR spectra (Fig. 7b) and the hydrophobicity mapping (Fig. 7i). The non-homogeneous surface polarity and low contact angles observed for such an apolar polymer (Fig. 7i) may be the result of surface oxidation (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O band at 1724 cm−1 in Fig. 7b) and/or of the presence of lubricant-rich areas (see amide bands between 1700 and 1590 in Fig. 7c. The addition of the antibacterial copolymer results in a slight increase in the roughness (Fig. 7h). The hydrophobicity is nevertheless not significantly modified and rich antibacterial copolymer areas on the film surface cannot be certainly attributed: the oxidation band and amide lubricant bands (green arrow in Fig. 7e)) are still observed, yet no other bands appear significantly, even if, for the film containing 2 wt% of the copolymer, small bands at around 1100 cm−1 (ether stretching band in Fig. 7g) are locally detected and may be the result of the presence of the copolymer but cannot be ascribed with certainty to the additive.
O band at 1724 cm−1 in Fig. 7b) and/or of the presence of lubricant-rich areas (see amide bands between 1700 and 1590 in Fig. 7c. The addition of the antibacterial copolymer results in a slight increase in the roughness (Fig. 7h). The hydrophobicity is nevertheless not significantly modified and rich antibacterial copolymer areas on the film surface cannot be certainly attributed: the oxidation band and amide lubricant bands (green arrow in Fig. 7e)) are still observed, yet no other bands appear significantly, even if, for the film containing 2 wt% of the copolymer, small bands at around 1100 cm−1 (ether stretching band in Fig. 7g) are locally detected and may be the result of the presence of the copolymer but cannot be ascribed with certainty to the additive.
The combination of all these techniques showed that the surface is not sufficiently modified to detect the copolymer. This is a striking difference with the materials whose antibacterial properties are conferred by various surface modification techniques.39
Antibacterial packaging application
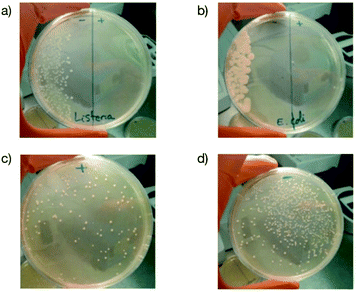
Finally, to demonstrate that our approach could be realistically envisioned as a solution to prevent bacterial infection in a real-world scenario, we prepared antibacterial LDPE films containing 2 wt% of the copolymer as an additive and test them as packaging for mozzarella as already reported.40The idea was to show whether such a plastic that is commonly used to prepare films used in food packaging applications could be useful to (i) avoid the development of pathogenic bacteria such as Listeria monocytogenes and enterohemorrhagic E. coli that are responsible every year for contamination widely reported in the media41,42 and (ii) preserve the bacteria within the cheese that are necessary to impart gustative properties.43Fig. 8a and b clearly prove that both Listeria monocytogenes and enterohemorrhagic E. coli were completely killed after contact with the additive containing film. In the case of bacteria already present in cheese, we observe that a large fraction of them survived (Fig. 8c and d) although with a lower amount than the pristine film. This result could be interesting for increasing the shelf-life of food and allows us to envision the possibility of using B(A10%)Me copolymer containing plastics as food packaging materials.
Conclusions
We demonstrate that simply adding an amphiphilic synthetic cationic diblock copolymer based on methacrylic monomers into common organic matrices (such as HDPE, LDPE, PLA and PETG) via an extrusion process confers antibacterial properties to the pristine materials after 1 h of exposure. The effect of the concentration for such additives has been studied and we demonstrate that even 0.5 wt% is sufficient to impart good antibacterial efficiency (>80% of bacterial killing) for both E. coli and S. aureus. The increase to 2 wt% led to complete inhibition of both bacteria on HDPE, LDPE and PLA and more than 90% of bacterial killing on PETG. The mechanism of action was also investigated and the permeabilization of the bacterial membrane was monitored via the propidium iodide assay, suggesting that the killing of bacteria occurred by contact and membrane destabilization. This action mechanism is expected to limit the development of bacterial resistance. In addition, the study on HaCaT and L929 fibroblast cell lines reveals no toxicity of the materials. Further analysis of the materials demonstrated that the mechanical properties over a wide range of temperatures are not altered. The stable mechanical properties are of prime importance for the utilization of these materials in their specific applications.For a better understanding of the mode of action of such materials, complete characterization of the surface has been carried out. ToF-SIMS, nano-IR spectroscopy and AFM did not reveal any specific modification of the surface, suggesting that there is a homogeneous incorporation of the additive in the materials without complete migration towards the surface. This is a critical advantage compared to antibacterial materials whose properties are given by a surface modification (antibacterial coatings). Lastly, to demonstrate its application in food packaging, a LDPE film containing 2 wt% of the additive was shown to inhibit the growth of Listeria monocytogenes and enterohemorrhagic E. coli. At the same time, the additive containing film slows down the development of cheese bacteria in mozzarella brine, which could increase the shelf-life of the food contained in the LDPE film.
Our methodology therefore constitutes a straightforward and cost-effective solution to generate intrinsically and long-lasting antibacterial materials for a plethora of applications, including packaging, disposable materials and medical devices.
Author contributions
Y. G. D. G. and C. L. conceived the original idea of the project. S. H. conducted the copolymer synthesis and material preparations. G. M. and A. M.-P. performed the measurements of material properties. S. F., J. S., A. D.-B. and N. Y. conducted the material characterization. S. H. and M. M. performed the antibacterial and innocuity tests. L. M. and C. B.-K. conducted the surface characterization of the materials by ToF-SIMS. C. B.-K., Y. G. and C. L. wrote the original manuscript. All authors contributed to the scientific planning and discussions.Conflicts of interest
D. G., M. M., Y. G. and C. L. are named inventors on patent applications (French Patent Application FR1756390 (2017) and PCT WO2019/008176 (2018)) filed by the CNRS, Ecole Centrale Marseille, and Aix-Marseille University on the antibacterial materials described in the current study.Acknowledgements
The authors thank the CNRS, SATT Sud-Est and Aix-Marseille University for support. C. B.-K. acknowledges the Australian Research Council (ARC) for funding in the context of a Laureate Fellowship and the Queensland University of Technology (QUT) for continued support including via its Centre for Materials Science.Notes and references
- M. Baym, L. K. Stone and R. Kishony, Science, 2016, 351, 5 CrossRef PubMed.
- M. F. Chellat, L. Raguz and R. Riedl, Angew. Chem., Int. Ed., 2016, 55, 6600–6626 CrossRef CAS PubMed.
- M. I. Hutchings, A. W. Truman and B. Wilkinson, Curr. Opin. Microbiol., 2019, 51, 72–80 CrossRef CAS.
- A. Munoz-Bonilla and M. Fernandez-Garcia, Prog. Polym. Sci., 2012, 37, 281–339 CrossRef CAS.
- M. M. Konai, B. Bhattacharjee, S. Ghosh and J. Haldar, Biomacromolecules, 2018, 19, 1888–1917 CrossRef CAS.
- K. G. Neoh, M. Li, E. T. Kang, E. Chiong and P. A. Tambyah, J. Mater. Chem. B, 2017, 5, 2045–2067 RSC.
- L. Karam, R. Roustom, M. G. Abiad, T. Ei-Obeid and I. N. Savvaidis, Int. J. Food Microbiol., 2019, 291, 42–47 CrossRef CAS.
- A. Orsuwan, S. Kwon, N. Bumbudsanpharoke and S. Ko, Food Control, 2019, 100, 176–182 CrossRef CAS.
- J. Bruenke, I. Roschke, S. Agarwal, T. Riemann and A. Greiner, Macromol. Biosci., 2016, 16, 647–654 CrossRef CAS PubMed.
- A. Kyzioł, W. Khan, V. Sebastian and K. Kyzioł, Chem. Eng. J., 2020, 385, 123888 CrossRef.
- G. N. Tew, D. H. Liu, B. Chen, R. J. Doerksen, J. Kaplan, P. J. Carroll, M. L. Klein and W. F. DeGrado, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5110–5114 CrossRef CAS.
- G. N. Tew, R. W. Scott, M. L. Klein and W. F. Degrado, Acc. Chem. Res., 2010, 43, 30–39 CrossRef CAS PubMed.
- D. Campoccia, L. Montanaro and C. R. Arciola, Biomaterials, 2013, 34, 8533–8554 CrossRef CAS PubMed.
- S. Ye, D. Wei, X. Xu, Y. Guan and A. Zheng, Polym. Adv. Technol., 2020, 31, 1847–1856 CrossRef CAS.
- H. Zuo, D. Wu and R. Fu, J. Appl. Polym. Sci., 2012, 125, 3537–3544 CrossRef CAS.
- B. T. Benkhaled, S. Hadiouch, H. Olleik, J. Perrier, C. Ysacco, Y. Guillaneuf, D. Gigmes, M. Maresca and C. Lefay, Polym. Chem., 2018, 9, 3127–3141 RSC.
- D. Gigmes, Y. Guillaneuf, C. Guillaneuf, M. Maresca and C. Ysacco, Solid organic antibacterial material, 2018, WO2019008176A1 Search PubMed.
- G. Liu, Q. Zhang, Y. Li, X. Wang, H. Wu, Y. Wei, Y. Zeng and L. Tao, iScience, 2020, 23, 100754 CrossRef CAS.
- W. Cao, D. Wei, A. Zheng and Y. Guan, Eur. Polym. J., 2019, 118, 231–238 CrossRef CAS.
- M. R. Badrossamay and G. Sun, React. Funct. Polym., 2008, 68, 1636–1645 CrossRef CAS.
- F. Chen, X. Ding, Y. Jiang, Y. Guan, D. Wei, A. Zheng and X. Xu, ACS Omega, 2020, 5, 10481–10488 CrossRef CAS PubMed.
- D. Wei, Y. Ding, T. Wang, J. Yang, Y. Guan and A. Zheng, J. Appl. Polym. Sci., 2017, 134, 44190 Search PubMed.
- R. Cuervo-Rodriguez, F. Lopez-Fabal, J. L. Gomez-Garces, A. Munoz-Bonilla and M. Fernandez-Garcia, Macromol. Biosci., 2017, 17, 1700258 CrossRef.
- J.-M. Thomassin, S. Lenoir, J. Riga, R. Jérôme and C. Detrembleur, Biomacromolecules, 2007, 8, 1171–1177 CrossRef CAS.
- C. Ergene, K. Yasuhara and E. F. Palermo, Polym. Chem., 2018, 9, 2407–2427 RSC.
- A. C. Engler, J. P. K. Tan, Z. Y. Ong, D. J. Coady, V. W. L. Ng, Y. Y. Yang and J. L. Hedrick, Biomacromolecules, 2013, 14, 4331–4339 CrossRef CAS PubMed.
- P. R. Judzewitsch, T. K. Nguyen, S. Shanmugam, E. H. H. Wong and C. Boyer, Angew. Chem., Int. Ed., 2018, 57, 4559–4564 CrossRef CAS PubMed.
- A. Kuroki, P. Sangwan, Y. Qu, R. Peltier, C. Sanchez-Cano, J. Moat, C. G. Dowson, E. G. L. Williams, K. E. S. Locock, M. Hartlieb and S. Perrier, ACS Appl. Mater. Interfaces, 2017, 9, 40117–40126 CrossRef CAS.
- A. Kuroki, A. K. Tchoupa, M. Hartlieb, R. Peltier, K. E. S. Locock, M. Unnikrishnan and S. Perrier, Biomaterials, 2019, 217, 119249 CrossRef CAS.
- J. Tan, J. Tay, J. Hedrick and Y. Y. Yang, Biomaterials, 2020, 252, 120078 CrossRef CAS PubMed.
- A. V. Ruzette and L. Leibler, Nat. Mater., 2005, 4, 19–31 CrossRef CAS.
- J. Nicolas, Y. Guillaneuf, C. Lefay, D. Bertin, D. Gigmes and B. Charleux, Prog. Polym. Sci., 2013, 38, 63–235 CrossRef CAS.
- E. Guegain, Y. Guillaneuf and J. Nicolas, Macromol. Rapid Commun., 2015, 36, 1227–1247 CrossRef CAS PubMed.
- I. Sovadinova, E. F. Palermo, M. Urban, P. Mpiga, G. A. Caputo and K. Kuroda, Polymers, 2011, 3, 1512–1532 CrossRef CAS.
- H. Takahashi, G. A. Caputo, S. Vemparala and K. Kuroda, Bioconjugate Chem., 2017, 28, 1340–1350 CrossRef CAS PubMed.
- W. A. Dengler, J. Schulte, D. P. Berger, R. Mertelsmann and H. H. Fiebig, Anti-Cancer Drugs, 1995, 6, 522–532 CrossRef CAS PubMed.
- L. B. Oyama, S. E. Girdwood, A. R. Cookson, N. Fernandez-Fuentes, F. Prive, H. E. Vallin, T. J. Wilkinson, P. N. Golyshin, O. V. Golyshina, R. Mikut, K. Hilpert, J. Richards, M. Wootton, J. E. Edwards, M. Maresca, J. Perrier, F. T. Lundy, Y. Luo, M. Zhou, M. Hess, H. C. Mantovani, C. J. Creevey and S. A. Huws, npj Biofilms Microbiomes, 2017, 3, 9 CrossRef PubMed.
- E. Di Pasquale, C. Salmi-Smail, J. M. Brunel, P. Sanchez, J. Fantini and M. Maresca, Chem. Phys. Lipids, 2010, 163, 131–140 CrossRef CAS PubMed.
- J. Hasan, R. J. Crawford and E. P. Lvanova, Trends Biotechnol., 2013, 31, 31–40 CrossRef.
- B. Agrillo, M. Balestrieri, M. Gogliettino, G. Palmieri, R. Moretta, Y. T. R. Proroga, I. Rea, A. Cornacchia, F. Capuano, G. Smaldone and L. De Stefano, Int. J. Mol. Sci., 2019, 20(3), 601 CrossRef CAS.
- A. Gerard, S. El-Hajjaji, E. Niyonzima, G. Daube and M. Sindic, Int. J. Dairy Technol., 2018, 71, 825–843 CrossRef CAS.
- M. Gouali and F. X. Weill, Presse Med., 2013, 42, 68–75 CrossRef PubMed.
- C. Pelaez and T. Requena, Int. Dairy J., 2005, 15, 831–844 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Copolymer synthesis, mechanical properties, membrane permeation assay, surface characterization and ToF-SIMS analyses. See DOI: 10.1039/d1py01344k |
| This journal is © The Royal Society of Chemistry 2022 |