Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Multi-responsive poly(oligo(ethylene glycol)methyl methacrylate)-co-poly(2-(diisopropylamino)ethyl methacrylate) hyperbranched copolymers via reversible addition fragmentation chain transfer polymerization†
Dimitrios
Selianitis
and
Stergios
Pispas
 *
*
Theoretical and Physical Chemistry Institute, National Hellenic Research Foundation, 48 Vassileos Constantinou Avenue, 11635 Athens, Greece. E-mail: pispas@eie.gr
First published on 3rd November 2021
Abstract
In this work, we report on the synthesis and self-organization in aqueous media of a series of novel multi-responsive poly(oligo(ethylene glycol)methyl methacrylate)-co-poly(2-(diisopropylamino)ethyl methacrylate), P(OEGMA-co-DIPAEMA), hyperbranched copolymers, using ethylene glycol dimethacrylate (EGDMA) units as branching points. Three P(OEGMA-co-DIPAEMA) hyperbranched copolymers with different ratios of hydrophilic to hydrophobic components are prepared by the reversible addition–fragmentation chain transfer (RAFT) polymerization, in order to study their properties in aqueous solutions. By using dynamic light scattering (DLS), the formation of aggregates with dimensions in the nanoscale, responsive to changes in solution temperature and pH, is observed. The hyperbranched copolymer with the lowest DIPAEMA hydrophobic component shows the largest dimensions at pH 7 and pH 10 compared to the other two copolymers. The hyperbranched copolymers are capable of encapsulating the hydrophobic drug curcumin (CUR) and demonstrate stability in the presence of serum proteins. These new hyperbranched copolymers could be potential candidates for drug delivery and bio-imaging applications.
Introduction
In recent years hyperbranched polymers have attracted the interest of polymer scientists as they exhibit remarkable structural properties.1–3 Hyperbranched polymers are macromolecules with a high degree of polymerization, which, like dendrimers, are characterized by macromolecular topologies similar to that of a tree, having a large number of internal/external functional groups. Dendrimers belong to the broad category of hyperbranched molecules, having a more symmetric topology.4–7 The multi-stage syntheses of dendrimers are performed by a series of different steps, requiring repetitive purification steps and resulting in expensive products with, sometimes, limited use in large-scale industrial applications. However, for the majority of applications where structural perfection is not required, hyperbranched polymers can replace dendrimers by overcoming the cost problem.8,9 In contrast to dendrimers, randomly branched,10 hyperbranched polymers can be easily synthesized by single-step polymerization. Therefore, they are economical products for both small and large-scale industrial applications.11,12 The hyperbranched structure imparts to the polymer unique properties, such as low viscosity and compact conformations, both in solutions and in melts, compared to the corresponding linear polymers.13,14 Hyperbranched polymers exhibit incomplete or irregularly branched structure and a large number of reactive/functional groups on their surface and their interior. These features make hyperbranched polymers potential candidates for a wide range of biomedical applications,15,16 such as in drug17,18 and gene delivery19,20 and bioimaging.21,22Most of hyperbranched polymers are formed by the condensation of multifunctional monomers of the type ABx, where A is the functional group reacting with group B and x ≥ 2.23,24 The technique creates three-dimensional polymers with an irregular structure. This structure provides some unique physical and chemical properties, such as the lower viscosity and lower glass transition temperatures, compared to their respective linear analogs.25,26 The synthesis of well-defined hyperbranched polymers is accomplished by controlled radical polymerization techniques (ATRP,27,28 NMP,29,30 RAFT31,32), which provide tremendous compatibility with a wide range of monomers. The RAFT technique allows the synthesis of copolymers with predicted molecular weight, narrow molecular weight distributions, and variable molecular architectures, namely linear,33,34 stars,35,36 hyperbranched37,38 (co)polymers, which can be responsive to pH39,40 and temperature41,42 changes. Perrier et al. reported for the first time the synthesis of hyperbranched methyl methacrylate (MMA) and ethylene glycol dimethacrylate (EGDMA) copolymers via reversible addition fragmentation chain transfer (RAFT) polymerization using 2-(2-cyanopropyl) dithiobenzoate as the chain transfer agent.43
Herein, we report on the synthesis of novel poly(oligo(ethylene glycol) methacrylate)-co-poly(2-(diisopropylamino)ethyl methacrylate), P(OEGMA-co-DIPAEMA), hyperbranched copolymers via RAFT polymerization, utilizing EGDMA as the branching agent. We also study their self-organization behavior in response to various physicochemical stimuli such as pH, temperature and ionic strength by light scattering techniques and fluorescence spectroscopy. Furthermore, the hyperbranched copolymers were utilized to encapsulate curcumin (CUR). The colloidal stability of the loaded nanocarriers in the presence of serum and the fluorescence properties of the loaded nanocarriers, were investigated. The 2-(diisopropylamino)ethyl methacrylate (PDIPAEMA) homopolymer is a double-responsive polymer, towards pH and temperature, with a pKa of around 6.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 44 and Tcpca. 28 °C.45 PDIPAEMA becomes a water-soluble polymer due to the protonation of its amino groups at pH range below neutral, where it behaves as a cationic polyelectrolyte.46 Close at and above neutral pH, where the deprotonation of amino groups takes place, PDIPAEMA is converted to a hydrophobic polymer. The oligo(ethylene glycol) methacrylate (OEGMA) homopolymer is a hydrophilic polymer and provides stealth properties to polymer nanostructures. This means that the drug loaded polymer structure is water soluble and undetected from the blood components and the immune system, as a result of the presence of the hydrophilic oligo(ethylene glycol) side chains. Therefore, three novel P(OEGMA-co-DIPAEMA) hyperbranched copolymers were synthesized, having different ratios of hydrophilic to hydrophobic segments, in order to study the structure–properties relations in this novel family of copolymers.
44 and Tcpca. 28 °C.45 PDIPAEMA becomes a water-soluble polymer due to the protonation of its amino groups at pH range below neutral, where it behaves as a cationic polyelectrolyte.46 Close at and above neutral pH, where the deprotonation of amino groups takes place, PDIPAEMA is converted to a hydrophobic polymer. The oligo(ethylene glycol) methacrylate (OEGMA) homopolymer is a hydrophilic polymer and provides stealth properties to polymer nanostructures. This means that the drug loaded polymer structure is water soluble and undetected from the blood components and the immune system, as a result of the presence of the hydrophilic oligo(ethylene glycol) side chains. Therefore, three novel P(OEGMA-co-DIPAEMA) hyperbranched copolymers were synthesized, having different ratios of hydrophilic to hydrophobic segments, in order to study the structure–properties relations in this novel family of copolymers.
Experimental
Materials
Monomers, 2-(diisopropylamino)ethyl methacrylate (DIPAEMA) (97%), oligo(ethylene glycol) methacrylate (OEGMA) (average Mn = 475), and crosslinker/branching agent ethylene glycol dimethacrylate (EGDMA, 98%) were obtained from Sigma-Aldrich. DIPAEMA, OEGMA and EGDMA monomers were passed through an inhibitor removing column for purification before polymerization. Azobis(isobutyronitrile) (AIBN) was used as radical initiator and was purified by recrystallization from methanol. 1,4-Dioxane (≥99.8% pure, Aldrich) was dried over molecular sieves before use. Tetrahydrofuran (THF 99.9% pure), n-hexane (≥97%), 4-cyano-4-(phenylcarbonothioylthio) pentanoic acid (CPAD) and other reagents from Sigma Aldrich were used as received.Synthesis of hyperbranched polymers
For the synthesis of P(OEGMA-co-DIPAEMA) hyperbranched copolymers, CPAD was utilized as the chain transfer agent (CTA), AIBN as the radical initiator and 1,4-dioxane as the solvent. A typical synthesis of hyperbranched copolymer (copolymer HB1) is described below: in a one-necked, round-bottom flask with a magnetic stirrer (50 mL), OEGMA (1.45 g, 3.05 mmol), DIPAEMA (0.24 g, 1.1 mmol), EGDMA (0.032 g, 0.16 mmol), CPAD (0.048 g, 0.17 mmol), AIBN (0.0056 g, 0.03 mmol) and 1,4-dioxane (18 mL, 10% w/w monomer concentration), were added. The solution was degassed by nitrogen gas flow for 15 min and then the round-bottom flask was heated in an oil bath at 70 °C for 24 h. After that, the mixture was placed at −20 °C for 20 min and subsequently was exposed to air. The reaction product was then precipitated in a large excess of n-hexane for removal of unreacted monomers. The P(OEGMA-co-DIPAEMA) hyperbranched copolymer was placed in a vacuum oven for 48 h for drying.Self-assembly studies of hyperbranched copolymers in aqueous media
Stock solutions of hyperbranched copolymers were prepared by dissolving directly the solid materials in acidic water (10 mL), adjusted to pH 3, with a proper quantity of HCl (polymer concentration, c = 1 × 10−3 g mL−1) and left overnight to bring the solutions in equilibrium. Afterwards, proper quantities of 1 M NaOH solution were added in the stock solutions in order to adjust the solution pH at pH 7 and pH 10. Solutions at pH 7.4 were prepared using a phosphate buffered saline (PBS) solution where the copolymers were dissolved directly. For fluorescence studies, pyrene was utilized as a probe and a 1 mM stock solution of the probe was added in appropriate quantity to each P(OEGMA-co-DIPAEMA) hyperbranched copolymer solution of different pH. The solutions were left overnight before measurements (final cpyrene = 10−7 M). For light scattering studies, the solutions were filtered through 0.45 μm hydrophilic PVDF filters. The effect of ionic strength on hyperbranched copolymer solutions was studied by adding proper amounts of 1 M NaCl in the aqueous copolymer solutions initially prepared. Simultaneous static and dynamic light scattering measurements were utilized to determine the changes in mass and the hydrodynamic radius, Rh, of the copolymers in aqueous solutions.Curcumin loading studies
The process followed for curcumin entrapment into hyperbranched copolymer nanostructures is described below. Initially, two separate stock solutions of copolymer in THF and curcumin in THF were prepared. For example, for the preparation of HB1–30% w/w CUR, 0.01 g of copolymer and 0.00029 g of drug were dissolved in THF. Subsequently, these solutions were mixed and then the copolymer/CUR mixture was injected directly in 10 mL phosphate buffered saline (PBS) under vigorous stirring. Then the organic solvent was evaporated by heating at 65 °C. The encapsulation of CUR was studied in two of the three hyperbranched copolymers, because the HB3 copolymer was impossible to encapsulate a large amount of drug, probably due to the high amount of hydrophobic component. The drug-loaded hyperbranched copolymers were measured by FTIR spectroscopy and by DLS (at 25 °C and at 90° angle) to verify the formation and stability of CUR-loaded nanostructures. Measurements on the first day of preparation and after up to 18 days in respect to the day of solution preparation were performed. Using UV-Vis spectroscopy, the reference curve of curcumin was constructed by measuring the absorbance at λmax = 420 nm, and was utilized to calculate the actual percentage of curcumin entrapped in the copolymer nanostructures.Characterization methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1. Hyperbranched polymers were dissolved in THF at concentrations of 2–4 mg mL−1.
000 g mol−1. Hyperbranched polymers were dissolved in THF at concentrations of 2–4 mg mL−1.
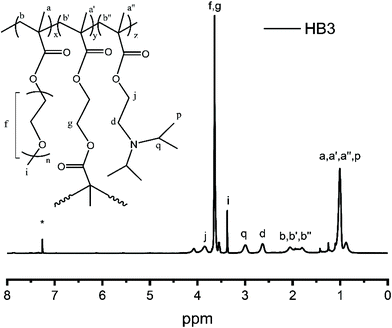
1H-NMR spectra peaks of P(OEGMA-co-DIPAEMA) (Fig. 2, 600 MHz, CDCl3, ppm): 0.86, 1.25 (9H, –CH2CCH3–), 1 (3H, CH3CH–), 1.79–2.04 (6H, –CH2C–), 2.63 (2H, –CH2N), 2.99 (H, –NCH), 3.37 (3H, –(CH2CH2O)9CH3–), 3.64 (36H, –(CH2CH2O)9), 2H, –COOH(CH2)CH2, 3.84 (2H, –COOCH2[CH2], 2H, –COOCH2[CH2]).
For fluorescence measurements on CUR-loaded hyperbranched copolymers the same instrument was utilized. Curcumin excitation wavelength was set at 405 nm.
For static light scattering studies the same instrument was used in the angular range of 30°–150° and at 25 °C. Toluene was utilized as the calibration standard. The dn/dc values were calculated from literature data, where available, also based on the refractive indices of the constituting blocks and THF. Static light scattering profiles were analyzed by the Zimm and Guinier models using the software available by the manufacturer.
Results and discussion
Synthesis and molecular characterization of P(OEGMA-co-DIPAEMA) hyperbranched copolymers
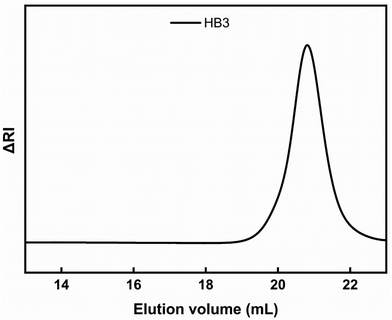
Three P(OEGMA-co-DIPAEMA) hyperbranched copolymers were synthesized with different molecular weights and chemical composition utilizing RAFT polymerization procedures. The ratio of EGDMA/CTA was kept constant at 1.1 for all experiments to avoid the formation of insoluble crosslinked polymers. AIBN was utilized as the radical initiator and 1,4-dioxane as the solvent of the polymerization reaction. The synthesis of P(OEGMA-co-DIPAEMA) was accomplished at 70 °C for 24 h (Scheme 1). CPAD was selected for its efficiency in RAFT polymerization of methacrylic monomers based on results from the literature.47,48 EGDMA led to the development of the branched polymer chains introducing branching points. Molecular characteristics of the copolymers were recorded by means of SEC and 1H-NMR spectroscopy methods. In Fig. 1, the chromatogram of HB3 is presented, where a narrow peak with symmetric molecular weight distribution and low dispersity is observed, a fact that demonstrates control of polymerization. It should be noted that molecular weights determined by SEC are apparent ones due to the different hydrodynamic properties of the hyperbranched methacrylate based copolymers in respect to those of the linear PS used for calibration. Table 1 presents the molecular characteristics of all hyperbranched copolymers synthesized.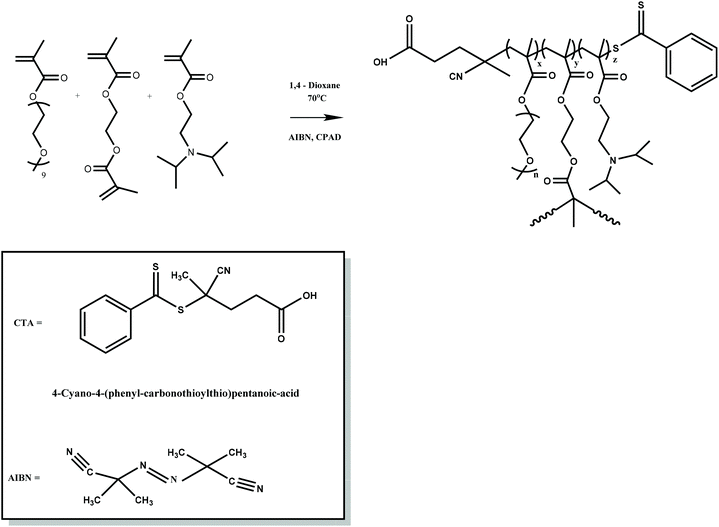 | ||
| Scheme 1 Synthesis scheme followed for the preparation of P(OEGMA-co-DIPAEMA) hyperbranched copolymers. | ||
1H-NMR spectroscopy was utilized to identify the chemical structure and to determine the composition of P(OEGMA-co-DIPAEMA) hyperbranched copolymers. Characteristic peaks which were chosen to calculate the composition of each copolymer are presented in Fig. 2. For OEGMA component the –CH3 protons at 3.35 ppm were selected (peak i)49 and for the PDIPAEMA component the –CH2 protons at 2.61 ppm were used (peak d).50 However, it should be noted that it is not possible to determine the exact composition of EGDMA due to the overlap of the peaks from the OEGMA constituent at 3.64 ppm (peak f and g) and the low content of the cross-linker (below 2 wt% in all cases).
By using SLS in THF it was possible to determine the apparent weight average molecular weight for the hyperbranched copolymers. It is observed that the Mw app determined by SLS are higher than Mw estimated by SEC, supporting the hyperbranched structure of the copolymers synthesized.
Hyperbranched copolymer self-assembly in aqueous solutions
P(OEGMA-co-DIPAEMA) hyperbranched copolymers are expected to be affected by physicochemical stimuli when dissolved in aqueous media, due to the chemical properties of the DIPAEMA component. Therefore, self-organization studies were undertaken utilizing different physicochemical methods. Aqueous solutions of P(OEGMA-co-DIPAEMA) hyperbranched copolymers were studied by fluorescence spectroscopy to determine if they present a critical aggregation concentration (CAC). The measurements were carried out by the use of pyrene as the fluorescence probe, which has the ability to be encapsulated into the hydrophobic domains of amphiphilic self-organized polymeric structures formed in aqueous media. CAC is defined as the concentration corresponding to the point where the ratio of the relative intensities of the first and third peaks (I1/I3) in the pyrene fluorescence spectrum changes abruptly. According to the literature, I1/I3 ratio values in the range 1–1.3 denote a hydrophobic microenvironment, while in polar environment the I1/I3 ratio attains values in the range 1.7–1.9. The values determined for the hyperbranched copolymers at different pH are shown in Table S1 in ESI.† The copolymer solutions were prepared in concentrations ranging from 10−8–10−3 g mL−1. In the case of P(OEGMA-co-DIPAEMA) hyperbranched copolymers containing DIPAEMA as a structural unit, the properties of the aggregates formed in aqueous solutions are expected to depend on solution pH. The measurements were made for pH 3, 7.4 and 10. At acidic conditions (pH 3) the amine groups of PDIPAEMA are fully protonated,39 and also due to the presence of the hydrophilic component POEGMA, aggregates were not observed. I1/I3 values vs. copolymer concentration from pyrene fluorescence spectra for pH 7.4 (PBS) and pH 10 are illustrated in Fig. 3.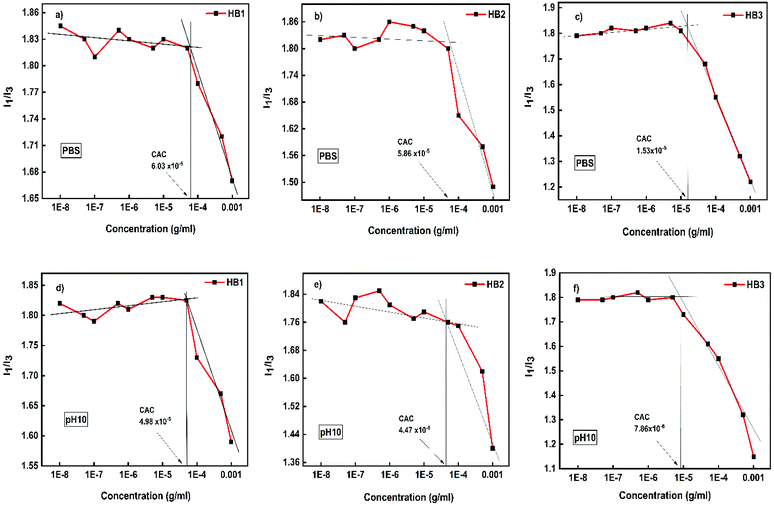 | ||
| Fig. 3 CAC determination for P(OEGMA-co-DIPAEMA) hyperbranched copolymers at pH 7.4 (PBS) (upper row) and pH 10 (lower row). | ||
In all cases a clear plateau is observed at low concentrations, where there are no aggregates present in the aqueous solutions, followed by the transition region in between lower and higher concentrations, where copolymer aggregates are formed. As shown, the chemical composition of the copolymers determines the actual critical aggregation concentration, with CAC being lower for copolymer HB3 with the higher DIPAEMA content. It is also observed that CAC always occurs at lower concentrations at pH 10 and at higher concentration in PBS. This is to be expected since the closer to neutral pH the more hydrophobic character is exhibited by the hyperbranched copolymer, due to the deprotonation of the amino groups of DIPAEMA segments. Therefore, the formation of aggregates is preferred in aqueous solution. When the DIPAEMA content increases the polymeric aggregates are formed at lower concentrations because of the hydrophobic behavior of DIPAEMA segments.
Complementary studies of P(OEGMA-co-DIPAEMA) hyperbranched copolymers were performed by light scattering methods and the results are presented in Table S2 (ESI†). Specifically, dynamic light scattering measurements were performed to determine the hydrodynamic radius (Rh) of the self-organized nanoaggregates, the change in the scattered light intensity (which is related to the mass of the formed nanoparticles) and their size polydispersity index (PDI), as a result of copolymer response to pH and temperature changes. Contin analysis of DLS results from HB1–HB3 copolymer solutions are illustrated in Fig. 4. Based on the results for the HB1 copolymer, two populations are observed with a broad size distribution, except for solutions in PBS (pH 7.4, which also contains 0.15 M salt). The small size peak indicates free polymeric chains (Rhca. 3 nm) while the larger in size represents nanoaggregates, confirming the formation of nanoparticles by spontaneous self-organization of the hyperbranched copolymers. The results also demonstrate that the OEGMA side chains create steric stabilization of the nanoparticles. Full protonation of DIPAEMA amino groups in acidic environment creates the tendency for rather extended macromolecular conformations. Protonation of DIPAEMA segments results in electrostatic interactions that leads to rather swollen self-assembled nanostructures, as is generally the case with polyelectrolytes. In PBS solutions, the presence of a small amount of salt disrupts the hydration structure around the polymer chains resulting in the formation of large aggregates, due to the predominance of polymer–polymer interactions. In the case of HB2 and HB3 copolymers, where the DIPAEMA content is higher than that of HB1, the same tendency seems to be maintained for the systems at acidic pH for the reasons explained above. However, an increase of pH (to pH 7 and 10) appears to create smaller nanoparticles with relatively wide size distributions, accompanied by the complete absence of large swollen nanostructures. This observation indicates that by increasing the hydrophobic component through the deprotonation of the amino groups of PDIPAEMA, the dimensions of the nanoparticles are significantly reduced (shrinking of nanostructures occurs). It is worth noting that the small size of the particles formed is in agreement with the architecture of the copolymers, because the hyperbranched molecules are distinguished by their more compact structures compared to linear ones. The self-assembly of HB2 and HB3 copolymers in PBS solution appears to be different from that of HB1. Specifically, for HB2 two peaks are observed in DLS experiments. This may be due to the higher content of DIPAEMA which gives the systems an extra impetus to form smaller, more compact nanoparticles. In the case of HB3, where the DIPAEMA content in the copolymer further increases, smaller nanoparticles are formed with narrower size distributions compared to HB1 and HB2.
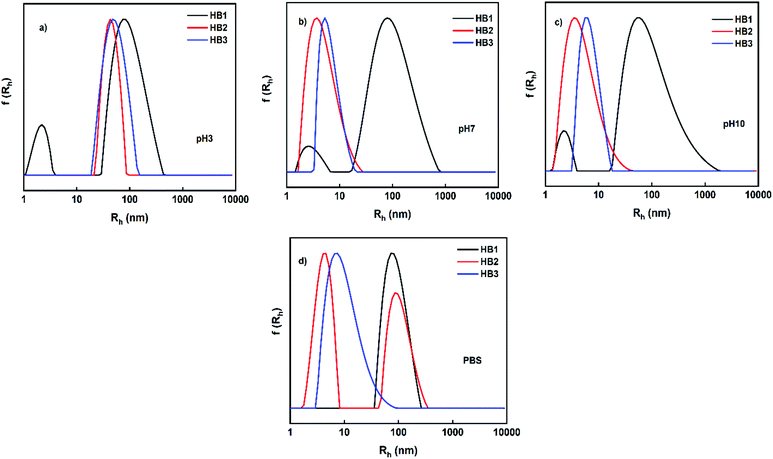 | ||
| Fig. 4 Size distributions from DLS for the HB1–HB3 hyperbranched copolymer solutions at different pHs. | ||
Dynamic light scattering experiments as a function of temperature were also performed for the HB1–HB3 copolymers due to the thermo-responsive character of DIPAEMA component.45 The results from Contin analysis are presented in Fig. 5. The measurements were recorded at pH 7 and in PBS solutions by changing solution temperature in the range 25–60 °C. Based on the results obtained at pH 7, it is observed that HB1 copolymer does not exhibit significant differences due to temperature increase, in contrast to the copolymers that have a higher content of DIPAEMA. In particular, HB2 and HB3 copolymers show the formation of larger aggregates at 60 °C, which demonstrates the increasing compactness of existing aggregates. In PBS solutions, the formation of more well-defined self-assembled nanoparticles is observed with narrower size distributions in contrast to pH 7. The presence of a small amount of salt in PBS solutions, together with the increase of temperature, seems to affect the structure of the aggregates in a different way compared to the solutions of similar pH which contain a lesser amount of salt. It is remarkable that HB3 copolymer solutions, which contains the highest content of hydrophobic DIPAEMA, exhibit large swollen particles with a significantly narrower size distribution.
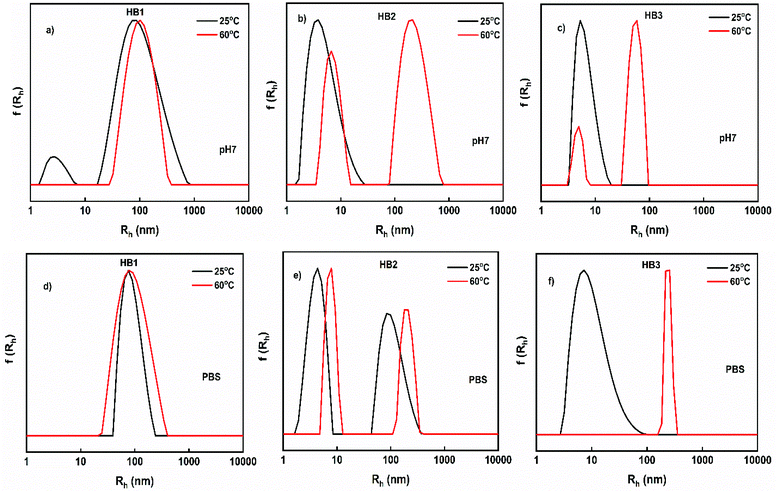 | ||
| Fig. 5 Size distributions from DLS for the HB1–HB3 hyperbranched copolymers solutions as a function of pH and temperature. | ||
The changes in scattering intensity (i.e. the changes in mass of the aggregates) as a function of pH and temperature are illustrated in Fig. 6. There is an increase in scattered intensity for all hyperbranched copolymers by increasing temperature, except for HB1 copolymer in PBS which shows a small decrease of scattered intensity with temperature. This observation agrees with the scenario that, due to DIPAEMA partial deprotonation at pH 7, the increase in temperature leads to enhanced hydrophobicity of the system and, therefore, to the formation of nanoparticles of constantly increasing mass. As for the hydrodynamic radius results, two populations of the aggregates are observed for the copolymers with larger DIPAEMA content. This observation is attributed to the formation of small (maybe unimolecular) species and larger multimolecular aggregates. In PBS solution and specifically for HB1 copolymer there is a relative decrease in the mass of aggregates by increasing temperature while no drastic changes are observed in terms of their dimensions. For HB2 copolymer solutions the mass of the polymeric nanoparticles increases by increasing temperature. However, there are two populations of species (most probably large multichain aggregates and isolated chains or aggregates with a lower number of chains) in the whole range of temperatures. This is followed by an increase of their dimensions. It is noteworthy that for HB3 in PBS (i.e. in the presence of a small amount of salt) the mass of the nanoparticles increases rapidly just from 40 °C in contrast to the other hyperbranched copolymers. The same is observed in terms of the dimensions of the nanoparticles which are five times larger than those at pH 7 (no salt added). These observations lead to the conclusion that the presence of salt contributes drastically to changes in the structure of the polymeric aggregates.
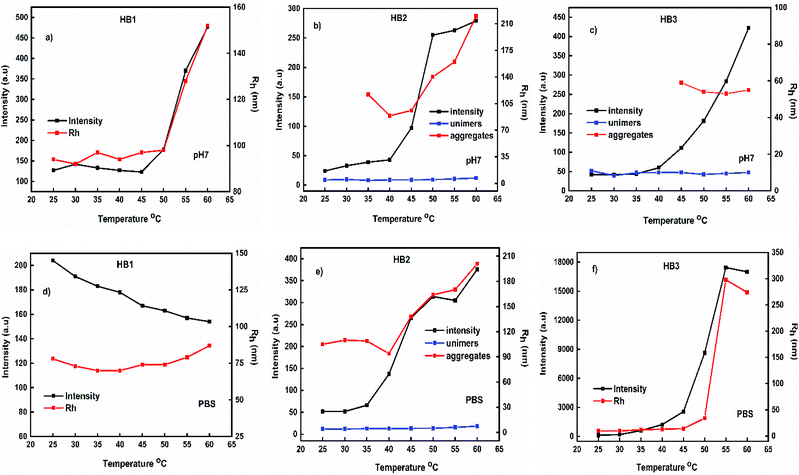 | ||
| Fig. 6 Variation of scattered intensity and Rh as a function of pH and temperature from light scattering experiments for the P(OEGMA-co-DIPAEMA) hyperbranched copolymers. | ||
A summary of the results on the self-organization of HB copolymers in aqueous media at different pHs and temperatures is presented in ESI (Table S2†). It is observed that HB2 and HB3 copolymers exhibit a low scattered intensity in most cases when the amino groups are partially or fully protonated, which is related to the small mass of the nanoparticles. Their relatively small dimensions indicate that the species in solution may be molecularly dissolved hyperbranched copolymers, as expected for hyperbranched amphiphilic structures.51
Also, ζ-potential measurements were performed to determine the surface charge and colloidal stability of the hyperbranched copolymers. Positive values of ζ-potential are observed at pH 3, as expected due to the protonation of DIPAEMA amino groups. At pH 7 and 10 where partial and full deprotonation of the amino groups takes place, respectively, negative ζ-potential values are obtained, which are rather attributed to the absorption of OH− anions at the particle surface and to the carboxyl groups coming from the chain transfer agent fragments attached to the macromolecules.
Effect of ionic strength on P(OEGMA-co-DIPAEMA) hyperbranched solution assembly
Taking into account that the hyperbranched copolymers are converted to hyperbranched polyelectrolytes at pH 3, light scattering measurements as a function of solution ionic strength were performed in order to detect possible changes in the structure of polymeric aggregates. Fig. 7 presents the variation of Rh and scattered light intensity as a function of NaCl concentration in copolymer aqueous solutions.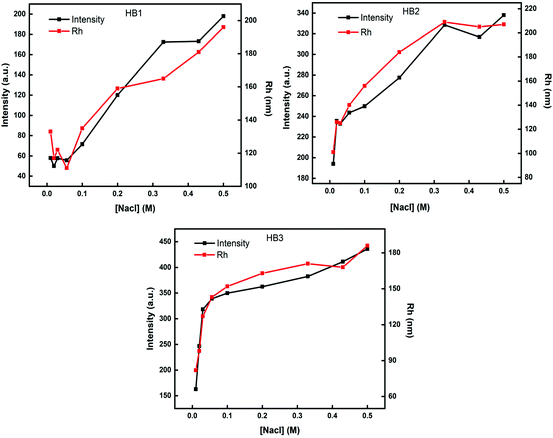 | ||
| Fig. 7 Light scattering measurements for P(OEGMA-co-DIPAEMA) hyperbranched copolymer solutions (c = 1 × 10−3 g mL−1) at pH 3 as a function of solution ionic strength. | ||
An increase in scattering intensity and hydrodynamic radius values occurs for all copolymers as ionic strength of the solution increases. In particular, by increasing the amount of hydrophobic component, a sharp increase in the mass and dimensions of the nanoparticles is observed by the first additions of NaCl. This observation is in agreement with the ζ-potential values which disclose the highly charged polymeric aggregates of HB2 and HB3 copolymers at this pH. The extended secondary aggregation of the initially formed species in solutions is attributed to electrostatic screening effects as salt concentration increases.52
Curcumin encapsulation in P(OEGMA-co-DIPAEMA) hyperbranched copolymers assemblies
In order to study the ability of P(OEGMA-co-DIPAEMA) hyperbranched copolymer aggregates to act as drug carriers, the model hydrophobic drug curcumin (CUR) was utilized. The properties of the mixed copolymer/drug nanoparticles were studied by light scattering and spectroscopy methods. Drug-loaded aggregates with 30% w/w targeted CUR encapsulation levels for HB1 and HB2 were prepared, according to the protocol described above. The final concentration of the copolymers was 1 × 10−3 g mL−1. Solutions of drug-loaded copolymer nanoassemblies were studied by DLS at 25 °C and 90° angle. UV-Vis and ATR-FTIR spectroscopy techniques were utilized to determine the successful encapsulation of curcumin in the aggregates of the copolymers, which are reported in the ESI.† Utilizing the constructed calibration curve of curcumin in THF the actual amount of entrapped drug could be determined as presented in Table 2. In Fig. 8, comparative size distributions of P(OEGMA-co-DIPAEMA) hyperbranched copolymers are presented, before and after curcumin encapsulation.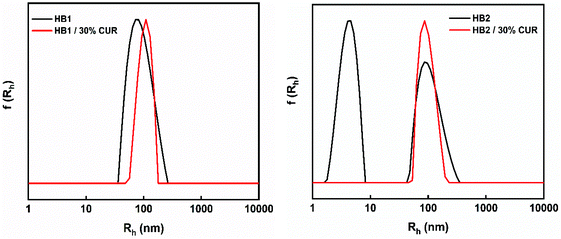 | ||
| Fig. 8 Comparison of size distributions from DLS for P(OEGMA-co-DIPAEMA) hyperbranched solutions before and after encapsulation of curcumin. | ||
From Fig. 8 it is clearly observed that by adding curcumin the size distributions become narrower. This means that the encapsulation of the drug enhanced the self-assembly of polymeric nanoparticles. Especially, for the HB2 copolymer the addition of curcumin results in the disappearance of small particles and only a sharp peak with nanoscale dimensions is present. This may declare that the interactions between copolymer and curcumin are prevailing and the drug was entrapped more efficiently in the polymeric aggregates. Presumably, preexisting free copolymer chains (or smaller aggregates) were incorporated to the mixed copolymer-drug nanoassemblies. It is worth noting that a significant increase in the mass of drug-loaded aggregates occurs (Table 2), which demonstrates the enhanced formation of mixed nanoparticles by addition of hydrophobic curcumin.
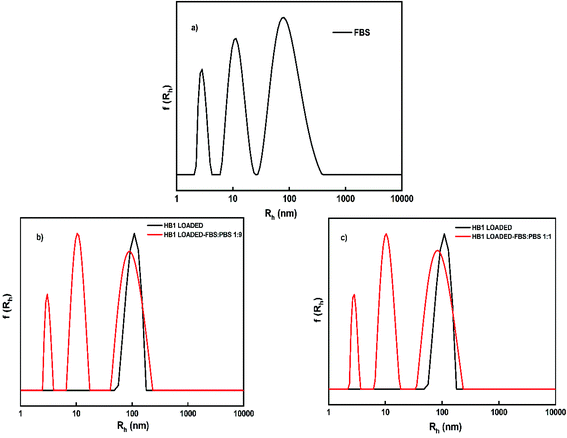
After entrapment of CUR in the aggregates of the HB1 and HB2 hyperbranched copolymers the stability of CUR-loaded nanoparticles in the presence of serum was studied by DLS measurements. DLS data were collected at 25 °C and at 90° angle after 1 h of adding the CUR-loaded nanoparticles into the FBS/PBS mixed solution. In Fig. 9, characteristic size distributions of CUR-loaded nanoparticles are presented before and after mixing with FBS/PBS media. The data on temporal stability studies are reported in ESI (Fig. S5†).
From the DLS data illustrated in Fig. 9 for the HB1 copolymer nanoparticles is evident that no further aggregation of the existing mixed copolymer-drug particles is taking place. Therefore, CUR-loaded aggregates exhibit substantial stability in the presence of serum proteins.
Based on our previous work53 and in general from the extensive literature that exists regarding the optical properties of curcumin, it has been proven that curcumin provides strong intrinsic fluorescence, a property that can be useful for bio-imaging applications.54 Therefore, fluorescence studies of CUR-loaded hyperbranched copolymers were performed under physiological conditions in PBS solutions and at pH 5.5 (the typical pH for tumor tissues). The measurements were contacted by UV-Vis and fluorescence techniques, in order to determine the optical properties of curcumin after its entrapment into the polymeric aggregates. UV-Vis studies are reported in ESI.† It is known the literature that the solubility of curcumin is very limited in water (4.2 μg mL−1).54,55 CUR solubility increases significantly by encapsulation in the hydrophobic domains of copolymer aggregates. CUR-loaded HB2 and HB1 nanoparticles exhibit a 2 and 7 times increase of curcumin solubility, respectively, compared to the case of pure water (real CUR concentrations are 9.7 and 29 μg mL−1, respectively). In contrast, in PBS solutions there is no increase of drug solubility for HB1 copolymer aggregates, whereas for HB2 this is 2 times greater (real CUR concentration for HB2 is 8.7 μg mL−1). The enhancement in solubility revealed at pH 5.5 compared to PBS solution may be related to the protonation of amino groups of DIPAEMA segments, as polymeric chains may acquire more extended conformations in aqueous media. After CUR entrapment, the loaded polymeric aggregates exhibit a significant fluorescence. In Fig. 10, the peak of curcumin at 489 nm in THF shifted to 505 nm (HB2) and 515 nm (HB1) in aqueous media at pH5.5, and in PBS solution the peak of curcumin shifted to 499 nm (HB2) and 503 nm (HB1). This shift is assumed to be due to the hydrophobic interactions that take place between the drug and the hydrophobic DIPAEMA component of the copolymer at these pH values.
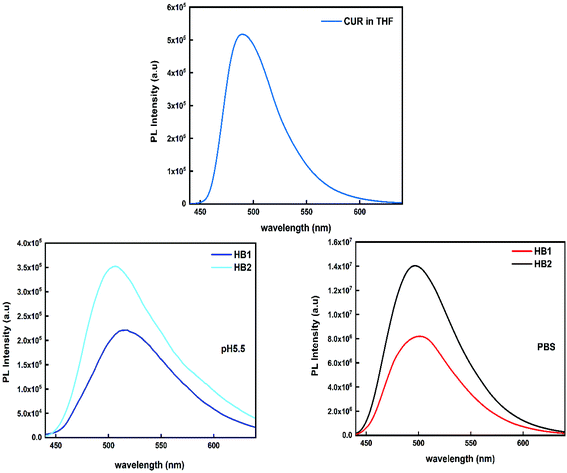 | ||
| Fig. 10 Upper: Fluorescence of CUR in THF solution (cCUR = 100 μg mL−1). Down: Fluorescence of CUR-loaded nanoparticles at pH 5.5 (left) and in PBS solution (right). | ||
Conclusions
Three novel, well-defined and multi-responsive P(OEGMA-co-DIPAEMA) hyperbranched copolymers, having different compositions of the potentially hydrophobic, pH and temperature responsive DIPAEMA component and the hydrophilic OEGMA component were synthesized by RAFT polymerization. EGDMA was utilized as the branching monomer to form hyperbranched copolymers. By using static light scattering it was possible to determine the molecular weights of all hyperbranched copolymers as evidence for the branched macromolecular structures, resulting from the synthetic procedures followed. Dynamic light scattering and fluorescence spectroscopy methods were used to determine the self-organization behavior of the copolymers in aqueous solution as a function of pH and temperature. From DLS results, it was observed that the copolymers form large aggregates in aqueous solutions, regardless of the chemical composition and temperature, when the amino groups of DIPAEMA are fully protonated at low pH. In contrast, when partial or total deprotonation of the amino groups takes place (at pH 7 and pH 10), nanoparticles of very small dimensions and mass appeared, while tending to increase in size and mass as solution temperature increases. Solution ionic strength also affects the structure of copolymers in aqueous solutions at pH 3 resulting in the formation of larger aggregates by increasing NaCl concentration. Neat and CUR-loaded polymeric aggregates present significant stability in the presence of serum. Furthermore, CUR-loaded nanoparticles exhibit significant fluorescence and, thus, they may show potential utilization in bio-imaging applications.Conflicts of interest
There are no conflicts to declare.Notes and references
- V. M. Garamus, T. V. Maksimova, H. Kautz, E. Barriau, H. Frey, U. Schlotterbeck, S. Mecking and W. Richtering, Macromolecules, 2004, 37, 8394–8399 CrossRef CAS.
- D. Wilms, S.-E. Stiriba and H. Frey, Acc. Chem. Res., 2010, 43, 129–141 CrossRef CAS PubMed.
- F. Wurm and H. Frey, in Polymer Science: A Comprehensive Reference, 10 Volume Set, Elsevier, 2012, pp. 177–198 Search PubMed.
- A. Hirao, Y. Tsunoda, A. Matsuo, K. Sugiyama and T. Watanabe, Macromol. Res., 2006, 14, 272–286 CrossRef CAS.
- D. Konkolewicz, M. J. Monteiro and S. Perrier, Macromolecules, 2011, 44, 7067–7087 CrossRef CAS.
- J. Li, J. Wang, Y. Zang, T. Aoki, T. Kaneko and M. Teraguchi, Chem. Lett., 2012, 41, 1462–1464 CrossRef CAS.
- I. N. Kurniasih, J. Keilitz and R. Haag, Chem. Soc. Rev., 2015, 44, 4145–4164 RSC.
- Y. H. Kim, J. Polym. Sci., Part A: Polym. Chem., 1998, 36, 1685–1698 CrossRef CAS.
- Y. Chen, L. Wang, H. Yu, Y. Zhao, R. Sun, G. Jing, J. Huang, H. Khalid, N. M. Abbasi and M. Akram, Prog. Polym. Sci., 2015, 45, 23–43 CrossRef CAS.
- D. Wilms, M. Schömer, F. Wurm, M. I. Hermanns, C. J. Kirkpatrick and H. Frey, Macromol. Rapid Commun., 2010, 31, 1811–1815 CrossRef CAS PubMed.
- C. Ma, S. Qiu, B. Yu, J. Wang, C. Wang, W. Zeng and Y. Hu, Chem. Eng. J., 2017, 322, 618–631 CrossRef CAS.
- M. Irfan and M. Seiler, Ind. Eng. Chem. Res., 2010, 49, 1169–1196 CrossRef CAS.
- Y. Segawa, T. Higashihara and M. Ueda, Polym. Chem., 2013, 4, 1746–1759 RSC.
- D. Zhang, D. Jia and Z. Zhou, Macromol. Res., 2009, 17, 289–295 CrossRef CAS.
- Y. Zhou, W. Huang, J. Liu, X. Zhu and D. Yan, Adv. Mater., 2010, 22, 4567–4590 CrossRef CAS PubMed.
- H. Jin, W. Huang, X. Zhu, Y. Zhou and D. Yan, Chem. Soc. Rev., 2012, 41, 5986–5997 RSC.
- S. Chen, X.-Z. Zhang, S.-X. Cheng, R.-X. Zhuo and Z.-W. Gu, Biomacromolecules, 2008, 9, 2578–2585 CrossRef CAS PubMed.
- Y. Deng, J. K. Saucier-Sawyer, C. J. Hoimes, J. Zhang, Y.-E. Seo, J. W. Andrejecsk and W. M. Saltzman, Biomaterials, 2014, 35, 6595–6602 CrossRef CAS PubMed.
- Q.-F. Zhang, Q.-Y. Yu, Y. Geng, J. Zhang, W.-X. Wu, G. Wang, Z. Gu and X.-Q. Yu, ACS Appl. Mater. Interfaces, 2014, 6, 15733–15742 CrossRef CAS PubMed.
- Q. Mou, Y. Ma, X. Jin and X. Zhu, Mol. Syst. Des. Eng., 2016, 1, 25–39 RSC.
- Q. Zhu, F. Qiu, B. Zhu and X. Zhu, RSC Adv., 2013, 3, 2071–2083 RSC.
- M. Jafari, S. S. Abolmaali, H. Najafi and A. M. Tamaddon, Int. J. Pharm., 2020, 576, 118959 CrossRef CAS PubMed.
- S. Wang, S. Tateyama, D. Kaneko, S.-y. Ohki and T. Kaneko, Polym. Degrad. Stab., 2011, 96, 2048–2054 CrossRef CAS.
- Y. Shi, R. W. Graff, X. Cao, X. Wang and H. Gao, Angew. Chem., 2015, 127, 7741–7745 CrossRef.
- D. Wang, T. Zhao, X. Zhu, D. Yan and W. Wang, Chem. Soc. Rev., 2015, 44, 4023–4071 RSC.
- Q. Zhu, J. Wu, C. Tu, Y. Shi, L. He, R. Wang, X. Zhu and D. Yan, J. Phys. Chem. B, 2009, 113, 5777–5780 CrossRef CAS PubMed.
- C. Liu, Y.-y. Fei, H.-l. Zhang, C.-y. Pan and C.-y. Hong, Macromolecules, 2018, 52, 176–184 CrossRef.
- T. Cuneo, X. Wang, Y. Shi and H. Gao, Macromol. Chem. Phys., 2020, 221, 2000008 CrossRef CAS.
- X. Wang and H. Gao, Polymers, 2017, 9, 188 CrossRef PubMed.
- T. Cuneo and H. Gao, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2020, 12, e1640 Search PubMed.
- C. Zhang, Y. Zhou, Q. Liu, S. Li, S. Perrier and Y. Zhao, Macromolecules, 2011, 44, 2034–2049 CrossRef CAS.
- J. Xu, L. Tao, J. Liu, V. Bulmus and T. P. Davis, Macromolecules, 2009, 42, 6893–6901 CrossRef CAS.
- M. Eberhardt and P. Théato, Macromol. Rapid Commun., 2005, 26, 1488–1493 CrossRef CAS.
- J. Zeng, K. Shi, Y. Zhang, X. Sun, L. Deng, X. Guo, Z. Du and B. Zhang, J. Colloid Interface Sci., 2008, 322, 654–659 CrossRef CAS PubMed.
- A. Skandalis and S. Pispas, J. Polym. Sci., Part A: Polym. Chem., 2019, 57, 1771–1783 CrossRef CAS.
- R. T. Mayadunne, J. Jeffery, G. Moad and E. Rizzardo, Macromolecules, 2003, 36, 1505–1513 CrossRef CAS.
- A. P. Vogt, S. R. Gondi and B. S. Sumerlin, Aust. J. Chem., 2007, 60, 396–399 CrossRef CAS.
- P. R. Bachler, K. E. Forry, C. A. Sparks, M. D. Schulz, K. B. Wagener and B. S. Sumerlin, Polym. Chem., 2016, 7, 4155–4159 RSC.
- X. Xu, A. E. Smith, S. E. Kirkland and C. L. McCormick, Macromolecules, 2008, 41, 8429–8435 CrossRef CAS.
- H. Tai, C. L. Duvall, A. S. Hoffman, P. S. Stayton and W. Wang, Macromol. Mater. Eng., 2012, 297, 1175–1183 CrossRef CAS.
- A. Ramírez-Jiménez, K. A. Montoya-Villegas, A. Licea-Claverie and M. A. Gónzalez-Ayón, Polymers, 2019, 11, 1657 CrossRef PubMed.
- Y. Wang, Y. Kotsuchibashi, Y. Liu and R. Narain, Langmuir, 2014, 30, 2360–2368 CrossRef CAS PubMed.
- B. Liu, A. Kazlauciunas, J. T. Guthrie and S. Perrier, Macromolecules, 2005, 38, 2131–2136 CrossRef CAS.
- Y. Q. Hu, M. S. Kim, B. S. Kim and D. S. Lee, Polymer, 2007, 48, 3437–3443 CrossRef CAS.
- T. Thavanesan, C. Herbert and F. A. Plamper, Langmuir, 2014, 30, 5609–5619 CrossRef CAS PubMed.
- M. Licciardi, Y. Tang, N. Billingham, S. Armes and A. Lewis, Biomacromolecules, 2005, 6, 1085–1096 CrossRef CAS PubMed.
- M. Kafetzi and S. Pispas, Macromol. Chem. Phys., 2021, 222, 2000358 CrossRef CAS.
- J. Coupris, S. Pascual, L. Fontaine, T. Lequeux and T. N. Pham, Polym. Chem., 2015, 6, 4597–4604 RSC.
- A. Skandalis and S. Pispas, Polym. Chem., 2017, 8, 4538–4547 RSC.
- D. Selianitis and S. Pispas, J. Polym. Sci., 2020, 58, 1867–1880 CrossRef CAS.
- L. Chen, J. D. Simpson, A. V. Fuchs, B. E. Rolfe and K. J. Thurecht, Mol. Pharm., 2017, 14, 4485–4497 CrossRef CAS PubMed.
- T. Sentoukas and S. Pispas, J. Polym. Sci., Part A: Polym. Chem., 2018, 56, 1962–1977 CrossRef CAS.
- D. Selianitis and S. Pispas, Polym. Int., 2021, 70, 1508–1522 CrossRef CAS.
- M. Liu, C. P. Teng, K. Y. Win, Y. Chen, X. Zhang, D. P. Yang, Z. Li and E. Ye, Macromol. Rapid Commun., 2019, 40, 1800216 CrossRef PubMed.
- H. N. Nguyen, P. T. Ha, A. Sao Nguyen, D. T. Nguyen, H. D. Do, Q. N. Thi and M. N. H. Thi, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2016, 7, 025001 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1py01320c |
| This journal is © The Royal Society of Chemistry 2021 |