Open Access Article
Open Access ArticleInhibition of off-target cleavage by RNase H using an artificial cationic oligosaccharide†
Rintaro Iwata
Hara
 *ab and
Takeshi
Wada
*ab and
Takeshi
Wada
 *a
*a
aDepartment of Medicinal and Life Sciences, Faculty of Pharmaceutical Sciences, Tokyo University of Science, 2641 Yamazaki, Noda, Chiba 278-8510, Japan. E-mail: twada@rs.tus.ac.jp
bDepartment of Neurology and Neurological Science, Graduate School of Medical and Dental Sciences, Tokyo Medical and Dental University, 1-5-45, Yushima, Bunkyo-Ku, Tokyo, 113-8519, Japan. E-mail: haranuro@tmd.ac.jp
First published on 23rd July 2021
Abstract
Sequence-dependent off-target effects are a serious problem of antisense oligonucleotide-based drugs. Some of these side effects are induced by ribonuclease H (RNase H)-mediated cleavage of non-target RNAs with base sequences similar to that of the target RNA. We found that an artificial cationic oligosaccharide, ODAGal4, improved single-base discrimination for RNase H cleavage.
Introduction
The development of nucleic acid drugs, which are composed of nucleic acid molecules, such as DNA and RNA, has been advancing over the past decade. Antisense oligonucleotides (ASOs) are currently the most advanced type of nucleic acid drug, with an increasing number of ASO drugs being developed for clinical use.1 ASOs are classified into two types, ribonuclease H-dependent ASOs (RNase H-dependent ASOs) and RNase H-independent ASOs, and appropriate chemical modifications are necessary for each type of ASO.2–5 RNase H is an endogenous nuclease that recognizes DNA/RNA hybrid duplexes to cleave their RNA strand. RNase H-dependent ASOs are at least partly composed of a consecutive DNA region.6–9 They bind to their target RNA by Watson–Crick base pairing to form an ASO/RNA duplex, and the DNA/RNA structure in the ASO/RNA duplex is recognized by RNase H to cleave the target RNA. RNase H-independent ASOs also bind to their target by Watson–Crick base pairing, but they work without the RNase H-mediated cleavage of RNA.2Sequence-dependent off-target effects are a serious problem of ASO-based drugs,10 in which non-target RNAs with base sequences similar to that of the target RNA are downregulated and/or functionally blocked. To reduce these side-effects, several effective approaches have been adopted including the screening of good target sequences, the use of longer ASOs, and chemical modifications of ASOs.10 However, it remains still difficult to abolish completely the risk of side effects of these drugs. Therefore, the development of other technologies aimed at controlling such side effects is required.
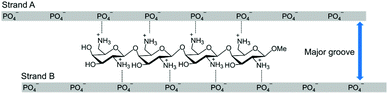
DNA/RNA hybrid duplexes mainly form an A-type duplex structure that is different from B-type DNA/DNA duplexes. The characteristics of the A-type duplexes include a narrow and deep major groove and a wide and shallow minor groove.11 We have developed artificial cationic oligosaccharides and oligopeptides that can bind within the major groove of the A-type nucleic acid duplexes by interacting with the phosphate groups of both strands of the duplexes (Fig. 1).12–18 We have also reported that, by binding to the duplexes, these cationic oligosaccharides increase the thermal stability of the duplexes and the enzymatic stability against RNase A without disturbing RNase H-mediated cleavage.12,16 Conversely, we have not studied the effect of these cationic molecules on the RNase H-mediated cleavage of non-target RNAs and the mismatch discrimination abilities of ASOs. Herein, we aimed to study these parameters by using diaminogalactose 4mer (ODAGal4, Fig. 1),12,18 which is an artificial cationic oligosaccharide with excellent affinity to DNA/RNA hybrid duplexes.
Results and discussion
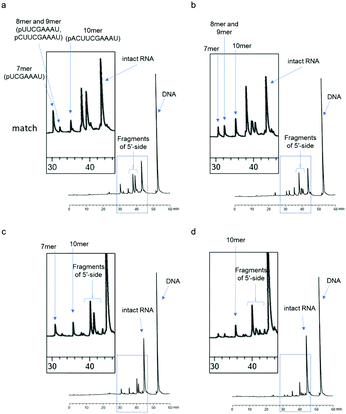
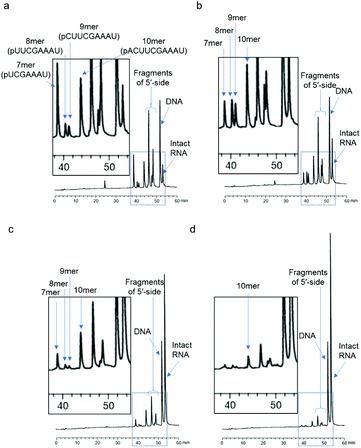
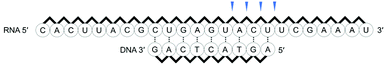
First, we used a 24mer RNA, CACUUACGCUGAGUACUUCGAAAU (R1, capital letters indicate RNA in this paper), as the target RNA, based on the firefly luciferase gene (GenBank accession no.: U47296) and a previous siRNA study.19 A short 9mer DNA, agtactcag (D1, small letters indicate DNA in this paper), was chosen to simplify the analysis of cleaved fragments and the study of ODAGal4 function. Equal amounts of the DNA and RNA were mixed and the mixture was treated with E. coli RNase H at 37 °C for 5 min. RNase H cleavage affords 3′-hydroxy and 5′-phosphate fragments.20 After the RNase H treatment, 86% of R1 was cleaved and 7mer–10mer complexes of 5′-phosphate RNA fragments (p-RNAs), pUCGAAAU (7mer) to pACUUCGAAAU (10mer), were detected by reverse-phase HPLC (RP-HPLC) (Fig. 2(a)). Therefore, the cleavage sites at the RNA were identified as shown in Fig. 3. Next, the same experiments were conducted in the presence of an equal amount of ODAGal4 to the DNA and RNA oligomers. While a similar amount of the RNA was also cleaved (76%, Fig. 2(b)), the amount of each fragment was different. Compared with the results obtained in the absence of ODAGal4, while the amount of 7mer p-RNA was decreased, the amounts of 8mer p-RNA and 9mer p-RNA were increased. These results can be explained by assuming that ODAGal4 bound to the 5′-terminal region of the DNA strand (Fig. S1†). On the other hand, although we tried to confirm the binding preference of ODAGal4 by UV melting (Tm) analysis of hybrid duplexes 9mer DNA and 24mer RNA, changes of UV absorbance derived from single stranded RNAs disturbed the analysis (Fig. S182†). Therefore, we used 12mer/12mer hybrid duplexes as a model sequence in Tm experiments. But there was little difference in ΔTm values with ODAGal4 due to the position of the mismatch base (Fig. S183 and Table S1†) in this case.Next, we used a single-base mismatch RNA oligomer, CACUUACGUUGAGUACUUCGAAAU (R2), in which a cytidine was replaced by a uridine (the bold character indicates the mismatch base). In the absence of ODAGal4, similar types of fragments to those obtained using matched RNA were detected in different amounts. Conversely, in this case, the amount of the four fragments was smaller in the presence of ODAGal4; consequently, the cleaved amount of RNAs was markedly decreased (31% to 12% in Fig. 2(c) and (d)). These results suggested that the binding region of ODAGal4 was not the same between the scenarios of the matched RNA and these mismatch RNA oligomers.
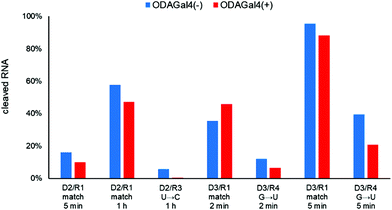
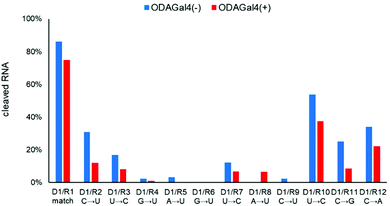
Next, we conducted the same experiments using D1 and 24mer RNA oligomers, which have other single-mismatch nucleobases. For the RNA oligomers, a U was replaced by a C or one of the other three types of nucleosides was replaced by a U from the matched RNA R1 (Fig. 4). In most cases, the addition of ODAGal4 reduced the amount of cleaved RNAs with a single-base mismatch sequence, with the exception of R8, in which RNA cleavage was slightly increased by ODAGal4. Among the results, the mismatch discrimination was found to be improved especially for the RNA oligomers with a mismatch base near the 5′-terminal (R2 and R3 in Fig. 4, 2(c) and (d), and Fig. S8–S10†). We conducted further experiments using RNA oligomers with a guanosine or an adenosine as a mismatch base (R11 and R12). Although the RNase H cleavage of the RNA with a G in place of a C was efficiently inhibited by the addition of ODAGal4 (R11, 25% to 8%), that of the RNA with an A in place of a C was inhibited only moderately by ODAGal4 (R12, 39% to 28%). These results suggest that the type of mismatch base also affects the inhibitory properties of ODAGal4 against RNase H-mediated cleavage of DNA/RNA hybrid duplexes with a mismatch base pair.
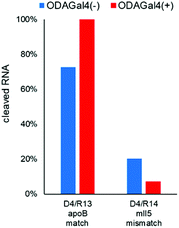
Next, we performed RNase H cleavage experiments using other DNA 9mers (Fig. 5 and 6). In Fig. 5, we used R1 as the matched RNA and two types of other 9mer DNA oligomers (D2 and D3), both of which bound to a slightly different region from D1. In both cases, the addition of ODAGal4 improved the mismatch discrimination in RNase H cleavage, similar to the results obtained for D1. As another example, we evaluated the mismatch discrimination between the RNA oligomer R13 (as the matched RNA), based on the mouse apoB mRNA (GenBank accession no.: NM_009693), and the RNA oligomer R14, the sequence of which was based on the mouse mll5 mRNA (GenBank accession no.: NM_026984), as the mismatch RNA (Fig. 6). We used a 9mer DNA, attggtatt, that was shown to be present within the apoB-targeted ASO oligonucleotides in previous studies.21,22 Mll5 knockdown was also observed as an off-target effect in the previous studies. In this case, the cleavage of the mll5-derived RNA oligomer was reduced and the cleavage of the apoB-derived RNA oligomer was enhanced by the addition of ODAGal4. It was also interesting to observe that the cleavage site in R13 seemed to be at a single position only in the presence of ODAGal4 (Fig. S57–S62†). In summary, the mismatch discrimination of RNase H cleavage was improved in multiple types of 9mer DNAs. Throughout the results shown in Fig. 4–6, it could be a drawback that there were some cases where the cleavage of matched RNAs was also decreased. We do not have a clear understanding of which types of sequences tend to result in the decrease or increase of the cleavage of matched RNAs at present.
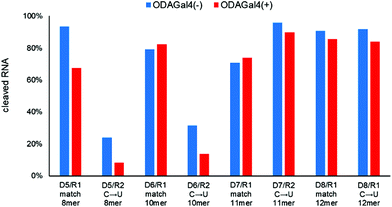
If we use a longer DNA oligomer in the RNase H cleavage experiments, the number of cleavage sites in both match and mismatch RNAs should increase and the variety of ODAGal4 binding sites is also expected to augment; consequently, mismatch discrimination improvement by ODAGal4 is expected to be less effective in this case. To confirm this, we conducted experiments using DNA oligomers with four different lengths, i.e., 8mer, 10mer, 11mer, and 12mer (Fig. 7). In the case of the 8mer DNA (D5), the cleavage of the mismatch RNA was decreased (24% to 8%), and that of the matched RNA was also reduced (93 to 68%). The 10mer DNA (D6) yielded similar results to the case of 9mer, and the mismatch discrimination for RNase H cleavage was markedly improved by the addition of ODAGal4. Conversely, in the cases of the 11mer and 12mer DNA oligomers (D7 and D8), the RNase H cleavage of both match and mismatch RNAs was not inhibited. These results showed that ODAGal4 could improve the mismatch discrimination of RNase H cleavage only in the cases of the 9mer and 10mer DNA oligomers; however, the effect sharply declined when using the 11mer or longer DNA oligomers. These findings are problematic for therapeutic applications, because the 10mer is obviously too short to be used as an ASO drug.
As mentioned above, ODAGal4 was not effective in improving mismatch discrimination in RNase H cleavage when we used 11mer or longer DNA oligomers, and the cause of the problem is at least partly that both RNase H recognition sites and ODAGal4 binding sites can be too many. To tackle this problem, we first used gapmers, which have a consecutive DNA region flanked by modified RNAs, to limit RNase H cleavage sites in longer oligonucleotides. We conducted an RNase H assay using the 13mer apoB LNA/DNA gapmer and 27mer RNAs of match and mismatch sequences to the gapmer. In this case, however, no effective inhibition of off-target cleavage by RNase H was observed using ODAGal4 (Fig. S169–S181†). We considered that the failure resulted because there are still many ODAGal4 binding sites, while RNase H recognition sites are limited by using the gapmer.
Next, we combined ODAGal4 technology with charge-neutral methylphosphonate (MP) backbone modification.23,24 An MP linkage can avoid RNase H recognition, and its partial introduction into a DNA oligomer can limit the RNase H recognition sites of the oligomer.25–27 In addition, the neutral type of modification is expected to be also less preferable for binding of ODAGal4, because ODAGal4 is considered to bind to DNA/RNA hybrid duplexes via electrostatic interactions between phosphate groups and amino groups. Therefore, it was expected to limit not only RNase H recognition but also ODAGal4 binding by partial introduction of an MP linkage to a DNA oligomer.
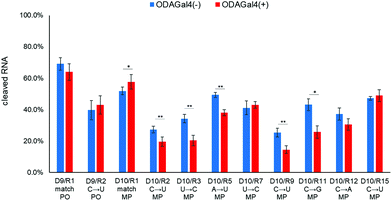
Based on these hypotheses, we designed a 12mer DNA containing a single MP linkage “agtactcagMPcgt” (D10; MP indicates a methylphosphonate linkage), in which the conventional 9mer DNA, D1, and another 3mer DNA (cgt) were linked via an MP linkage. Because of this linkage, both RNase H and ODAGal4 are expected to bind mainly to a region of phosphate groups of the conventional 9mer structure. HPLC profiles obtained after the RNase H treatment of R1 or R2 with D10 are shown in Fig. 8. In the case of R1, it yielded similar results to those of the mixture of D1 and R1 (Fig. 8a and b); while the amount of the 7mer fragment was decreased, the amount of the 8mer and 9mer was increased by the addition of ODAGal4 (Fig. 8a and b). Conversely, in the case of R2, the amount of 7mer was decreased without an increase in any of the other fragments (Fig. 8c and d). As a result, while the total amount of cleaved R1 was slightly increased, the cleavages of R2 were inhibited by the addition of ODAGal4 (Fig. 9). On the other hand, when using an unmodified 12mer DNA (D9: agtactcagcgt) instead of D10, the cleavage amounts of both R1 and R2 were not significantly changed by ODAGal4 (Fig. 9 and Fig. S87–S102†). In addition, Fig. 9 also shows the results of the RNase H cleavage experiments using D10 and a variety of mismatch RNA 24mers. The cleavage of mismatch RNAs was significantly lower in the presence of ODAGal4 than that in the absence of ODAGal4, not only in the case of R2, but also for R3, R5, R9, and R11. Conversely, as expected, ODAGal4 did not inhibit the cleavage of R15, in which the mismatch base pair is located in the region of the additional 3mer DNA in the D10/R15 hybrid duplex. Collectively, ODAGal4 enhanced the mismatch discrimination of the 12mer DNA in RNase H cleavage in a wide variety of mismatch RNA 24mers with mismatch base pairs located within the region of the conventional 9mer DNA.
Conclusions
We proposed a novel approach to improve mismatch discrimination for E. coli RNase H cleavage using ODAGal4 combined with a neutral P-modification of oligonucleotides. ODAGal4 selectively inhibited the RNase H cleavage of mismatch RNA oligomers when using 9mer and 10mer DNAs. While there are several studies about improving mismatch discrimination using an appropriate design of oligonucleotides themselves,7,28–30 this study showed the possibility of improving mismatch discrimination by using an additive molecule. In addition, by combining this technology with a single methylphosphonate modification in a DNA strand, the range of application of ODAGal4 was extended to longer oligomers. Although D10 itself, with many unmodified phosphate linkages, must be too unstable to be used as therapeutics, our results will be useful for showing the novel concept of reducing sequence-dependent off-target effects of ASO-based drugs.Experimental section
General information
All DNA and RNA oligomers were purchased from Japan Bio Services Co., Ltd. E. coli RNase H was purchased from Takara Bio Inc. A PROTEOSAVE™ SS 0.5 mL Microtube (Sumitomo Bakelite Co., Ltd) was used as the reaction container for the RNase H assay. RP-HPLC was performed using a μBondasphere column (5 μm, C18, 100 Å, 3.9 × 150 mm) (Waters).RNase H assay
A DNA oligomer (5 μM) and an RNA oligomer (5 μM) were dissolved in 10 mM Tris-HCl buffer containing 102 mM NaCl and 0.51 mM MgCl2 (pH 7.2 at 37 °C). To 40 μL of the solution, water (2 μL) or 0.1 mM ODAGal4 solution (2 μL) was added. Subsequently, RNase H (E. coli RNase H, 5 U in 8 μL of 10 mM Tris-HCl buffer containing 100 mM NaCl and 0.5 mM MgCl2) was added at 37 °C; i.e., the final concentrations of the DNA, RNA, and ODAGal4 were 4 μM (or 0 μM for ODAGal4) in 10 mM Tris-HCl buffer containing 100 mM NaCl and 0.5 mM MgCl2. After 2 min, 5 min, or 1 h, the mixture was rapidly heated to 95 °C, left for 3 min, and then rapidly cooled to 4 °C. The mixture was analyzed by RP-HPLC. Each cleavage amount of the intact RNA was calculated by comparing the peak area ratio of the DNA peak and the remaining intact RNA peak to the DNA and RNA peaks of each corresponding RNase H-untreated sample.The RNase H assay for gapmers was also conducted under the same conditions except for lower final concentrations of RNase H, oligonucleotides, and ODAGal4; RNase H: 1 U/200 μL; gapmer: 1 μM; RNA: 1 μM; and ODAGal4: 0 or 1 μM.
Author contributions
R. I. H. and T. W. established the research plans, R. I. H. conducted the experiments, and both wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by JST-CREST (JPMJCR12L4), JSPS KAKENHI (19K06993), the Takeda Science Foundation, and the Tokyo Medical and Dental University Presidential Young Investigators Award.Notes and references
- S. T. Crooke, J. L. Witztum, C. F. Bennett and B. F. Baker, Cell Metab., 2018, 27, 714–739 CrossRef CAS PubMed.
- J. Kurreck, Eur. J. Biochem., 2003, 270, 1628–1644 CrossRef CAS PubMed.
- A. M. Quemener, L. Bachelot, A. Forestier, E. Donnou-Fournet, D. Gilot and M. Galibert, Wiley Interdiscip. Rev.: RNA, 2020, 11, e1594 Search PubMed.
- T. C. Roberts, R. Langer and M. J. A. Wood, Nat. Rev. Drug Discovery, 2020, 19, 673–694 CrossRef CAS PubMed.
- L. K. McKenzie, R. El-Khoury, J. D. Thorpe, M. J. Damha and M. Hollenstein, Chem. Soc. Rev., 2021, 50, 5126–5164 RSC.
- H. Donis-Keller, Nucleic Acids Res., 1979, 7, 179–192 CrossRef CAS.
- H. Inoue, Y. Hayase, S. Iwai and E. Ohtsuka, FEBS Lett., 1987, 215, 327–330 CrossRef CAS.
- M. Nowotny, S. A. Gaidamakov, R. J. Crouch and W. Yang, Cell, 2005, 121, 1005–1016 CrossRef CAS PubMed.
- M. Nowotny, S. A. Gaidamakov, R. Ghirlando, S. M. Cerritelli, R. J. Crouch and W. Yang, Mol. Cell, 2007, 28, 264–276 CrossRef CAS.
- P. J. Kamola, J. D. A. Kitson, G. Turner, K. Maratou, S. Eriksson, A. Panjwani, L. C. Warnock, G. A. D. Guilloux, K. Moores, E. L. Koppe, W. E. Wixted, P. A. Wilson, N. J. Gooderham, T. W. Gant, K. L. Clark, S. A. Hughes, M. R. Edbrooke and J. D. Parry, Nucleic Acids Res., 2015, 43, 8638–8650 CrossRef CAS PubMed.
- T. E. Cheatham and P. A. Kollman, J. Am. Chem. Soc., 1997, 119, 4805–4825 CrossRef CAS.
- R. I. Hara, Y. Hisada, Y. Maeda, T. Yokota and T. Wada, Sci. Rep., 2018, 8, 4323 CrossRef PubMed.
- R. Iwata, A. Doi, Y. Maeda and T. Wada, Org. Biomol. Chem., 2015, 13, 9504–9515 RSC.
- R. Iwata, M. Sudo, K. Nagafuji and T. Wada, J. Org. Chem., 2011, 76, 5895–5906 CrossRef CAS PubMed.
- Y. Maeda, R. I. Hara, K. Nishina, K. Yoshida-Tanaka, T. Sakamoto, T. Yokota and T. Wada, Nucleosides, Nucleotides Nucleic Acids, 2019, 1–11 Search PubMed.
- R. I. Hara, Y. Maeda, H. Fujimaki and T. Wada, Chem. Commun., 2018, 54, 8526–8529 RSC.
- Y. Maeda, R. Iwata and T. Wada, Bioorg. Med. Chem., 2013, 21, 1717–1723 CrossRef CAS PubMed.
- R. I. Hara, Y. Maeda, T. Sakamoto and T. Wada, Org. Biomol. Chem., 2017, 15, 1710–1717 RSC.
- S. M. Elbashir, J. Harborth, W. Lendeckel, A. Yalcin, K. Weber and T. Tuschl, Nature, 2001, 411, 494–498 CrossRef CAS PubMed.
- A. Krakowiak, A. Owczarek, M. Koziołkiewicz and W. J. Stec, ChemBioChem, 2002, 3, 1242–1250 CrossRef CAS PubMed.
- E. M. Straarup, N. Fisker, M. Hedtjärn, M. W. Lindholm, C. Rosenbohm, V. Aarup, H. F. Hansen, H. Ørum, J. B. R. Hansen and T. Koch, Nucleic Acids Res., 2010, 38, 7100–7111 CrossRef CAS PubMed.
- K. Nishina, W. Piao, K. Yoshida-Tanaka, Y. Sujino, T. Nishina, T. Yamamoto, K. Nitta, K. Yoshioka, H. Kuwahara, H. Yasuhara, T. Baba, F. Ono, K. Miyata, K. Miyake, P. P. Seth, A. Low, M. Yoshida, C. F. Bennett, K. Kataoka, H. Mizusawa, S. Obika and T. Yokota, Nat. Commun., 2015, 6, 7969 CrossRef CAS PubMed.
- S. Agrawal and J. Goodchild, Tetrahedron Lett., 1987, 28, 3539–3542 CrossRef CAS.
- P. S. Miller, J. Yano, E. Yano, C. Carroll, K. Jayaraman and P. O. P. Ts'o, Biochemistry, 1979, 18, 5134–5143 CrossRef CAS PubMed.
- M. T. Migawa, W. Shen, W. B. Wan, G. Vasquez, M. E. Oestergaard, A. Low, C. L. De Hoyos, R. Gupta, S. Murray, M. Tanowitz, M. Bell, J. G. Nichols, H. Gaus, X. Liang, E. E. Swayze, S. T. Crooke and P. P. Seth, Nucleic Acids Res., 2019, 47, 5465–5479 CrossRef CAS PubMed.
- P. J. Furdon, Z. Dominski and R. Kole, Nucleic Acids Res., 1989, 17, 9193–9204 CrossRef CAS PubMed.
- S. Agrawal, S. H. Mayrand, P. C. Zamecnik and T. Pederson, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 1401–1405 CrossRef CAS PubMed.
- D. Magner, E. Biala, J. Lisowiec-Wachnicka and R. Kierzek, Sci. Rep., 2017, 7, 12532 CrossRef.
- T. A. Vickers and S. T. Crooke, PLoS One, 2014, 9, e108625 CrossRef PubMed.
- W. F. Lima and S. T. Crooke, J. Biol. Chem., 1997, 272, 27513–27516 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ob00983d |
| This journal is © The Royal Society of Chemistry 2021 |

![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif)
![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)