Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis, structure and midkine binding of chondroitin sulfate oligosaccharide analogues†
Myriam
Torres-Rico
 ,
Susana
Maza
,
José L.
de Paz
,
Susana
Maza
,
José L.
de Paz
 * and
Pedro M.
Nieto
* and
Pedro M.
Nieto
 *
*
Glycosystems Laboratory, Instituto de Investigaciones Químicas (IIQ), cicCartuja, CSIC and Universidad de Sevilla, Americo Vespucio, 49, 41092 Sevilla, Spain. E-mail: pedro.nieto@iiq.csic.es; jlpaz@iiq.csic.es
First published on 25th May 2021
Abstract
The preparation of chondroitin sulfate (CS) oligosaccharide mimetics, more easily synthesized than natural sequences, is a highly interesting task because these compounds pave the way for modulation of the biological processes in which CS is involved. Herein, we report the synthesis of CS type E analogues which present easily accessible glucose units instead of glucuronic acid (GlcA) moieties. NMR experiments and molecular dynamics simulations showed that the 3D structure of these compounds is similar to the structure of the natural CS-E oligosaccharides. In addition, fluorescence polarization (FP) and saturation transfer difference NMR (STD-NMR) experiments revealed that the synthesized CS-like derivatives were able to interact with midkine, a model heparin-binding growth factor, suggesting that the presence of the GlcA carboxylate groups is not essential for the binding. Overall, our results indicate that the synthesized glucose-containing oligosaccharides can be considered as functional and structural CS mimetics.
Introduction
Chondroitin sulfate (CS) is a structurally heterogeneous polysaccharide belonging to the glycosaminoglycan (GAG) family that participates in a wide range of biological processes.1 Chondroitin sulfate E (CS-E) is a particular type of this polysaccharide, characterized by the disulfated disaccharide unit GalNAc(4,6-di-OSO3)-β(1 → 4)-GlcA-β(1 → 3). Although CS-E sequences constitute a minor part of naturally existing CS, this sulfation motif plays a relevant role in central nervous system development, growth factor signaling and pathogen attachment.2 For example, it has been demonstrated that CS-E tetrasaccharides specifically recognize certain proteins, modulating neuronal growth.3,4 One of these proteins is midkine, a heparin binding growth factor that is also involved in cancer and inflammation.5,6Structurally defined CS oligosaccharides are required to establish structure–activity relationships and study CS-protein interactions at the molecular level. Thus, several research groups have reported the chemical synthesis of CS oligomers,7,8 including CS-E domains.9–14 However, despite recent impressive advances,15 the preparation of CS sequences is still a complicated task. The synthesis of well-defined CS analogues, which mimic the natural CS sequences and are more easily synthesized, is an attractive alternative. Reported CS/GAG mimetics16 include multivalent systems,17–20 sulfated carbohydrate derivatives21–26 and non-carbohydrate sulfated compounds.27–29 These mimetics could potentially modulate the CS-protein interactions that are responsible for critical biological processes. In this context, Mallet and coworkers published in 2013 the synthesis of CS-E oligosaccharide analogues where the galactosamine units were replaced by galactose building blocks.30 This modification facilitated the access to these compounds because lactose was employed as starting material, avoiding the use of expensive galactosamine moieties. In vitro and in vivo biological experiments demonstrated the ability of these analogues to mimic natural CS-E.
Inspired by this, we report here the preparation of a novel type of CS-E oligosaccharide mimetics containing glucose instead of glucuronic acid (GlcA). In particular, the synthesis of tetrasaccharide 1 and pentasaccharide 2 is described (Fig. 1). The synthetic access to natural CS-E sequences is hampered by the use of GlcA units that are considered as disarmed building blocks due to the presence of the electron-withdrawing carboxylic acid at position 5. Thus, glycosyl donors derived from GlcA are often associated with low-yielding glycosylation reactions.31,32 We envisioned that the replacement of GlcA by easily accessible glucose units would facilitate the synthesis of CS-E like oligosaccharides. In this manuscript, we also study the 3D structure of the synthesized analogues by NMR and molecular dynamics (MD) simulations. Moreover, the interaction between these compounds and midkine was analyzed by fluorescence polarization (FP) and saturation transfer difference NMR (STD-NMR) experiments. The preparation of derivatives such as 1 and 2 enabled us to study the importance of carboxylic acid functions in the interactions between CS and midkine by comparison with the natural GlcA containing oligomers.
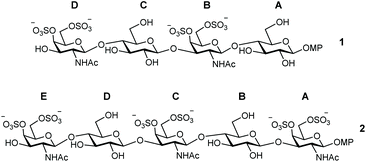 | ||
| Fig. 1 Structures of CS oligosaccharide mimetics 1 and 2. Pyranose rings labelling with letters A–D (or A–E) is employed throughout the text. MP = 4-methoxyphenyl. | ||
Results and discussion
Synthesis of CS mimetics
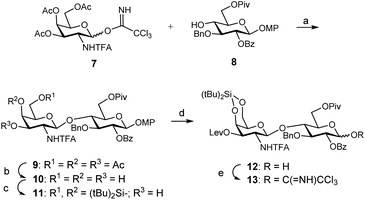
For the synthesis of these CS mimetics, we employed known monosaccharide building blocks 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14 and 8
14 and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33 as starting materials (Scheme 1), easily prepared from D-galactosamine hydrochloride and 1,2;5,6-di-O-isopropylidene-α-D-glucofuranose, respectively. Disaccharide 9 was obtained in high yield (82%) by glycosylation between 8 and trichloroacetimidate donor 7. 1,2-trans Glycosidic linkage was exclusively formed due to the presence of the N-trifluoroacetyl (N-TFA) participating group. Disaccharide 9 was then transformed into adequate donor and acceptor to prepare the tetrasacharide derivative by 2 + 2 coupling. Acetate groups were selectively removed by using p-toluenesulfonic acid in a CH2Cl2/MeOH mixture. Introduction of the silylene group on triol 10 using di-tert-butylsilyl bis(trifluoromethanesulfonate) (tBu2Si(OTf)2) in pyridine afforded disaccharide acceptor 11 in 94% yield. The presence of this orthogonal silylene protecting group at positions 4 and 6 of the galactosamine units allowed the selective sulfation of these positions at the end of the synthetic route to generate CS-E related sequences. Levulinoylation of the remaining 3-OH followed by oxidative removal of the 4-methoxyphenyl group using ceric ammonium nitrate (CAN) provided the 1-hydroxy sugar 12. Trichloroacetimidate 13 was prepared in high yield by treatment with trichloroacetonitrile and K2CO3.
33 as starting materials (Scheme 1), easily prepared from D-galactosamine hydrochloride and 1,2;5,6-di-O-isopropylidene-α-D-glucofuranose, respectively. Disaccharide 9 was obtained in high yield (82%) by glycosylation between 8 and trichloroacetimidate donor 7. 1,2-trans Glycosidic linkage was exclusively formed due to the presence of the N-trifluoroacetyl (N-TFA) participating group. Disaccharide 9 was then transformed into adequate donor and acceptor to prepare the tetrasacharide derivative by 2 + 2 coupling. Acetate groups were selectively removed by using p-toluenesulfonic acid in a CH2Cl2/MeOH mixture. Introduction of the silylene group on triol 10 using di-tert-butylsilyl bis(trifluoromethanesulfonate) (tBu2Si(OTf)2) in pyridine afforded disaccharide acceptor 11 in 94% yield. The presence of this orthogonal silylene protecting group at positions 4 and 6 of the galactosamine units allowed the selective sulfation of these positions at the end of the synthetic route to generate CS-E related sequences. Levulinoylation of the remaining 3-OH followed by oxidative removal of the 4-methoxyphenyl group using ceric ammonium nitrate (CAN) provided the 1-hydroxy sugar 12. Trichloroacetimidate 13 was prepared in high yield by treatment with trichloroacetonitrile and K2CO3.
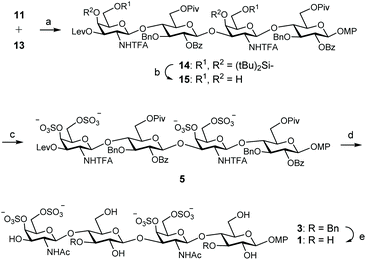
With donor and acceptor disaccharides in hand, we studied the 2 + 2 coupling to yield tetrasaccharide 14 (Scheme 2). Building block 11 was first reacted with 1.5 equiv. of donor 13 under trimethylsilyl trifluoromethanesulfonate (TMSOTf) catalysis at 0 °C. The corresponding tetramer 14 was obtained in only 22% yield. 19F NMR spectrum showed two singlets at δ = −75.6 and −75.7 ppm corresponding to the two trifluoroacetyl moieties. When this glycosylation was performed at room temperature with 2.5 equiv. of donor, using TMSOTf as the promoter, tetrasaccharide 14 was isolated in 44% yield. A similar result (47%) was obtained by using 3 equiv. of donor 13 and tert-butyldimethylsilyl trifluoromethanesulfonate (TBSOTf) activation at room temperature (Scheme 2). Therefore, an excess of glycosyl donor (2.5–3 equiv.) at room temperature was required to produce tetrasaccharide 14 in an acceptable yield due to the low reactivity shown by disaccharide 11. A similar glycosylation outcome was previously obtained by us when using the equivalent GlcA disaccharide building blocks.14
Next, silylene groups were selectively removed by treatment with an excess of (HF)n·Py complex in THF at 0 °C (Scheme 2). Microwave-assisted sulfation34 using SO3·Me3N complex in DMF gave tetrasulfated derivative 5 as sodium salt after purification by size exclusion and silica gel chromatographies and treatment with Dowex-Na+ ion-exchange resin. 1H and 13C NMR spectra showed the typical downfield shifts of the signals at sulfated positions. Basic hydrolysis of esters and trifluoroacetyl amides was carried out by treatment with H2O2/LiOH and NaOH. The released amino groups were then selectively acetylated to afford 3 in good yield after size exclusion chromatography. Finally, the benzyl ethers were removed by hydrogenation over Pd(OH)2/C to give fully deprotected compound 1. The structure of 1 was confirmed by NMR and mass spectroscopic analysis.
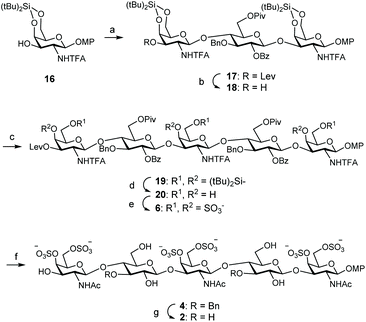
For the preparation of CS related pentamer 2, we first accomplished the synthesis of trisaccharide 17 by 2 + 1 coupling of trichloroacetimidate 13 and known monosaccharide acceptor 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 (Scheme 3). Remarkably, this glycosylation proceeded better than the condensation between 13 and 11, and the trisaccharide was isolated in 56% yield by employing only 1.5 equiv. of donor at room temperature under TMSOTf activation. Increasing the amount of 13 (up to 2.5 equiv.) did not lead to a better result. Treatment of 17 with hydrazine monohydrate gave trisaccharide 18, ready for elongation at 3-OH. Glycosylation of 18 with trichloroacetimidate 13 at room temperature afforded pentasaccharide 19 (Table 1). When using 1.5 equiv. of donor, pentasaccharide was isolated in low yield (entries 1 and 2). Increasing the amount of activator (up to 60 mol% with respect to the donor)36 did not improve the result (entry 3). Interestingly, silylene functionalities proved to be stable under these highly acidic reaction conditions. The yield of 19 was finally raised to 52% (+24% of unreacted acceptor 18) by using 3 equiv. of donor and TBSOTf activation at room temperature (entry 4). 1H and 13C NMR spectra were assigned by using 2D experiments (COSY and HSQC). Three 19F NMR signals appeared at δ = −75.5, −75.8 and −76.0 ppm for the trifluoroacetyl moieties. Cleavage of the silylene groups followed by extensive sulfation under microwave heating afforded hexasulfated, water soluble pentamer 6 (Scheme 3). The NMR chemical shifts of the H-4 and H-6 protons in galactosamine residues confirmed the presence of sulfate groups at these positions (Table 2). The 4,6-di-O-sulfation was also supported by the changes in the chemical shifts of C-4 and C-6 carbons in galactosamine units (Table 2). Hydrolysis of esters and amides involved treatment with H2O2/LiOH followed by aqueous NaOH. Although this two-step saponification procedure is typical for deprotection of uronic acid containing oligosaccharides, avoiding the β-elimination side reaction,37 we also obtained a better yield using this method in comparison with the direct treatment with aqueous NaOH in MeOH. The three amino groups were then selectively acetylated with acetic anhydride and triethylamine in MeOH to afford dibenzylated 4 in excellent yield. Finally, hydrogenolysis of 4 gave the fully deprotected pentasaccharide 2. The NMR and mass spectra were in good agreement with the structure.
35 (Scheme 3). Remarkably, this glycosylation proceeded better than the condensation between 13 and 11, and the trisaccharide was isolated in 56% yield by employing only 1.5 equiv. of donor at room temperature under TMSOTf activation. Increasing the amount of 13 (up to 2.5 equiv.) did not lead to a better result. Treatment of 17 with hydrazine monohydrate gave trisaccharide 18, ready for elongation at 3-OH. Glycosylation of 18 with trichloroacetimidate 13 at room temperature afforded pentasaccharide 19 (Table 1). When using 1.5 equiv. of donor, pentasaccharide was isolated in low yield (entries 1 and 2). Increasing the amount of activator (up to 60 mol% with respect to the donor)36 did not improve the result (entry 3). Interestingly, silylene functionalities proved to be stable under these highly acidic reaction conditions. The yield of 19 was finally raised to 52% (+24% of unreacted acceptor 18) by using 3 equiv. of donor and TBSOTf activation at room temperature (entry 4). 1H and 13C NMR spectra were assigned by using 2D experiments (COSY and HSQC). Three 19F NMR signals appeared at δ = −75.5, −75.8 and −76.0 ppm for the trifluoroacetyl moieties. Cleavage of the silylene groups followed by extensive sulfation under microwave heating afforded hexasulfated, water soluble pentamer 6 (Scheme 3). The NMR chemical shifts of the H-4 and H-6 protons in galactosamine residues confirmed the presence of sulfate groups at these positions (Table 2). The 4,6-di-O-sulfation was also supported by the changes in the chemical shifts of C-4 and C-6 carbons in galactosamine units (Table 2). Hydrolysis of esters and amides involved treatment with H2O2/LiOH followed by aqueous NaOH. Although this two-step saponification procedure is typical for deprotection of uronic acid containing oligosaccharides, avoiding the β-elimination side reaction,37 we also obtained a better yield using this method in comparison with the direct treatment with aqueous NaOH in MeOH. The three amino groups were then selectively acetylated with acetic anhydride and triethylamine in MeOH to afford dibenzylated 4 in excellent yield. Finally, hydrogenolysis of 4 gave the fully deprotected pentasaccharide 2. The NMR and mass spectra were in good agreement with the structure.
| Entry | Donor 13 (equiv.) | Promotor/amount | Isolated yield 19 (%) | Recovered acceptor 18 (%) |
|---|---|---|---|---|
| a The glycosylations were run in CH2Cl2 at room temperature. b mol% with respect to the donor. | ||||
| 1a | 1.5 | TMSOTf, 20 mol%b | 30 | 49 |
| 2a | 1.5 | TBSOTf, 20 mol%b | 34 | 40 |
| 3a | 1.5 | TBSOTf, 60 mol%b | 34 | 42 |
| 4a | 3 | TBSOTf, 20 mol%b | 52 | 24 |
| Compound | H-4 | H-6 | C-4 | C-6 |
|---|---|---|---|---|
| 20 | 3.94–4.04 | 3.44–3.94 | 66.3–68.2 | 61.6–62.5 |
| 6 | 4.87–4.89 | 4.08–4.44 | 72.4–76.8 | 67.8–68.9 |
Fluorescence polarization (FP) experiments
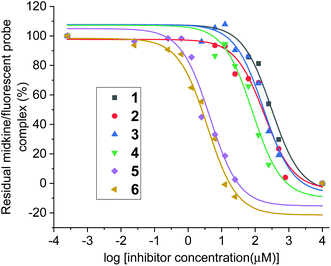
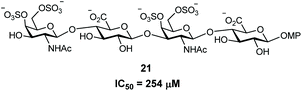
With compounds 1 and 2 in hand, their interactions with midkine were analysed in order to demonstrate that these derivatives are mimetics of the natural products. As mentioned before, midkine is a relevant protein target that binds heparin and CS-E. Sulfated intermediates 3–6 were also included in this study because we have previously demonstrated that compounds displaying hydrophobic functionalities strongly bind to midkine protein.38 In order to calculate relative binding affinities and obtain IC50 values, a FP competition experiment was used.33,35 As previously reported, the FP of samples containing a fixed amount of protein and fluorescent probe and increasing concentrations of the different CS mimetics was measured (Fig. 2 and ESI†). The resulting inhibition curves were fitted and IC50 values, defined as the CS mimetic concentration required for 50% inhibition, were determined (Table 3 and ESI†). These binding affinities were compared with those previously obtained for CS-E tetrasaccharide derivatives bearing GlcA units (Fig. 3).38 Fully deprotected tetra- and pentasaccharides 1 and 2 bound to midkine in the high micromolar range (IC50 = 189 and 130 μM, respectively). Previously synthesized CS-E tetrasaccharide 21 showed a similar affinity (IC50 = 254 μM, Fig. 3).38 Therefore, our data indicated that glucose containing oligosaccharides 1–2 mimicked the natural CS-E sequences and were able to recognize midkine, with binding affinities in the same range. Regarding partially deprotected compounds 3 and 4 (Table 3), our results indicated that the presence of hydrophobic benzyl groups enhanced the binding to midkine giving valuable information for the design of high-affinity ligands (compare IC50 values of 1–2 and 3–4). These results are in good agreement with previous reports pointing out interesting biological activities and protein binding properties of GAG mimetics containing hydrophobic scaffolds or groups.39–41 Thus, fully protected derivatives 5 and 6 showed a ≈30–50 fold inhibition increase in comparison with the fully deprotected oligosaccharides 1 and 2 (Table 3). Importantly, these molecules were soluble in water at 100 μM concentration (5 in the presence of 1% of DMSO). These results suggest that compounds such as 3–6 can be useful for the modulation of CS-protein interactions and the subsequent biological activities.Three-dimensional structure of CS mimetics by NMR and molecular dynamics (MD)
In order to determine the 3D structure of the compounds previously described, we decided to perform an NMR analysis.42 This study would allow us to compare the structure of the CS mimetics and the natural oligosaccharides, with particular emphasis in potential changes coming from the replacement of the carboxylate moieties by hydroxymethyl groups.For this purpose, we recorded NMR spectra of the synthesized oligosaccharides in pure water. The 3JHH coupling constants for 1–4 were calculated (see Tables S1 and S3†) to analyse the ring conformations. On the other hand, we also obtained the interprotonic experimental distances for 1–4 (see Tables S2 and S4†) from NOESY experiments (Fig. 4 and S7†) at variable mixing times by using the initial rate approximation.43 In particular, the interglycosidic NOE were analysed to estimate the interglycosidic distances and define the glycosidic linkage geometry. Internal ring distances between non-consecutive protons were used to confirm the conformation estimated by 3JHH.
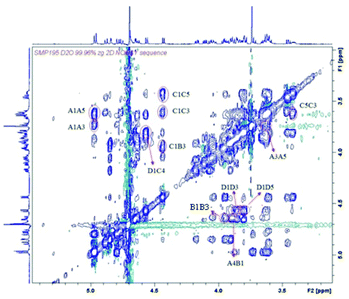 | ||
| Fig. 4 NOESY spectrum of tetrasaccharide 3 at 1.5 mM, 300 K and 600 MHz, in D2O, mixing time 1000 ms. | ||
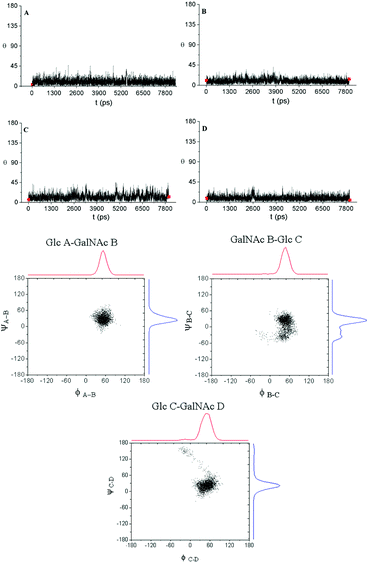
In all the cases measured, the coupling constants were compatible with the 4C1 conformation of the pyranose rings (see Tables S1 and S3†). We also confirmed the 4C1 conformation by analysing the intra-ring H1–H3–H5 distances; in all cases they were compatible with the expected chair (see Tables S2 and S4†). Once evaluated the conformations of the individual sugar rings, we studied the interprotonic distances between both sides of the glycosidic linkages. The arrangement of all the glycosidic linkages can be detected by the observation of NOE between the anomeric proton and the proton ipso relative to the glycosidic linkage for syn-Ψ arrangements (H1–H4 distance for GalNAcβ(1 → 4)Glc linkages and H1–H3 for Glcβ(1 → 3)GalNAc bonds; for example, see Fig. 4: interglycosidic peaks H1D–H4C, H1C–H3B, H1B–H4A). In all the cases, they were compatible with a prevalence of the syn-Ψ rotamer while signs of the presence of the anti-Ψ conformer were absent beyond the limit of sensitivity (5%).
The NMR analysis of fully protected derivatives 5 and 6 in D2O was severely hampered due to solubility problems (5) and signals overlapping and broadening (6). In any case, no evidences for different ring conformations or other glycosidic linkage arrangements were detected for 6.
Overall, the NMR results qualitatively agree with the observations for natural CS oligosaccharides.14 For deprotected oligosaccharides 1–2, and also for partially protected 3–4 and fully substituted 6, in spite of the large steric hindrance of the substituents, the arrangement of the glycosidic linkages and the sugar ring conformations were the same as in the CS case.14 Therefore, the replacement of GlcA units by glucose moieties, in spite of changing the charges balance of the oligosaccharide, does not have influence on the global three dimensional arrangement.
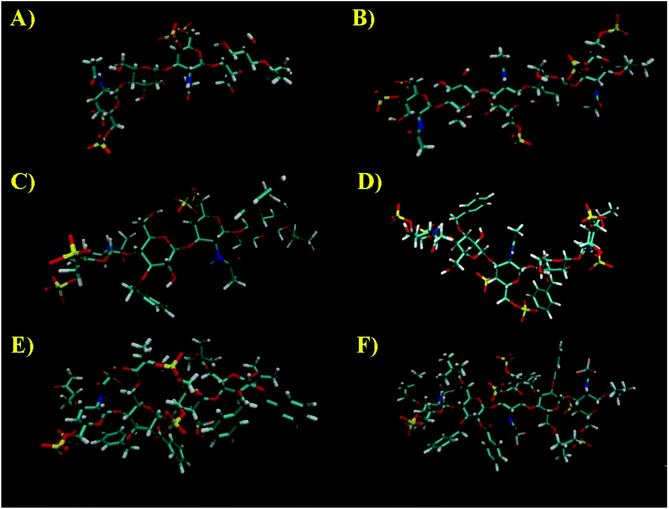
Next, MD simulations were carried out and the results were compared with the NMR experimental data. We calculated the MD trajectories using AMBER software with GLYCAM parameters for pyranoses (see experimental part for details). Tetrasaccharide 1 and pentasaccharide 2, the two compounds with no benzylic substituents in their backbone, yielded averaged 3D structures compatible with the experimental observations, 3JHH and NOEs (Tables S1–S4†). Their 3D shapes can be described as a high pitch helical structure, almost quasilinear, with four residues per turn, which is virtually a linear and elongated shape (Fig. 5). All the sugar rings have a monoconformational behavior being in 4C1 conformation, and the glycosidic linkages are in a syn-Ψ disposition (see ESI† for puckering coordinates and Φ,Ψ trajectories). The introduction of an extra GalNAc residue at the reducing end in 2 does not influence the behaviour of the glycosidic linkages nor the ring pucker. This is in full agreement with the previous NMR analysis.
When we performed a similar study for 3 and 4, we found theoretical results in disagreement with the experimental observations (Tables S1–S4†). The conformations of the glucose rings displaying benzyl groups at position 3, became incompatible with the experimental observations with frequent visits to the equator of the Cremer-Pople puckering sphere. Then, we decided to explore the time-averaged restrained MD (tar-MD) solution to solve this artefact by restraining the intra-ring H1–H3–H5 distances to the experimental ones.44 This strategy only should affect to the conformation of the corresponding ring, fixing the conformation without affecting any other structural issues. Then, we back-calculated the NMR magnitudes from the tar-MD structures and they showed an excellent agreement with the experimental observations (Tables S1–S4†). All the coupling constants were compatible with 4C1 chair conformations of the sugar rings. Furthermore, the calculated interprotonic distances were also compatible with the experimental ones.
In particular, in the case of 3, when we run a short tar-MD of 8 ns, the simulations became compatible with the experimental results with all the rings in 4C1 conformation (Fig. 6, top). The introduction of these restrictions not only fixed the six membered ring conformations but also yielded a glycosidic linkages behavior compatible with the NOESY observations, that corresponds with a syn-Ψ geometry (Fig. 6, bottom). We do not have an explanation for this performance as the restriction at the ring are not directly related with the glycosidic linkage geometry. In addition, a reduction on the flexibility of the glycosidic linkages was observed. For 4, the tar-MD needed more time (50 ns) to reach the agreement with the experimental data (see ESI†).
STD-NMR experiments
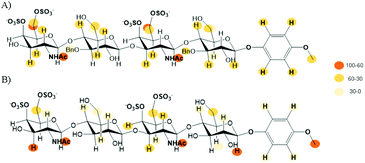
Finally, we performed an NMR analysis on the complexes between midkine and the synthesized CS mimetics.45–47 Thus, we recorded NOESY and STD-NMR experiments at different saturation times for 1–4 in the presence of 20 μM midkine. The reference experiments (in the absence of protein) were recorded in the same buffer (10 mM phosphate, 150 mM NaCl pH = 7.5) used to measure the interactions in order to guarantee the same motional behaviour with or without midkine, avoiding the potential effects in the NMR spectrum of the salt concentration. The signals of the sugar ligands in the complexes appear at the same chemical shift that they were assigned in the absence of the protein. The formation of the complex was confirmed by an appreciable broadening, sign of complex formation as the sign of the NOE effect cannot be used because it is already negative for the free compound.All the compounds 1–4 gave STD signals compatible with the binding to midkine (see Fig. 7 and ESI†). They suggest a disordered complex, as a single complex cannot justify the results obtained. The STD signals were compatible with magnetization arising from all the spatial directions: top, down, left and right. This can be explained when several complex geometries are coexisting in the binding site. Similar STD results were obtained for the interaction between natural CS tetrasaccharides and midkine.14
Conclusions
We have developed a synthetic approach for the preparation of a new type of CS analogues where the GlcA units were replaced by more easily accessible glucose moieties. The designed protecting group strategy allowed us to control the stereochemistry of the glycosidic bonds and introduce the sulfate groups at positions corresponding to biologically active CS-E. Although glucose containing building blocks showed only moderate reactivity in 2 + 2 and 2 + 3 coupling reactions, tetrasaccharide 1 and pentasaccharide 2 were successfully synthesized.Experimental 3JHH coupling constants and NOE-derived interprotonic distances for the synthesized oligosaccharides were consistent with a 3D structure in which all the monosaccharide rings adopt 4C1 chair conformations and the glycosidic linkages present syn-Ψ dispositions. MD simulations supported the NMR experimental data, indicating a 3D shape similar to the natural CS-E oligosaccharide structure.
On the other hand, FP competition assays demonstrated that the synthesized oligosaccharides lacking the carboxylate functions of GlcA units were able to mimic CS-E sequences and interact with growth factor midkine in the micromolar range, suggesting that the CO2− group is not essential for the interaction. Our data also showed that sulfated intermediates 3–6 were more potent inhibitors, confirming that the presence of hydrophobic substituents strongly enhanced the binding. The interaction was also demonstrated by STD-NMR experiments. We found that the STD effects were extended along the entire sugar chain. These results are compatible with the coexistence of random sugar orientations, without a dominant one, in the protein binding site.
In summary, the reported oligosaccharides can be considered as functional and structural CS-E mimetics with potential applications in the study of CS-protein interactions and the modulation of the corresponding biological processes.
Experimental
General synthetic procedures
Thin layer chromatography (TLC) analyses were performed on silica gel 60 F254 precoated on aluminium plates (Merck) and the compounds were detected by staining with cerium(IV) sulfate (1 g)/ammonium molybdate tetrahydrate (21 g)/sulfuric acid (30 mL) solution in water (0.47 L) or with anisaldehyde solution [anisaldehyde (25 mL) with sulfuric acid (25 mL), ethanol (450 mL) and acetic acid (1 mL)], followed by heating at over 200 °C. Column chromatography was carried out on silica gel 60 (0.2–0.063 mm or 0.040–0.015 mm; Merck). Optical rotations were determined with a PerkinElmer 341 polarimeter. 1H-, 19F- and 13C-NMR spectra were acquired on a Bruker Avance III-400 spectrometer. Unit A refers to the reducing end monosaccharide in the NMR data. Electrospray mass spectra (ESI MS) were carried out with an Esquire 6000 ESI-Ion Trap from Bruker Daltonics. High resolution mass spectra (HR MS) were carried out by CITIUS (Universidad de Sevilla). Microwave-based sulfation reactions were performed using a Biotage Initiator Eight synthesizer in sealed reaction vessels.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 9 as a colorless oil (141 mg, 82%). TLC (toluene–EtOAc 3
1) to afford 9 as a colorless oil (141 mg, 82%). TLC (toluene–EtOAc 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Rf 0.25; [α]20D +5° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3): δ 7.99 (d, 2H, Ar), 7.68 (d, 1H, JNH,2 = 9.6 Hz, NH), 7.60 (t, 1H, Ar), 7.46 (m, 2H, Ar), 7.29–7.15 (m, 5H, Ar), 6.92 (m, 2H, Ar), 6.75 (m, 2H, Ar), 5.47 (t, 1H, J1,2 = J2,3 = 8.3 Hz, H-2), 5.36 (d, 1H, J3,4 = 2.6 Hz, H-4′), 5.10 (dd, 1H, J2,3 = 11.2 Hz, H-3′), 5.02 (d, 1H, H-1), 4.93–4.68 (2d, 2H, CH2(Bn)), 4.61 (d, 1H, J1,2 = 8.3 Hz, H-1′), 4.41–4.31 (m, 3H, H-2′, 2 × H-6), 4.03–3.85 (m, 5H, H-3, H-4, H-5′, 2 × H-6′), 3.75 (m, 4H, H-5, Me (OMP)), 2.16–2.03 (3s, 9H, OAc), 1.27 (s, 9H, C(CH3)3); 13C-NMR (100 MHz, CDCl3): δ 179.3, 170.5, 170.3, 170.1, 165.1 (5 × CO), 157.8 (q, JC,F = 37.9 Hz, COCF3), 155.6–114.4 (Ar), 115.8 (q, COCF3), 100.9 (C-1′), 100.6 (C-1), 79.9, 77.5 (C-3, C-4), 74.8 (CH2(Bn)), 73.5 (C-5), 72.8 (C-2), 71.0 (C-5′), 69.9 (C-3′), 66.1 (C-4′), 62.7 (C-6), 60.8 (C-6′), 55.6 (Me (OMP)), 51.5 (C-2′), 39.0 (C(CH3)3), 27.1 (C(CH3)3), 20.6–20.4 (3 × CH3); HR MS: m/z: calcd for C46H52F3NO17Na: 970.3080; found: 970.3088 [M + Na]+.
1) Rf 0.25; [α]20D +5° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3): δ 7.99 (d, 2H, Ar), 7.68 (d, 1H, JNH,2 = 9.6 Hz, NH), 7.60 (t, 1H, Ar), 7.46 (m, 2H, Ar), 7.29–7.15 (m, 5H, Ar), 6.92 (m, 2H, Ar), 6.75 (m, 2H, Ar), 5.47 (t, 1H, J1,2 = J2,3 = 8.3 Hz, H-2), 5.36 (d, 1H, J3,4 = 2.6 Hz, H-4′), 5.10 (dd, 1H, J2,3 = 11.2 Hz, H-3′), 5.02 (d, 1H, H-1), 4.93–4.68 (2d, 2H, CH2(Bn)), 4.61 (d, 1H, J1,2 = 8.3 Hz, H-1′), 4.41–4.31 (m, 3H, H-2′, 2 × H-6), 4.03–3.85 (m, 5H, H-3, H-4, H-5′, 2 × H-6′), 3.75 (m, 4H, H-5, Me (OMP)), 2.16–2.03 (3s, 9H, OAc), 1.27 (s, 9H, C(CH3)3); 13C-NMR (100 MHz, CDCl3): δ 179.3, 170.5, 170.3, 170.1, 165.1 (5 × CO), 157.8 (q, JC,F = 37.9 Hz, COCF3), 155.6–114.4 (Ar), 115.8 (q, COCF3), 100.9 (C-1′), 100.6 (C-1), 79.9, 77.5 (C-3, C-4), 74.8 (CH2(Bn)), 73.5 (C-5), 72.8 (C-2), 71.0 (C-5′), 69.9 (C-3′), 66.1 (C-4′), 62.7 (C-6), 60.8 (C-6′), 55.6 (Me (OMP)), 51.5 (C-2′), 39.0 (C(CH3)3), 27.1 (C(CH3)3), 20.6–20.4 (3 × CH3); HR MS: m/z: calcd for C46H52F3NO17Na: 970.3080; found: 970.3088 [M + Na]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 10 as a white amporphous solid (1.99 g, 69%). TLC (CH2Cl2–MeOH 20
1) to afford 10 as a white amporphous solid (1.99 g, 69%). TLC (CH2Cl2–MeOH 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Rf 0.27; [α]20D +41° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3/CD3OD 9
1) Rf 0.27; [α]20D +41° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3/CD3OD 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): δ 7.97 (d, 2H, Ar), 7.61 (t, 1H, Ar), 7.46 (m, 2H, Ar), 7.20–7.05 (m, 5H, Ar), 6.87 (m, 2H, Ar), 6.71 (m, 2H, Ar), 5.33 (t, 1H, J1,2 = J2,3 = 9.0 Hz, H-2), 5.00 (m, 2H, H-1, CH2(Bn)), 4.60 (d, 1H, CH2(Bn)), 4.42 (d, 1H, J1,2 = 8.2 Hz, H-1′), 4.35 (br d, 1H, J6,6 = 11.4 Hz, H-6), 4.13 (m, 2H, H-6, H-2′), 3.98 (t, 1H, J3,4 = J4,5 = 9.3 Hz, H-4), 3.90–3.80 (m, 3H, H-3, H-4′, H-6′), 3.75–3.71 (m, 4H, H-5, Me (OMP)), 3.64 (dd, 1H, J3,4 = 2.9 Hz, J2,3 = 10.7 Hz, H-3′), 3.56 (br d, 1H, H-6′), 3.44 (m, 1H, H-5′), 1.24 (s, 9H, C(CH3)3); 13C-NMR (100 MHz, CDCl3/CD3OD 9
1): δ 7.97 (d, 2H, Ar), 7.61 (t, 1H, Ar), 7.46 (m, 2H, Ar), 7.20–7.05 (m, 5H, Ar), 6.87 (m, 2H, Ar), 6.71 (m, 2H, Ar), 5.33 (t, 1H, J1,2 = J2,3 = 9.0 Hz, H-2), 5.00 (m, 2H, H-1, CH2(Bn)), 4.60 (d, 1H, CH2(Bn)), 4.42 (d, 1H, J1,2 = 8.2 Hz, H-1′), 4.35 (br d, 1H, J6,6 = 11.4 Hz, H-6), 4.13 (m, 2H, H-6, H-2′), 3.98 (t, 1H, J3,4 = J4,5 = 9.3 Hz, H-4), 3.90–3.80 (m, 3H, H-3, H-4′, H-6′), 3.75–3.71 (m, 4H, H-5, Me (OMP)), 3.64 (dd, 1H, J3,4 = 2.9 Hz, J2,3 = 10.7 Hz, H-3′), 3.56 (br d, 1H, H-6′), 3.44 (m, 1H, H-5′), 1.24 (s, 9H, C(CH3)3); 13C-NMR (100 MHz, CDCl3/CD3OD 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): δ 179.2, 165.5 (2 × CO), 158.6 (q, JC,F = 37.9 Hz, COCF3), 155.5–114.4 (Ar), 116.1 (q, JC,F = 287.9 Hz, COCF3), 100.6 (C-1′), 100.3 (C-1), 80.5 (C-3), 76.6 (C-5′), 76.2 (CH2(Bn)), 75.7 (C-4), 73.5 (C-5), 72.6 (C-2), 70.6 (C-3′), 68.5 (C-4′), 62.9 (C-6), 62.1 (C-6′), 55.5 (Me (OMP)), 53.8 (C-2′), 38.9 (C(CH3)3), 26.9 (C(CH3)3); HR MS: m/z: calcd for C40H46F3NO14Na: 844.2763; found: 844.2754 [M + Na]+.
1): δ 179.2, 165.5 (2 × CO), 158.6 (q, JC,F = 37.9 Hz, COCF3), 155.5–114.4 (Ar), 116.1 (q, JC,F = 287.9 Hz, COCF3), 100.6 (C-1′), 100.3 (C-1), 80.5 (C-3), 76.6 (C-5′), 76.2 (CH2(Bn)), 75.7 (C-4), 73.5 (C-5), 72.6 (C-2), 70.6 (C-3′), 68.5 (C-4′), 62.9 (C-6), 62.1 (C-6′), 55.5 (Me (OMP)), 53.8 (C-2′), 38.9 (C(CH3)3), 26.9 (C(CH3)3); HR MS: m/z: calcd for C40H46F3NO14Na: 844.2763; found: 844.2754 [M + Na]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 11 as a white amporphous solid (1.17 g, 94%). TLC (toluene–EtOAc 3
1) to afford 11 as a white amporphous solid (1.17 g, 94%). TLC (toluene–EtOAc 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Rf 0.28; [α]20D +9° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3): δ 7.91 (m, 2H, Ar), 7.56 (t, 1H, Ar), 7.49 (d, 1H, J2,NH = 9.2 Hz, NH), 7.41 (t, 2H, Ar), 7.23–7.04 (m, 5H, Ar), 6.87 (m, 2H, Ar), 6.73 (m, 2H, Ar), 5.41 (t, 1H, H-2), 4.98 (d, 1H, J1,2 = 7.6 Hz, H-1), 4.95 (d, 1H, CH2(Bn)), 4.66 (m, 2H, H-6a, CH2(Bn)), 4.39 (d, 1H, J3,4 = 3.3 Hz, H-4′), 4.36 (d, 1H, J1,2 = 8.5 Hz, H-1′), 4.29 (m, 1H, H-2′), 4.24–4.15 (m, 3H, H-6b, H-6′a, H-6′b), 3.88 (t, 1H, J2,3 = J3,4 = 8.0 Hz, H-3), 3.78–3.68 (m, 5H, H-4, Me (OMP), H-5), 3.55 (ddd, J2,3 = 10.8 Hz, H-3′), 3.33 (br s, 1H, H-5′), 2.73 (d, 1H, J3,OH = 11.7 Hz, OH), 1.22 (s, 9H, C(CH3)3), 1.05, 1.04 (2s, 18H, Si(C(CH3)3)2); 13C-NMR (100 MHz, CDCl3): δ 179.7, 165.3 (2 × CO), 158.4 (q, JC,F = 37.0 Hz, COCF3), 155.7–114.6 (Ar), 116.1 (q, JC,F = 288.0 Hz, COCF3), 102.6 (C-1′), 100.4 (C-1), 80.7 (C-3), 79.8 (C-4), 75.1 (CH2(Bn)), 73.7 (C-5), 73.1 (C-2), 72.2 (C-3′, C-5′), 71.8 (C-4′), 66.6 (C-6′), 62.6 (C-6), 55.7 (Me (OMP)), 53.7 (C-2′), 39.2 (C(CH3)3), 27.7, 27.5 (Si(C(CH3)3)2), 27.3 (C(CH3)3), 23.5, 20.7 (Si(C(CH3)3)2); HR MS: m/z: calcd for C48H62F3NO14NaSi: 984.3784; found: 984.3776 [M + Na]+.
1) Rf 0.28; [α]20D +9° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3): δ 7.91 (m, 2H, Ar), 7.56 (t, 1H, Ar), 7.49 (d, 1H, J2,NH = 9.2 Hz, NH), 7.41 (t, 2H, Ar), 7.23–7.04 (m, 5H, Ar), 6.87 (m, 2H, Ar), 6.73 (m, 2H, Ar), 5.41 (t, 1H, H-2), 4.98 (d, 1H, J1,2 = 7.6 Hz, H-1), 4.95 (d, 1H, CH2(Bn)), 4.66 (m, 2H, H-6a, CH2(Bn)), 4.39 (d, 1H, J3,4 = 3.3 Hz, H-4′), 4.36 (d, 1H, J1,2 = 8.5 Hz, H-1′), 4.29 (m, 1H, H-2′), 4.24–4.15 (m, 3H, H-6b, H-6′a, H-6′b), 3.88 (t, 1H, J2,3 = J3,4 = 8.0 Hz, H-3), 3.78–3.68 (m, 5H, H-4, Me (OMP), H-5), 3.55 (ddd, J2,3 = 10.8 Hz, H-3′), 3.33 (br s, 1H, H-5′), 2.73 (d, 1H, J3,OH = 11.7 Hz, OH), 1.22 (s, 9H, C(CH3)3), 1.05, 1.04 (2s, 18H, Si(C(CH3)3)2); 13C-NMR (100 MHz, CDCl3): δ 179.7, 165.3 (2 × CO), 158.4 (q, JC,F = 37.0 Hz, COCF3), 155.7–114.6 (Ar), 116.1 (q, JC,F = 288.0 Hz, COCF3), 102.6 (C-1′), 100.4 (C-1), 80.7 (C-3), 79.8 (C-4), 75.1 (CH2(Bn)), 73.7 (C-5), 73.1 (C-2), 72.2 (C-3′, C-5′), 71.8 (C-4′), 66.6 (C-6′), 62.6 (C-6), 55.7 (Me (OMP)), 53.7 (C-2′), 39.2 (C(CH3)3), 27.7, 27.5 (Si(C(CH3)3)2), 27.3 (C(CH3)3), 23.5, 20.7 (Si(C(CH3)3)2); HR MS: m/z: calcd for C48H62F3NO14NaSi: 984.3784; found: 984.3776 [M + Na]+.
Lev2O (13 mL of a 0.19 M solution in CH2Cl2) was added at room temperature to a mixture of 11 (811 mg, 0.843 mmol) and DMAP (16 mg, 0.13 mmol). The mixture was stirred for 1.5 h, diluted with CH2Cl2, and washed with saturated aqueous NaHCO3, and brine. The organic phase was dried (MgSO4), filtered and concentrated to dryness to give the corresponding levulinated disaccharide that was directly used in the next step without further purification.
CAN (5.0 mL of a 0.67 M solution in H2O) was added to a solution of the levulinated compound (0.843 mmol) in CH2Cl2/MeCN (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2; 45 mL). After stirring for 2 h at room temperature, the reaction mixture was diluted with EtOAc, washed with H2O, saturated aqueous NaHCO3, and brine. The organic phase was dried (MgSO4), filtered and concentrated to dryness. The residue was purified by column chromatography (CH2Cl2–MeOH 80
2; 45 mL). After stirring for 2 h at room temperature, the reaction mixture was diluted with EtOAc, washed with H2O, saturated aqueous NaHCO3, and brine. The organic phase was dried (MgSO4), filtered and concentrated to dryness. The residue was purified by column chromatography (CH2Cl2–MeOH 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 → CH2Cl2–MeOH 70
1 → CH2Cl2–MeOH 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 12 as light yellow foam (704 mg, 88%, two steps, mixture of α/β anomers). TLC (CH2Cl2–MeOH 30
1) to afford 12 as light yellow foam (704 mg, 88%, two steps, mixture of α/β anomers). TLC (CH2Cl2–MeOH 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Rf 0.31, 0.41; 1H-NMR (400 MHz, CDCl3) (data for α anomer): δ 7.93 (m, 2H, Ar), 7.55 (m, 1H, Ar), 7.46 (d, 1H, J2,NH = 9.0 Hz, NH), 7.41 (m, 2H, Ar), 7.24–7.10 (m, 5H, Ar), 5.48 (t, 1H, J1,2 = J1,OH = 3.8 Hz, H-1), 5.09–4.99 (m, 2H, CH2(Bn), H-2), 4.76–4.59 (m, 3H, CH2(Bn), H-3′, H-6a), 4.59–4.50 (m, 3H, H-2′, H-4′, H-1′), 4.21 (dd, 1H, H-3), 4.18–4.05 (m, 4H, H-5, H-6b, H-6′a, H-6′b), 3.63 (m, 1H, H-4), 3.28 (br s, 1H, H-5′), 2.90 (d, 1H, OH), 2.76–2.57 (m, 4H, CH2(Lev)), 2.18 (s, 3H, CH3(Lev)), 1.25 (s, 9H, C(CH3)3), 1.06, 1.01 (2s, 18H, Si(C(CH3)3)2); 13C-NMR (100 MHz, CDCl3) (data for α anomer): δ 206.5, 179.6, 172.6, 165.9 (4 × CO), 157.8 (q, JC,F = 37.2 Hz, COCF3), 138.6–127.4 (Ar), 116.0 (q, JC,F = 288.1 Hz, COCF3), 102.4 (C-1′), 90.1 (C-1), 79.8 (C-4), 77.9 (C-3), 75.0 (CH2(Bn)), 73.7, 73.5 (C-2 and C-3′), 71.8 (C-5′), 69.5 (C-4′), 69.1 (C-5), 66.6 (C-6′), 62.1 (C-6), 50.6 (C-2′), 39.3 (C(CH3)3), 37.9 (CH2(Lev)), 29.8 (CH3(Lev)), 28.3 (CH2(Lev)), 27.7, 27.5 (Si(C(CH3)3)2), 27.4 (C(CH3)3), 23.4, 20.7 (Si(C(CH3)3)2); HR MS: m/z: calcd for C46H62F3NO15NaSi: 976.3733; found: 976.3725 [M + Na]+.
1) Rf 0.31, 0.41; 1H-NMR (400 MHz, CDCl3) (data for α anomer): δ 7.93 (m, 2H, Ar), 7.55 (m, 1H, Ar), 7.46 (d, 1H, J2,NH = 9.0 Hz, NH), 7.41 (m, 2H, Ar), 7.24–7.10 (m, 5H, Ar), 5.48 (t, 1H, J1,2 = J1,OH = 3.8 Hz, H-1), 5.09–4.99 (m, 2H, CH2(Bn), H-2), 4.76–4.59 (m, 3H, CH2(Bn), H-3′, H-6a), 4.59–4.50 (m, 3H, H-2′, H-4′, H-1′), 4.21 (dd, 1H, H-3), 4.18–4.05 (m, 4H, H-5, H-6b, H-6′a, H-6′b), 3.63 (m, 1H, H-4), 3.28 (br s, 1H, H-5′), 2.90 (d, 1H, OH), 2.76–2.57 (m, 4H, CH2(Lev)), 2.18 (s, 3H, CH3(Lev)), 1.25 (s, 9H, C(CH3)3), 1.06, 1.01 (2s, 18H, Si(C(CH3)3)2); 13C-NMR (100 MHz, CDCl3) (data for α anomer): δ 206.5, 179.6, 172.6, 165.9 (4 × CO), 157.8 (q, JC,F = 37.2 Hz, COCF3), 138.6–127.4 (Ar), 116.0 (q, JC,F = 288.1 Hz, COCF3), 102.4 (C-1′), 90.1 (C-1), 79.8 (C-4), 77.9 (C-3), 75.0 (CH2(Bn)), 73.7, 73.5 (C-2 and C-3′), 71.8 (C-5′), 69.5 (C-4′), 69.1 (C-5), 66.6 (C-6′), 62.1 (C-6), 50.6 (C-2′), 39.3 (C(CH3)3), 37.9 (CH2(Lev)), 29.8 (CH3(Lev)), 28.3 (CH2(Lev)), 27.7, 27.5 (Si(C(CH3)3)2), 27.4 (C(CH3)3), 23.4, 20.7 (Si(C(CH3)3)2); HR MS: m/z: calcd for C46H62F3NO15NaSi: 976.3733; found: 976.3725 [M + Na]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 + 1% Et3N) to afford 13 as a white foam (545 mg, 94%, mixture of α/β anomers). TLC (CH2Cl2–MeOH 50
1 + 1% Et3N) to afford 13 as a white foam (545 mg, 94%, mixture of α/β anomers). TLC (CH2Cl2–MeOH 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Rf 0.36 and 0.31; 1H-NMR (400 MHz, CDCl3) (data for β anomer): δ 8.58 (s, 1H, NH), 7.91 (m, 2H, Ar), 7.55 (m, 1H, Ar), 7.43–7.36 (m, 3H, Ar, NH), 7.23–7.10 (m, 5H, Ar), 5.97 (d, 1H, J1,2 = 6.6 Hz, H-1), 5.50 (t, 1H, J2,3 = 6.9 Hz, H-2), 4.87 (d, 1H, CH2(Bn)), 4.79 (dd, 1H, J2,3 = 11.0 Hz, J3,4 = 3.1 Hz, H-3′), 4.74 (d, 1H, CH2(Bn)), 4.61–4.45 (m, 4H, H-1′, H-4′, H-6a, H-2′), 4.26 (dd, 1H, J5,6a = 2.3 Hz, J6a,6b = 11.9 Hz, H-6b), 4.14–4.05 (m, 2H, H-6′a, H-6′b), 3.95 (t, 1H, J3,4 = 6.8 Hz, H-3), 3.87–3.82 (m, 2H, H-4, H-5), 3.30 (br s, 1H, H-5′), 2.74, 2.58 (2 m, 4H, CH2(Lev)), 2.17 (s, 3H, CH3(Lev)), 1.22 (s, 9H, C(CH3)3), 1.01, 1.00 (2s, 18H, Si(C(CH3)3)2); ESI MS: m/z: calcd for C48H62Cl3F3N2O15SiNa: 1119.3; found: 1119.3 [M + Na]+.
1) Rf 0.36 and 0.31; 1H-NMR (400 MHz, CDCl3) (data for β anomer): δ 8.58 (s, 1H, NH), 7.91 (m, 2H, Ar), 7.55 (m, 1H, Ar), 7.43–7.36 (m, 3H, Ar, NH), 7.23–7.10 (m, 5H, Ar), 5.97 (d, 1H, J1,2 = 6.6 Hz, H-1), 5.50 (t, 1H, J2,3 = 6.9 Hz, H-2), 4.87 (d, 1H, CH2(Bn)), 4.79 (dd, 1H, J2,3 = 11.0 Hz, J3,4 = 3.1 Hz, H-3′), 4.74 (d, 1H, CH2(Bn)), 4.61–4.45 (m, 4H, H-1′, H-4′, H-6a, H-2′), 4.26 (dd, 1H, J5,6a = 2.3 Hz, J6a,6b = 11.9 Hz, H-6b), 4.14–4.05 (m, 2H, H-6′a, H-6′b), 3.95 (t, 1H, J3,4 = 6.8 Hz, H-3), 3.87–3.82 (m, 2H, H-4, H-5), 3.30 (br s, 1H, H-5′), 2.74, 2.58 (2 m, 4H, CH2(Lev)), 2.17 (s, 3H, CH3(Lev)), 1.22 (s, 9H, C(CH3)3), 1.01, 1.00 (2s, 18H, Si(C(CH3)3)2); ESI MS: m/z: calcd for C48H62Cl3F3N2O15SiNa: 1119.3; found: 1119.3 [M + Na]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and Sephadex LH 20 chromatography (CH2Cl2–MeOH 1
1) and Sephadex LH 20 chromatography (CH2Cl2–MeOH 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to obtain 14 as a white amorphous solid (37 mg, 47%). TLC (toluene–acetone 6
1) to obtain 14 as a white amorphous solid (37 mg, 47%). TLC (toluene–acetone 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Rf 0.49; [α]20D +24° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3): δ 7.94 (m, 4H, Ar), 7.55 (m, 2H, Ar), 7.40 (m, 4H, Ar), 7.20–7.07 (m, 12H, 2NH, Ar), 6.86 (m, 2H, Ar), 6.70 (m, 2H, Ar), 5.44 (t, 1H, J1,2 = J2,3 = 7.6 Hz, H-2A or C), 5.15 (t, 1H, J1,2 = J2,3 = 7.4 Hz, H-2A or C), 5.00 (m, 2H, H-1A, H-1C), 4.84 (m, 4H, H-1B, H-3D, CH2(Bn)), 4.71 (d, 1H, CH2(Bn)), 4.61 (m, 2H, H-1D, CH2(Bn)), 5.54 (d, 1H, J3,4 = 3.1 Hz, H-4D), 4.48 (d, 1H, J3,4 = 2.5 Hz, H-4B), 4.42 (m, 3H, H-2D, H-6aA, H-6aC), 4.32 (dd, 1H, J5,6b = 4.7 Hz, J6a,6b = 12.1 Hz, H-6bA or C), 4.20–4.02 (m, 4H, H-6bA or C, H-6aD, H-2B, H-6bD), 3.99–3.92 (m, 4H, H-6aB, H-6bB, H-3B, H-3A or C), 3.84–3.79 (m, 2H, H-3A or C, H-4A or C), 3.77–3.71 (m, 5H, H-4A or C, H-5A or C, Me (OMP)), 3.62 (m, 1H, H-5A or C), 3.29 (br s, 1H, H-5D), 3.07 (br s, 1H, H-5B), 2.75 (m, 2H, CH2(Lev)), 2.59 (m, 2H, CH2(Lev)), 2.17 (s, 3H, CH3(Lev)), 1.21, 1.17 (2s, 18H, C(CH3)3), 1.00, 0.97, 0.92 (3s, 36H, Si(C(CH3)3)2); 13C-NMR (100 MHz, CDCl3): δ 206.5, 179.1, 178.3, 172.8, 165.3 (6 × CO), 157.8 (2q, COCF3), 155.6–114.5 (Ar), 115.9, 115.7 (2q, COCF3), 101.6 (C-1D), 101.1 (C-1B), 100.2, 100.0 (C-1A, C-1C), 80.9, 80.5 (C-3A, C-3C), 78.6, 78.0 (C-4A, C-4C), 75.5 (C-3B), 74.6 (C-5A or C), 74.2 (CH2(Bn)), 73.8, 73.7 (C-5A or C, CH2(Bn)), 73.4, 73.1 (C-2A, C-2C), 72.8 (C-3D), 72.3 (C-4B), 71.9 (C-5B, C-5D), 69.5 (C-4D), 66.8, 66.6 (C-6B, C-6D), 63.5, 62.8 (C-6A, C-6C), 55.7 (Me (OMP)), 52.9 (C-2B), 51.0 (C-2D), 39.1, 38.9 (C(CH3)3), 38.0 (CH2(Lev)), 29.8 (CH3(Lev)), 28.3 (CH2(Lev)), 27.7, 27.6, 27.4, 27.2 (Si(C(CH3)3)2, C(CH3)3), 23.4, 23.3, 20.7, 20.6 (Si(C(CH3)3)2); 19F NMR (376 MHz, CDCl3): δ −75.6 (s, 3F), −75.7 (s, 3F); HR MS: m/z: calcd for C94H122F6N2O28Si2Na: 1919.7519; found: 1919.7500 [M + Na]+.
1) Rf 0.49; [α]20D +24° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3): δ 7.94 (m, 4H, Ar), 7.55 (m, 2H, Ar), 7.40 (m, 4H, Ar), 7.20–7.07 (m, 12H, 2NH, Ar), 6.86 (m, 2H, Ar), 6.70 (m, 2H, Ar), 5.44 (t, 1H, J1,2 = J2,3 = 7.6 Hz, H-2A or C), 5.15 (t, 1H, J1,2 = J2,3 = 7.4 Hz, H-2A or C), 5.00 (m, 2H, H-1A, H-1C), 4.84 (m, 4H, H-1B, H-3D, CH2(Bn)), 4.71 (d, 1H, CH2(Bn)), 4.61 (m, 2H, H-1D, CH2(Bn)), 5.54 (d, 1H, J3,4 = 3.1 Hz, H-4D), 4.48 (d, 1H, J3,4 = 2.5 Hz, H-4B), 4.42 (m, 3H, H-2D, H-6aA, H-6aC), 4.32 (dd, 1H, J5,6b = 4.7 Hz, J6a,6b = 12.1 Hz, H-6bA or C), 4.20–4.02 (m, 4H, H-6bA or C, H-6aD, H-2B, H-6bD), 3.99–3.92 (m, 4H, H-6aB, H-6bB, H-3B, H-3A or C), 3.84–3.79 (m, 2H, H-3A or C, H-4A or C), 3.77–3.71 (m, 5H, H-4A or C, H-5A or C, Me (OMP)), 3.62 (m, 1H, H-5A or C), 3.29 (br s, 1H, H-5D), 3.07 (br s, 1H, H-5B), 2.75 (m, 2H, CH2(Lev)), 2.59 (m, 2H, CH2(Lev)), 2.17 (s, 3H, CH3(Lev)), 1.21, 1.17 (2s, 18H, C(CH3)3), 1.00, 0.97, 0.92 (3s, 36H, Si(C(CH3)3)2); 13C-NMR (100 MHz, CDCl3): δ 206.5, 179.1, 178.3, 172.8, 165.3 (6 × CO), 157.8 (2q, COCF3), 155.6–114.5 (Ar), 115.9, 115.7 (2q, COCF3), 101.6 (C-1D), 101.1 (C-1B), 100.2, 100.0 (C-1A, C-1C), 80.9, 80.5 (C-3A, C-3C), 78.6, 78.0 (C-4A, C-4C), 75.5 (C-3B), 74.6 (C-5A or C), 74.2 (CH2(Bn)), 73.8, 73.7 (C-5A or C, CH2(Bn)), 73.4, 73.1 (C-2A, C-2C), 72.8 (C-3D), 72.3 (C-4B), 71.9 (C-5B, C-5D), 69.5 (C-4D), 66.8, 66.6 (C-6B, C-6D), 63.5, 62.8 (C-6A, C-6C), 55.7 (Me (OMP)), 52.9 (C-2B), 51.0 (C-2D), 39.1, 38.9 (C(CH3)3), 38.0 (CH2(Lev)), 29.8 (CH3(Lev)), 28.3 (CH2(Lev)), 27.7, 27.6, 27.4, 27.2 (Si(C(CH3)3)2, C(CH3)3), 23.4, 23.3, 20.7, 20.6 (Si(C(CH3)3)2); 19F NMR (376 MHz, CDCl3): δ −75.6 (s, 3F), −75.7 (s, 3F); HR MS: m/z: calcd for C94H122F6N2O28Si2Na: 1919.7519; found: 1919.7500 [M + Na]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) Rf 0.26; 1H-NMR (400 MHz, CDCl3/CD3OD 4
2) Rf 0.26; 1H-NMR (400 MHz, CDCl3/CD3OD 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): δ 7.95 (m, 4H, Ar), 7.58 (m, 2H, Ar), 7.43 (m, 4H, Ar), 7.16–7.01 (m, 10H, Ar), 6.83 (m, 2H, Ar), 6.67 (m, 2H, Ar), 5.28 (dd, 1H, J1,2 = 8.0 Hz, J2,3 = 9.7 Hz, H-2A), 5.06 (dd, 1H, J1,2 = 7.9 Hz, J2,3 = 9.6 Hz, H-2C), 4.98–4.93 (m, 3H, CH2(Bn), H-1A), 4.86 (dd, 1H, J2,3 = 11.1 Hz, J3,4 = 3.1 Hz, H-3D), 4.72 (d, 1H, H-1C), 4.53–4.41 (m, 5H, H-1D, CH2(Bn), H-6aA or C, H-2D), 4.40 (d, 1H, J1,2 = 8.2 Hz, H-1B), 4.22 (br d, 1H, H-6aA or C), 4.11 (br t, 1H, H-2B), 4.05–3.90 (m, 6H, H-6bA, H-6bC, H-4D, H-4B, H-4A, H-4C), 3.87–3.75 (m, 5H, H-6aB, H-6aD, H-3C, H-3A, H-3B), 3.68 (s, 3H, Me (OMP)), 3.64–3.58 (m, 2H, H-5A, H-5C), 3.52–3.45 (m, 3H, H-6bB, H-6bD, H-5B or D), 3.39 (m, 1H, H-5B or D), 2.75 (m, 2H, CH2(Lev)), 2.53 (m, 2H, CH2(Lev)), 2.15 (s, 3H, CH3(Lev)), 1.21, 1.17 (2s, 18H, C(CH3)3); 13C-NMR (100 MHz, CDCl3/CD3OD 4
1): δ 7.95 (m, 4H, Ar), 7.58 (m, 2H, Ar), 7.43 (m, 4H, Ar), 7.16–7.01 (m, 10H, Ar), 6.83 (m, 2H, Ar), 6.67 (m, 2H, Ar), 5.28 (dd, 1H, J1,2 = 8.0 Hz, J2,3 = 9.7 Hz, H-2A), 5.06 (dd, 1H, J1,2 = 7.9 Hz, J2,3 = 9.6 Hz, H-2C), 4.98–4.93 (m, 3H, CH2(Bn), H-1A), 4.86 (dd, 1H, J2,3 = 11.1 Hz, J3,4 = 3.1 Hz, H-3D), 4.72 (d, 1H, H-1C), 4.53–4.41 (m, 5H, H-1D, CH2(Bn), H-6aA or C, H-2D), 4.40 (d, 1H, J1,2 = 8.2 Hz, H-1B), 4.22 (br d, 1H, H-6aA or C), 4.11 (br t, 1H, H-2B), 4.05–3.90 (m, 6H, H-6bA, H-6bC, H-4D, H-4B, H-4A, H-4C), 3.87–3.75 (m, 5H, H-6aB, H-6aD, H-3C, H-3A, H-3B), 3.68 (s, 3H, Me (OMP)), 3.64–3.58 (m, 2H, H-5A, H-5C), 3.52–3.45 (m, 3H, H-6bB, H-6bD, H-5B or D), 3.39 (m, 1H, H-5B or D), 2.75 (m, 2H, CH2(Lev)), 2.53 (m, 2H, CH2(Lev)), 2.15 (s, 3H, CH3(Lev)), 1.21, 1.17 (2s, 18H, C(CH3)3); 13C-NMR (100 MHz, CDCl3/CD3OD 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): δ 208.7, 179.3, 172.6, 165.8, 165.7 (6 × CO), 158.0 (2q, COCF3), 155.7–114.6 (Ar), 116.2 (q, JC,F = 287.1 Hz, COCF3), 115.9 (q, JC,F = 287.7 Hz, COCF3), 101.3 (C-1C), 100.5 (C-1A, C-1D), 100.3 (C-1B), 80.9 (C-3A), 80.6 (C-3C), 79.3 (C-3B), 76.7, 76.6, 76.5 (2 × CH2(Bn), C-5B, C-5D), 75.4, 75.1 (C-4A, C-4C), 73.8, 73.6 (C-5A, C-5C), 73.1 (C-3D), 72.9 (C-2C), 72.8 (C-2A), 68.1 (C-4B), 66.2 (C-4D), 62.9, 62.5, 62.3, 62.2 (C-6A, C-6B, C-6C, C-6D), 55.7 (Me (OMP)), 52.3 (C-2B), 51.2 (C-2D), 39.2, 39.1 (C(CH3)3), 38.1 (CH2(Lev)), 29.8 (CH3(Lev)), 28.1 (CH2(Lev)), 27.1 (C(CH3)3); HR MS: m/z: calcd for C78H90F6N2O28Na: 1639.5477; found: 1639.5465 [M + Na]+.
1): δ 208.7, 179.3, 172.6, 165.8, 165.7 (6 × CO), 158.0 (2q, COCF3), 155.7–114.6 (Ar), 116.2 (q, JC,F = 287.1 Hz, COCF3), 115.9 (q, JC,F = 287.7 Hz, COCF3), 101.3 (C-1C), 100.5 (C-1A, C-1D), 100.3 (C-1B), 80.9 (C-3A), 80.6 (C-3C), 79.3 (C-3B), 76.7, 76.6, 76.5 (2 × CH2(Bn), C-5B, C-5D), 75.4, 75.1 (C-4A, C-4C), 73.8, 73.6 (C-5A, C-5C), 73.1 (C-3D), 72.9 (C-2C), 72.8 (C-2A), 68.1 (C-4B), 66.2 (C-4D), 62.9, 62.5, 62.3, 62.2 (C-6A, C-6B, C-6C, C-6D), 55.7 (Me (OMP)), 52.3 (C-2B), 51.2 (C-2D), 39.2, 39.1 (C(CH3)3), 38.1 (CH2(Lev)), 29.8 (CH3(Lev)), 28.1 (CH2(Lev)), 27.1 (C(CH3)3); HR MS: m/z: calcd for C78H90F6N2O28Na: 1639.5477; found: 1639.5465 [M + Na]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The residue was then purified by silica gel column choromatography (EtOAc–MeOH–H2O 36
1). The residue was then purified by silica gel column choromatography (EtOAc–MeOH–H2O 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 → EtOAc–MeOH–H2O 24
3 → EtOAc–MeOH–H2O 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) and finally eluted from a Dowex 50WX2-Na+ column (MeOH) to obtain 5 as sodium salt (40 mg, 84%, white amorphous solid). TLC (EtOAc–MeOH–H2O 24
3) and finally eluted from a Dowex 50WX2-Na+ column (MeOH) to obtain 5 as sodium salt (40 mg, 84%, white amorphous solid). TLC (EtOAc–MeOH–H2O 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) Rf 0.28; 1H-NMR (400 MHz, CD3OD): δ 7.97 (m, 4H, Ar), 7.62 (m, 2H, Ar), 7.46 (m, 4H, Ar), 7.20–7.02 (m, 10H, Ar), 6.84 (m, 2H, Ar), 6.72 (m, 2H, Ar), 5.27 (br t, 1H, H-2A), 5.25 (t, 1H, J2,3 = 9.8 Hz, H-2C), 5.13 (d, 1H, J1,2 = 7.9 Hz, H-1A), 5.05 (dd, 1H, J2,3 = 11.3 Hz, J3,4 = 3.3 Hz, H-3D), 4.99 (2d, 2H, CH2(Bn)), 4.90 (m, 2H, H-4B, H-4D), 4.78 (d, 1H, J1,2 = 7.8 Hz, H-1C), 4.75 (d, 1H, J1,2 = 8.3 Hz, H–1D), 4.70 (d, 1H, J1,2 = 8.1 Hz, H-1B), 4.61 (d, 1H, CH2(Bn)), 4.55 (d, 1H, CH2(Bn)), 4.45–4.30 (m, 5H, H-6aA, H-6aB, H-6aC, H-6aD, H-2D), 4.30–4.07 (m, 5H, H-6bA, H-6bB, H-6bC, H-6bD, H-2B), 4.07–3.96 (m, 5H, H-4A, H-4C, H-3A, H-3B, H-5B or D), 3.90 (t, 1H, H-3C), 3.88–3.82 (m, 2H, H-5B or D, H-5A or C), 3.76 (m, 1H, H-5A or C), 3.68 (s, 3H, Me (OMP)), 3.31 (m, 2H, CH2(Lev)), 2.59 (m, 2H, CH2(Lev)), 2.13 (s, 3H, CH3(Lev)), 1.25, 1.20 (2s, 18H, C(CH3)3); 13C-NMR (100 MHz, CD3OD): δ 209.7, 180.1, 179.7, 173.9, 167.6, 167.1 (6 × CO), 159.4 (q, JC,F = 37.1 Hz, COCF3), 159.2 (q, JC,F = 37.4 Hz, COCF3), 156.9–115.4 (Ar), 117.4 (q, JC,F = 287.3 Hz, COCF3), 117.2 (q, JC,F = 288.2 Hz, COCF3), 102.5 (C-1C), 101.3 (C-1D), 101.1 (C-1A), 101.0 (C-1B), 81.1 (C-3C), 80.9 (C-3A), 77.9 (C-4A), 77.2 (C-4C), 76.8 (C-4B), 76.4 (C-3B), 76.1, 75.4 (2 × CH2(Bn), 75.0 (C-5A or C), 74.7, 74.6, 74.4 (C-5A or C, C-5B or D, C-2C, C-2A), 74.0 (C-5B or D), 72.4 (C-4D), 71.9 (C-3D), 68.5, 67.9 (C-6B, C-6D), 64.4, 64.2 (C-6A, C-6C), 56.0 (Me (OMP)), 54.1 (C-2B), 52.5 (C-2D), 40.0, 39.9 (C(CH3)3), 38.5 (CH2(Lev)), 29.6 (CH3(Lev)), 29.2 CH2(Lev)), 27.8, 27.6 (C(CH3)3); 19F NMR (376 MHz, CD3OD): δ −76.5 (s, 3F), −77.1 (s, 3F); ESI MS: m/z: calcd for C78H86F6N2O40S4Na3: 2001.3; found: 2000.7 [M + 3Na]−.
3) Rf 0.28; 1H-NMR (400 MHz, CD3OD): δ 7.97 (m, 4H, Ar), 7.62 (m, 2H, Ar), 7.46 (m, 4H, Ar), 7.20–7.02 (m, 10H, Ar), 6.84 (m, 2H, Ar), 6.72 (m, 2H, Ar), 5.27 (br t, 1H, H-2A), 5.25 (t, 1H, J2,3 = 9.8 Hz, H-2C), 5.13 (d, 1H, J1,2 = 7.9 Hz, H-1A), 5.05 (dd, 1H, J2,3 = 11.3 Hz, J3,4 = 3.3 Hz, H-3D), 4.99 (2d, 2H, CH2(Bn)), 4.90 (m, 2H, H-4B, H-4D), 4.78 (d, 1H, J1,2 = 7.8 Hz, H-1C), 4.75 (d, 1H, J1,2 = 8.3 Hz, H–1D), 4.70 (d, 1H, J1,2 = 8.1 Hz, H-1B), 4.61 (d, 1H, CH2(Bn)), 4.55 (d, 1H, CH2(Bn)), 4.45–4.30 (m, 5H, H-6aA, H-6aB, H-6aC, H-6aD, H-2D), 4.30–4.07 (m, 5H, H-6bA, H-6bB, H-6bC, H-6bD, H-2B), 4.07–3.96 (m, 5H, H-4A, H-4C, H-3A, H-3B, H-5B or D), 3.90 (t, 1H, H-3C), 3.88–3.82 (m, 2H, H-5B or D, H-5A or C), 3.76 (m, 1H, H-5A or C), 3.68 (s, 3H, Me (OMP)), 3.31 (m, 2H, CH2(Lev)), 2.59 (m, 2H, CH2(Lev)), 2.13 (s, 3H, CH3(Lev)), 1.25, 1.20 (2s, 18H, C(CH3)3); 13C-NMR (100 MHz, CD3OD): δ 209.7, 180.1, 179.7, 173.9, 167.6, 167.1 (6 × CO), 159.4 (q, JC,F = 37.1 Hz, COCF3), 159.2 (q, JC,F = 37.4 Hz, COCF3), 156.9–115.4 (Ar), 117.4 (q, JC,F = 287.3 Hz, COCF3), 117.2 (q, JC,F = 288.2 Hz, COCF3), 102.5 (C-1C), 101.3 (C-1D), 101.1 (C-1A), 101.0 (C-1B), 81.1 (C-3C), 80.9 (C-3A), 77.9 (C-4A), 77.2 (C-4C), 76.8 (C-4B), 76.4 (C-3B), 76.1, 75.4 (2 × CH2(Bn), 75.0 (C-5A or C), 74.7, 74.6, 74.4 (C-5A or C, C-5B or D, C-2C, C-2A), 74.0 (C-5B or D), 72.4 (C-4D), 71.9 (C-3D), 68.5, 67.9 (C-6B, C-6D), 64.4, 64.2 (C-6A, C-6C), 56.0 (Me (OMP)), 54.1 (C-2B), 52.5 (C-2D), 40.0, 39.9 (C(CH3)3), 38.5 (CH2(Lev)), 29.6 (CH3(Lev)), 29.2 CH2(Lev)), 27.8, 27.6 (C(CH3)3); 19F NMR (376 MHz, CD3OD): δ −76.5 (s, 3F), −77.1 (s, 3F); ESI MS: m/z: calcd for C78H86F6N2O40S4Na3: 2001.3; found: 2000.7 [M + 3Na]−.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to obtain 3. This compound was then dissolved in H2O (2 mL) and Amberlite IR-120 H+ resin was added (pH 3.0). The mixture was immediately filtered, treated with 0.04 M NaOH (pH 7.1) and lyophilised. The white solid was finally eluted from a column of Dowex 50WX4-Na+ (H2O) to obtain 3 as sodium salt (12.8 mg, 85%). TLC (EtOAc–MeOH–H2O 12
1) to obtain 3. This compound was then dissolved in H2O (2 mL) and Amberlite IR-120 H+ resin was added (pH 3.0). The mixture was immediately filtered, treated with 0.04 M NaOH (pH 7.1) and lyophilised. The white solid was finally eluted from a column of Dowex 50WX4-Na+ (H2O) to obtain 3 as sodium salt (12.8 mg, 85%). TLC (EtOAc–MeOH–H2O 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) Rf 0.40; 1H-NMR (400 MHz, D2O): δ 7.61–7.38 (m, 10H, Ar), 7.13 (m, 2H, Ar), 7.00 (m, 2H, Ar), 5.05 (m, 2H, H-1A, CH2(Bn)), 4.98 (d, 1H, CH2(Bn)), 4.94 (d, 1H, J3,4 = 2.8 Hz, H-4B), 4.88, 4.85 (2d, 2H, CH2(Bn)), 4.74 (d, 1H, J3,4 = 3.3 Hz, H-4D), 4.73 (d, 1H, J1,2 = 8.5 Hz, H-1B), 4.65 (d, 1H, J1,2 = 8.2 Hz, H-1D), 4.53 (d, 1H, J1,2 = 7.9 Hz, H-1C), 4.26 (dd, 1H, J5,6a = 4.7 Hz, J6a,6b = 11.0 Hz, H-6aB or D), 4.21 (dd, 1H, J5,6a = 4.0 Hz, J6a,6b = 11.2 Hz, H-6aB or D), 4.17–4.11 (m, 2H, H-2B, H-6bB or D), 4.08–3.87 (m, 10H, H-6bB or D, H-3B, H-2D, H-5B, H-5D, H-4A, H-4C, H-6aA, H-6aC, H-3D), 3.83 (s, 3H, Me (OMP)), 3.81 (t, 1H, J1,2 = J2,3 = 9.1 Hz, H-3A), 3.76–3.67 (m, 5H, H-6bA, H-6bC, H-2A, H-5A, H-3C), 3.54 (br t, 1H, H-2C), 3.51 (m, 1H, H-5C), 2.07, 2.06 (2s, 6H, NHAc); 13C–NMR (100 MHz, D2O; selected data from HSQC experiment): δ 104.0 (C-1C), 100.8 (C–1A), 100.5 (C-1D), 100.0 (C-1B), 81.3 (C-3A, C-3C), 76.4 (C-3B), 76.1 (C-4B), 75.2 (C-4D), 75.1 (C-5A), 75.0 (C-4C), 74.9 (C-4A), 74.8 (C-5C), 73.9, 73.3 (2 × CH2(Bn), 72.3 (C–2A), 72.0 (C-5B, C-2C), 71.7 (C-5D), 69.6 (C-3D), 67.4, 66.8 (C-6B, C-6D), 60.0 (C-6A, C-6C), 55.6 (Me (OMP)), 52.9 (C-2D), 51.9 (C-2B); ESI MS: m/z: calcd for C49H62N2O34S4Na22−: 698.1; found: 697.7 [M + 2Na]2−.
3) Rf 0.40; 1H-NMR (400 MHz, D2O): δ 7.61–7.38 (m, 10H, Ar), 7.13 (m, 2H, Ar), 7.00 (m, 2H, Ar), 5.05 (m, 2H, H-1A, CH2(Bn)), 4.98 (d, 1H, CH2(Bn)), 4.94 (d, 1H, J3,4 = 2.8 Hz, H-4B), 4.88, 4.85 (2d, 2H, CH2(Bn)), 4.74 (d, 1H, J3,4 = 3.3 Hz, H-4D), 4.73 (d, 1H, J1,2 = 8.5 Hz, H-1B), 4.65 (d, 1H, J1,2 = 8.2 Hz, H-1D), 4.53 (d, 1H, J1,2 = 7.9 Hz, H-1C), 4.26 (dd, 1H, J5,6a = 4.7 Hz, J6a,6b = 11.0 Hz, H-6aB or D), 4.21 (dd, 1H, J5,6a = 4.0 Hz, J6a,6b = 11.2 Hz, H-6aB or D), 4.17–4.11 (m, 2H, H-2B, H-6bB or D), 4.08–3.87 (m, 10H, H-6bB or D, H-3B, H-2D, H-5B, H-5D, H-4A, H-4C, H-6aA, H-6aC, H-3D), 3.83 (s, 3H, Me (OMP)), 3.81 (t, 1H, J1,2 = J2,3 = 9.1 Hz, H-3A), 3.76–3.67 (m, 5H, H-6bA, H-6bC, H-2A, H-5A, H-3C), 3.54 (br t, 1H, H-2C), 3.51 (m, 1H, H-5C), 2.07, 2.06 (2s, 6H, NHAc); 13C–NMR (100 MHz, D2O; selected data from HSQC experiment): δ 104.0 (C-1C), 100.8 (C–1A), 100.5 (C-1D), 100.0 (C-1B), 81.3 (C-3A, C-3C), 76.4 (C-3B), 76.1 (C-4B), 75.2 (C-4D), 75.1 (C-5A), 75.0 (C-4C), 74.9 (C-4A), 74.8 (C-5C), 73.9, 73.3 (2 × CH2(Bn), 72.3 (C–2A), 72.0 (C-5B, C-2C), 71.7 (C-5D), 69.6 (C-3D), 67.4, 66.8 (C-6B, C-6D), 60.0 (C-6A, C-6C), 55.6 (Me (OMP)), 52.9 (C-2D), 51.9 (C-2B); ESI MS: m/z: calcd for C49H62N2O34S4Na22−: 698.1; found: 697.7 [M + 2Na]2−.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 → CH2Cl2–MeOH 30
1 → CH2Cl2–MeOH 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to obtain 17 as amorphous solid (123 mg, 56%) and unreacted 16 (20 mg, 26%). TLC (toluene–acetone 5
1) to obtain 17 as amorphous solid (123 mg, 56%) and unreacted 16 (20 mg, 26%). TLC (toluene–acetone 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Rf 0.58; [α]20D +21° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3): δ 7.91 (m, 2H, Ar), 7.56 (m, 1H, Ar), 7.41 (m, 2H, Ar), 7.32 (d, 1H, J2,NH = 9.0 Hz, NH-C), 7.15–7.07 (m, 5H, Ar), 6.92–6.88 (m, 3H, Ar, NH-A), 6.77 (m, 2H, Ar), 5.34 (d, 1H, J1,2 = 8.3 Hz, H-1A), 5.24 (t, 1H, J1,2 = J2,3 = 8.0 Hz, H-2B), 5.17 (d, 1H, J1,2 = 7.6 Hz, H-1B), 4.88 (d, 1H, CH2(Bn)), 4.86 (dd, 1H, J2,3 = 11.0 Hz, J3,4 = 3.0 Hz, H-3C), 4.64 (d, 1H, J1,2 = 8.4 Hz, H-1C), 4.62 (d, 1H, CH2(Bn)), 4.60 (d, 1H, J3,4 = 2.6 Hz, H-4A), 4.55 (d, 1H, H-4C), 4.47–4.39 (m, 2H, H-2C, H–6aB), 4.31–4.24 (m, 2H, H-3A, H-6bB), 4.17–4.08 (m, 4H, H-6aA, H-6bA, H-6aC, H-6bC), 3.99 (m, 1H, H-2A), 3.83 (t, 1H, H-3B), 3.75 (m, 4H, H-4B, Me (OMP)), 3.62 (m, 1H, H-5B), 3.37 (br s, 1H, H-5A), 3.30 (br s, 1H, H-5C), 2.74 (m, 2H, CH2(Lev)), 2.58 (m, 2H, CH2(Lev)), 2.17 (s, 3H, CH3(Lev)), 1.22 (s, 9H, C(CH3)3), 1.07, 1.00 (2s, 36H, Si(C(CH3)3)2); 13C-NMR (100 MHz, CDCl3): δ 206.7, 179.1, 172.7, 165.2 (4 × CO), 157.8 (q, JC,F = 37.5 Hz, COCF3), 157.7 (q, JC,F = 36.9 Hz, COCF3), 156.0–114.6 (Ar), 115.9 (q, JC,F = 288.4 Hz, COCF3), 115.6 (q, JC,F = 288.4 Hz, COCF3), 101.7 (C-1C), 100.0 (C-1B), 99.4 (C-1A), 80.7 (C-3B), 78.7 (C-4B), 75.0 (C-3A), 74.4 (CH2(Bn)), 74.1 (C-5B), 73.2 (C-2B, C-4A), 72.8 (C-3C), 71.9 (C-5C), 71.6 (C-5A), 69.5 (C-4C), 67.1 (C-6A), 66.7 (C-6C), 63.2 (C-6B), 55.7 (Me (OMP)), 53.9 (C-2A), 51.0 (C-2C), 39.1 (C(CH3)3), 38.0 (CH2(Lev)), 29.8 (CH3(Lev)), 28.3 (CH2(Lev)), 27.7, 27.6, 27.4, 27.3 (Si(C(CH3)3)2, C(CH3)3), 23.4, 20.9, 20.6 (Si(C(CH3)3)2); HR MS: m/z: calcd for C69H94F6N2O21Si2Na: 1479.5690; found: 1479.5671 [M + Na]+.
1) Rf 0.58; [α]20D +21° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3): δ 7.91 (m, 2H, Ar), 7.56 (m, 1H, Ar), 7.41 (m, 2H, Ar), 7.32 (d, 1H, J2,NH = 9.0 Hz, NH-C), 7.15–7.07 (m, 5H, Ar), 6.92–6.88 (m, 3H, Ar, NH-A), 6.77 (m, 2H, Ar), 5.34 (d, 1H, J1,2 = 8.3 Hz, H-1A), 5.24 (t, 1H, J1,2 = J2,3 = 8.0 Hz, H-2B), 5.17 (d, 1H, J1,2 = 7.6 Hz, H-1B), 4.88 (d, 1H, CH2(Bn)), 4.86 (dd, 1H, J2,3 = 11.0 Hz, J3,4 = 3.0 Hz, H-3C), 4.64 (d, 1H, J1,2 = 8.4 Hz, H-1C), 4.62 (d, 1H, CH2(Bn)), 4.60 (d, 1H, J3,4 = 2.6 Hz, H-4A), 4.55 (d, 1H, H-4C), 4.47–4.39 (m, 2H, H-2C, H–6aB), 4.31–4.24 (m, 2H, H-3A, H-6bB), 4.17–4.08 (m, 4H, H-6aA, H-6bA, H-6aC, H-6bC), 3.99 (m, 1H, H-2A), 3.83 (t, 1H, H-3B), 3.75 (m, 4H, H-4B, Me (OMP)), 3.62 (m, 1H, H-5B), 3.37 (br s, 1H, H-5A), 3.30 (br s, 1H, H-5C), 2.74 (m, 2H, CH2(Lev)), 2.58 (m, 2H, CH2(Lev)), 2.17 (s, 3H, CH3(Lev)), 1.22 (s, 9H, C(CH3)3), 1.07, 1.00 (2s, 36H, Si(C(CH3)3)2); 13C-NMR (100 MHz, CDCl3): δ 206.7, 179.1, 172.7, 165.2 (4 × CO), 157.8 (q, JC,F = 37.5 Hz, COCF3), 157.7 (q, JC,F = 36.9 Hz, COCF3), 156.0–114.6 (Ar), 115.9 (q, JC,F = 288.4 Hz, COCF3), 115.6 (q, JC,F = 288.4 Hz, COCF3), 101.7 (C-1C), 100.0 (C-1B), 99.4 (C-1A), 80.7 (C-3B), 78.7 (C-4B), 75.0 (C-3A), 74.4 (CH2(Bn)), 74.1 (C-5B), 73.2 (C-2B, C-4A), 72.8 (C-3C), 71.9 (C-5C), 71.6 (C-5A), 69.5 (C-4C), 67.1 (C-6A), 66.7 (C-6C), 63.2 (C-6B), 55.7 (Me (OMP)), 53.9 (C-2A), 51.0 (C-2C), 39.1 (C(CH3)3), 38.0 (CH2(Lev)), 29.8 (CH3(Lev)), 28.3 (CH2(Lev)), 27.7, 27.6, 27.4, 27.3 (Si(C(CH3)3)2, C(CH3)3), 23.4, 20.9, 20.6 (Si(C(CH3)3)2); HR MS: m/z: calcd for C69H94F6N2O21Si2Na: 1479.5690; found: 1479.5671 [M + Na]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) was added. After stirring at room temperature for 2 h, the reaction mixture was quenched with acetone (0.8 mL). The mixture was diluted with CH2Cl2 and washed with 1 M HCl aqueous solution, saturated NaHCO3 aqueous solution and H2O. The organic layer was dried (MgSO4), filtered and concentrated in vacuo. The residue was purified by column chromatography (toluene–acetone 9
2) was added. After stirring at room temperature for 2 h, the reaction mixture was quenched with acetone (0.8 mL). The mixture was diluted with CH2Cl2 and washed with 1 M HCl aqueous solution, saturated NaHCO3 aqueous solution and H2O. The organic layer was dried (MgSO4), filtered and concentrated in vacuo. The residue was purified by column chromatography (toluene–acetone 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to afford 18 (168 mg, 95%) as a white amorphous solid. TLC (toluene–acetone 9
1) to afford 18 (168 mg, 95%) as a white amorphous solid. TLC (toluene–acetone 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Rf 0.15; [α]20D +10° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3): δ 7.89 (m, 2H, Ar), 7.56 (m, 1H, Ar), 7.40 (m, 2H, Ar), 7.32 (d, 1H, J2,NH = 9.2 Hz, NH-C), 7.15–7.04 (m, 5H, Ar), 6.90 (m, 2H, Ar), 6.83 (d, 1H, J2,NH = 6.9 Hz, NH-A), 6.77 (m, 2H, Ar), 5.35 (d, 1H, J1,2 = 8.2 Hz, H-1A), 5.23 (t, 1H, J1,2 = J2,3 = 8.1 Hz, H-2B), 5.18 (d, 1H, J1,2 = 7.8 Hz, H-1B), 4.92 (d, 1H, CH2(Bn)), 4.59 (m, 2H, H-4A, CH2(Bn)), 4.45–4.33 (m, 4H, H-6aB, H-1C, H-4C, H-6bB), 4.31 (dd, 1H, J2,3 = 11.2 Hz, J3,4 = 2.6 Hz, H-3A), 4.26–4.13 (m, 5H, H-6aC, H-2C, H-6aA, H-6bA, H-6bC), 3.98 (m, 1H, H-2A), 3.80 (t, 1H, J3,4 = 8.2 Hz, H-3B), 3.75 (s, 3H, Me (OMP)), 3.70 (t, 1H, J4,5 = 7.9 Hz, H–4B), 3.63–3.57 (m, 2H, H-5B, H-3C), 3.38 (br s, 1H, H-5A), 3.32 (br s, 1H, H-5C), 2.79 (d, 1H, J3,OH = 11.6 Hz, OH), 1.23 (s, 9H, C(CH3)3), 1.07, 1.04, 1.01 (4s, 36H, Si(C(CH3)3)2); 13C-NMR (100 MHz, CDCl3): δ 179.4, 165.2 (2 × CO), 158.3 (q, JC,F = 37.4 Hz, COCF3), 157.7 (q, JC,F = 36.9 Hz, COCF3), 156.0–114.6 (Ar), 116.03 (q, JC,F = 288.6 Hz, COCF3), 115.6 (q, JC,F = 289.1 Hz, COCF3), 102.2 (C-1C), 100.0 (C-1B), 99.3 (C-1A), 80.7 (C-3B), 79.2 (C-4B), 75.1 (C-3A), 75.0 (CH2(Bn)), 74.2 (C-5B), 73.3 (C-4A), 73.1 (C-2B), 72.3 (C-5C), 71.9 (C-3C), 71.8 (C-4C), 71.6 (C-5A), 67.1 (C-6A), 66.5 (C-6C), 62.8 (C-6B), 55.7 (Me (OMP)), 54.0 (C-2A), 53.9 (C-2C), 39.2 (C(CH3)3), 27.7, 27.6, 27.4, 27.3 (Si(C(CH3)3)2, C(CH3)3), 23.5, 23.4, 20.9, 20.7 (Si(C(CH3)3)2); HR MS: m/z: calcd for C64H88F6N2O19Si2Na: 1381.5322; found: 1381.5305 [M + Na]+.
1) Rf 0.15; [α]20D +10° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3): δ 7.89 (m, 2H, Ar), 7.56 (m, 1H, Ar), 7.40 (m, 2H, Ar), 7.32 (d, 1H, J2,NH = 9.2 Hz, NH-C), 7.15–7.04 (m, 5H, Ar), 6.90 (m, 2H, Ar), 6.83 (d, 1H, J2,NH = 6.9 Hz, NH-A), 6.77 (m, 2H, Ar), 5.35 (d, 1H, J1,2 = 8.2 Hz, H-1A), 5.23 (t, 1H, J1,2 = J2,3 = 8.1 Hz, H-2B), 5.18 (d, 1H, J1,2 = 7.8 Hz, H-1B), 4.92 (d, 1H, CH2(Bn)), 4.59 (m, 2H, H-4A, CH2(Bn)), 4.45–4.33 (m, 4H, H-6aB, H-1C, H-4C, H-6bB), 4.31 (dd, 1H, J2,3 = 11.2 Hz, J3,4 = 2.6 Hz, H-3A), 4.26–4.13 (m, 5H, H-6aC, H-2C, H-6aA, H-6bA, H-6bC), 3.98 (m, 1H, H-2A), 3.80 (t, 1H, J3,4 = 8.2 Hz, H-3B), 3.75 (s, 3H, Me (OMP)), 3.70 (t, 1H, J4,5 = 7.9 Hz, H–4B), 3.63–3.57 (m, 2H, H-5B, H-3C), 3.38 (br s, 1H, H-5A), 3.32 (br s, 1H, H-5C), 2.79 (d, 1H, J3,OH = 11.6 Hz, OH), 1.23 (s, 9H, C(CH3)3), 1.07, 1.04, 1.01 (4s, 36H, Si(C(CH3)3)2); 13C-NMR (100 MHz, CDCl3): δ 179.4, 165.2 (2 × CO), 158.3 (q, JC,F = 37.4 Hz, COCF3), 157.7 (q, JC,F = 36.9 Hz, COCF3), 156.0–114.6 (Ar), 116.03 (q, JC,F = 288.6 Hz, COCF3), 115.6 (q, JC,F = 289.1 Hz, COCF3), 102.2 (C-1C), 100.0 (C-1B), 99.3 (C-1A), 80.7 (C-3B), 79.2 (C-4B), 75.1 (C-3A), 75.0 (CH2(Bn)), 74.2 (C-5B), 73.3 (C-4A), 73.1 (C-2B), 72.3 (C-5C), 71.9 (C-3C), 71.8 (C-4C), 71.6 (C-5A), 67.1 (C-6A), 66.5 (C-6C), 62.8 (C-6B), 55.7 (Me (OMP)), 54.0 (C-2A), 53.9 (C-2C), 39.2 (C(CH3)3), 27.7, 27.6, 27.4, 27.3 (Si(C(CH3)3)2, C(CH3)3), 23.5, 23.4, 20.9, 20.7 (Si(C(CH3)3)2); HR MS: m/z: calcd for C64H88F6N2O19Si2Na: 1381.5322; found: 1381.5305 [M + Na]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 → CH2Cl2–acetone 30
1 → CH2Cl2–acetone 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to obtain 19 as a white amorphous solid (81 mg, 52%), and unreacted 18 (22 mg, 24%). TLC (toluene–acetone 9
1) to obtain 19 as a white amorphous solid (81 mg, 52%), and unreacted 18 (22 mg, 24%). TLC (toluene–acetone 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Rf 0.31; [α]20D +25° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3): δ 7.92 (m, 4H, Ar), 7.54 (m, 2H, Ar), 7.39 (m, 4H, Ar), 7.22 (m, 2H, NH), 7.15–7.06 (m, 10H, Ar), 7.00 (d, 1H, J2,NH = 7.2 Hz, NH), 6.91 (m, 2H, Ar), 6.77 (m, 2H, Ar), 5.34 (d, 1H, J1,2 = 8.3 Hz, H-1A), 5.21 (m, 2H, H-1B or D, H-2B or D), 5.16 (t, 1H, J1,2 = J2,3 = 7.5 Hz, H-2B or D), 4.99 (d, 1H, J1,2 = 7.4 Hz, H-1B or D), 4.89–4.82 (m, 3H, H-1C, H-3E, CH2(Bn)), 4.80 (d, 1H, CH2(Bn)), 4.64–4.59 (m, 4H, CH2(Bn), H-1E, H-4A), 4.54 (br s, 1H, H-4E), 4.52 (m, 1H, H-6aB or D), 4.49 (br s, 1H, H-4C), 4.45–4.37 (m, 2H, H-2E, H-6aB or D), 4.31 (dd, 1H, J5,6 = 4.8 Hz, J6a,6b = 12.1 Hz, H-6bB or D), 4.25 (dd, 1H, J2,3 = 11.3 Hz, J3,4 = 2.5 Hz, H-3A), 4.17–4.13 (m, 3H, H-6aA, H-6bA, H-6aE), 4.09–4.03 (m, 2H, H-6bE, H-6bB or D), 4.01–3.95 (m, 5H, H-2A, H-2C, H-3C, H-6aC, H-6bC), 3.83–3.79 (m, 2H, H-3B, H-3D), 3.76–3.71 (m, 5H, Me (OMP), H-4B, H–4D), 3.64–3.57 (m, 2H, H-5B, H-5D), 3.35 (br s, 1H, H-5A), 3.28 (br s, 1H, H-5E), 3.07 (br s, 1H, H-5C), 2.75 (m, 2H, CH2(Lev)), 2.59 (m, 2H, CH2(Lev)), 2.17 (s, 3H, CH3(Lev)), 1.20, 1.18 (2s, 18H, C(CH3)3), 1.06, 1.00, 0.98, 0.96, 0.95, 0.93 (6s, 54H, Si(C(CH3)3)2); 13C-NMR (100 MHz, CDCl3): δ 206.6, 179.0, 178.5, 172.7, 165.3, 165.2 (6 × CO), 157.9 (q, JC,F = 36.9 Hz, COCF3), 157.8 (q, JC,F = 36.9 Hz, COCF3), 157.7 (q, JC,F = 36.9 Hz, COCF3), 155.9–114.5 (Ar), 115.9 (q, JC,F = 288.1 Hz, COCF3), 115.7 (q, JC,F = 288.6 Hz, COCF3), 115.6 (q, JC,F = 288.6 Hz, COCF3), 101.6 (C-1E), 100.6 (C-1C), 100.1, 99.5 (C–1B, C-1D), 99.4 (C–1A), 81.1, 80.6 (C-3B, C-3D), 78.1, 78.0 (C-4B, C-4D), 75.4 (C-3C), 74.8 (C-3A), 74.5, 74.4 (C-5B, C-5D), 74.2, 73.7 (2 × CH2(Bn)), 73.4 (C-2B), 73.3, 73.2 (C-2D, C-4A), 72.8 (C-3E), 72.4 (C-4C), 71.9 (C-5E, C-5C), 71.6 (C-5A), 69.5 (C-4E), 67.1 (C-6A), 66.8, 66.6 (C-6C, C-6E), 63.6, 62.9 (C-6B, C-6D), 55.7 (Me (OMP)), 53.8, 53.3 (C-2A, C-2C), 51.0 (C-2E), 39.1, 39.0 (C(CH3)3), 38.0 (CH2(Lev)), 29.8 (CH3(Lev)), 28.3 (CH2(Lev)), 27.7, 27.6, 27.5, 27.4, 27.3 (Si(C(CH3)3)2, C(CH3)3), 23.4, 23.3, 20.9, 20.6 (Si(C(CH3)3)2); 19F NMR (376 MHz, CDCl3): δ −75.5 (s, 3F), −75.8 (s, 3F), −76.0 (s, 3F); HR MS: m/z: calcd for C110H148F9N3O33Si3Na: 2316.9062; found: 2316.9160 [M + Na]+.
1) Rf 0.31; [α]20D +25° (c 1.0, CHCl3); 1H-NMR (400 MHz, CDCl3): δ 7.92 (m, 4H, Ar), 7.54 (m, 2H, Ar), 7.39 (m, 4H, Ar), 7.22 (m, 2H, NH), 7.15–7.06 (m, 10H, Ar), 7.00 (d, 1H, J2,NH = 7.2 Hz, NH), 6.91 (m, 2H, Ar), 6.77 (m, 2H, Ar), 5.34 (d, 1H, J1,2 = 8.3 Hz, H-1A), 5.21 (m, 2H, H-1B or D, H-2B or D), 5.16 (t, 1H, J1,2 = J2,3 = 7.5 Hz, H-2B or D), 4.99 (d, 1H, J1,2 = 7.4 Hz, H-1B or D), 4.89–4.82 (m, 3H, H-1C, H-3E, CH2(Bn)), 4.80 (d, 1H, CH2(Bn)), 4.64–4.59 (m, 4H, CH2(Bn), H-1E, H-4A), 4.54 (br s, 1H, H-4E), 4.52 (m, 1H, H-6aB or D), 4.49 (br s, 1H, H-4C), 4.45–4.37 (m, 2H, H-2E, H-6aB or D), 4.31 (dd, 1H, J5,6 = 4.8 Hz, J6a,6b = 12.1 Hz, H-6bB or D), 4.25 (dd, 1H, J2,3 = 11.3 Hz, J3,4 = 2.5 Hz, H-3A), 4.17–4.13 (m, 3H, H-6aA, H-6bA, H-6aE), 4.09–4.03 (m, 2H, H-6bE, H-6bB or D), 4.01–3.95 (m, 5H, H-2A, H-2C, H-3C, H-6aC, H-6bC), 3.83–3.79 (m, 2H, H-3B, H-3D), 3.76–3.71 (m, 5H, Me (OMP), H-4B, H–4D), 3.64–3.57 (m, 2H, H-5B, H-5D), 3.35 (br s, 1H, H-5A), 3.28 (br s, 1H, H-5E), 3.07 (br s, 1H, H-5C), 2.75 (m, 2H, CH2(Lev)), 2.59 (m, 2H, CH2(Lev)), 2.17 (s, 3H, CH3(Lev)), 1.20, 1.18 (2s, 18H, C(CH3)3), 1.06, 1.00, 0.98, 0.96, 0.95, 0.93 (6s, 54H, Si(C(CH3)3)2); 13C-NMR (100 MHz, CDCl3): δ 206.6, 179.0, 178.5, 172.7, 165.3, 165.2 (6 × CO), 157.9 (q, JC,F = 36.9 Hz, COCF3), 157.8 (q, JC,F = 36.9 Hz, COCF3), 157.7 (q, JC,F = 36.9 Hz, COCF3), 155.9–114.5 (Ar), 115.9 (q, JC,F = 288.1 Hz, COCF3), 115.7 (q, JC,F = 288.6 Hz, COCF3), 115.6 (q, JC,F = 288.6 Hz, COCF3), 101.6 (C-1E), 100.6 (C-1C), 100.1, 99.5 (C–1B, C-1D), 99.4 (C–1A), 81.1, 80.6 (C-3B, C-3D), 78.1, 78.0 (C-4B, C-4D), 75.4 (C-3C), 74.8 (C-3A), 74.5, 74.4 (C-5B, C-5D), 74.2, 73.7 (2 × CH2(Bn)), 73.4 (C-2B), 73.3, 73.2 (C-2D, C-4A), 72.8 (C-3E), 72.4 (C-4C), 71.9 (C-5E, C-5C), 71.6 (C-5A), 69.5 (C-4E), 67.1 (C-6A), 66.8, 66.6 (C-6C, C-6E), 63.6, 62.9 (C-6B, C-6D), 55.7 (Me (OMP)), 53.8, 53.3 (C-2A, C-2C), 51.0 (C-2E), 39.1, 39.0 (C(CH3)3), 38.0 (CH2(Lev)), 29.8 (CH3(Lev)), 28.3 (CH2(Lev)), 27.7, 27.6, 27.5, 27.4, 27.3 (Si(C(CH3)3)2, C(CH3)3), 23.4, 23.3, 20.9, 20.6 (Si(C(CH3)3)2); 19F NMR (376 MHz, CDCl3): δ −75.5 (s, 3F), −75.8 (s, 3F), −76.0 (s, 3F); HR MS: m/z: calcd for C110H148F9N3O33Si3Na: 2316.9062; found: 2316.9160 [M + Na]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) Rf 0.32; 1H-NMR (400 MHz, CDCl3/CD3OD 4
1) Rf 0.32; 1H-NMR (400 MHz, CDCl3/CD3OD 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): δ 7.93 (m, 4H, Ar), 7.59 (m, 2H, Ar), 7.43 (m, 4H, Ar), 7.12–6.99 (m, 10H, Ar), 6.85 (m, 2H, Ar), 6.71 (m, 2H, Ar), 5.07–5.00 (m, 2H, H-2B, H-2D), 4.99 (d, 1H, J1,2 = 8.5 Hz, H-1A), 4.97, 4.91 (2d, 2H, CH2(Bn)), 4.86 (dd, 1H, J2,3 = 11.1 Hz, J3,4 = 3.2 Hz, H-3E), 4.71 (d, 1H, J1,2 = 7.9 Hz, H-1B or D), 4.66 (d, 1H, J1,2 = 7.9 Hz, H-1B or D), 4.53–4.46 (m, 5H, H-1C, H-1E, CH2(Bn), H-6aB, H-6aD), 4.43–4.38 (m, 2H, H-2E, CH2(Bn)), 4.13 (dd, 1H, J2,3 = 11.1 Hz, J3,4 = 2.9 Hz, H-3A), 4.04 (d, 1H, H-4A), 4.03–3.94 (m, 4H, H-2C, H-4E, H-4C, H-6bB or D), 3.94–3.76 (m, 8H, H-4B, H-4D, H-2A, H-6bB or D, H-3C, H-3B or D, H-6aA or C or E), 3.74–3.68 (m, 6H, H-3B or D, H-6aA or C or E, H-6bA or C or E, Me (OMP)), 3.59 (ddd, 1H, J4,5 = 9.5 Hz, J5,6 = 1.9 and 5.7 Hz, H-5B or D), 3.55–3.44 (m, 5H, H-5A, H-5C, H-5E, H-6bA or C or E), 2.84–2.45 (m, 4H, CH2(Lev)), 2.15 (s, 3H, CH3(Lev)), 1.20, 1.17 (2s, 18H, C(CH3)3); 13C-NMR (100 MHz, CDCl3/CD3OD 4
1): δ 7.93 (m, 4H, Ar), 7.59 (m, 2H, Ar), 7.43 (m, 4H, Ar), 7.12–6.99 (m, 10H, Ar), 6.85 (m, 2H, Ar), 6.71 (m, 2H, Ar), 5.07–5.00 (m, 2H, H-2B, H-2D), 4.99 (d, 1H, J1,2 = 8.5 Hz, H-1A), 4.97, 4.91 (2d, 2H, CH2(Bn)), 4.86 (dd, 1H, J2,3 = 11.1 Hz, J3,4 = 3.2 Hz, H-3E), 4.71 (d, 1H, J1,2 = 7.9 Hz, H-1B or D), 4.66 (d, 1H, J1,2 = 7.9 Hz, H-1B or D), 4.53–4.46 (m, 5H, H-1C, H-1E, CH2(Bn), H-6aB, H-6aD), 4.43–4.38 (m, 2H, H-2E, CH2(Bn)), 4.13 (dd, 1H, J2,3 = 11.1 Hz, J3,4 = 2.9 Hz, H-3A), 4.04 (d, 1H, H-4A), 4.03–3.94 (m, 4H, H-2C, H-4E, H-4C, H-6bB or D), 3.94–3.76 (m, 8H, H-4B, H-4D, H-2A, H-6bB or D, H-3C, H-3B or D, H-6aA or C or E), 3.74–3.68 (m, 6H, H-3B or D, H-6aA or C or E, H-6bA or C or E, Me (OMP)), 3.59 (ddd, 1H, J4,5 = 9.5 Hz, J5,6 = 1.9 and 5.7 Hz, H-5B or D), 3.55–3.44 (m, 5H, H-5A, H-5C, H-5E, H-6bA or C or E), 2.84–2.45 (m, 4H, CH2(Lev)), 2.15 (s, 3H, CH3(Lev)), 1.20, 1.17 (2s, 18H, C(CH3)3); 13C-NMR (100 MHz, CDCl3/CD3OD 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): δ 208.7, 179.3, 179.2, 172.7, 166.0, 165.9 (6 × CO), 158.4 (m, 3 × COCF3), 155.7–114.7 (Ar), 116.3 (q, JC,F = 287.5 Hz, COCF3), 115.9 (q, JC,F = 287.5 Hz, COCF3), 115.8 (q, JC,F = 287.5 Hz, COCF3), 101.3 (C-1B, C-1D), 100.5, 100.1 (C-1C, C-1E), 99.8 (C-1A), 80.7, 80.6 (C-3B, C-3D), 79.0 (C-3C), 78.2 (C-3A), 76.9, 76.8, 76.7 (2 × CH2(Bn), C-5B or D, C-5A or C or E), 75.2, 75.1 (C-5A or C or E, C-4B or D), 74.3 (C-4B or D), 73.8, 73.7 (C-5B or D, C-5A or C or E), 73.2 (C-3E), 73.0 (C-2B, C-2D), 68.2 (C-4A, C-4C), 66.3 (C-4E), 62.5, 62.2, 61.8, 61.6 (C–6A, C–6B, C-6C, C-6D, C-6E), 55.8 (Me (OMP)), 52.9 (C–2A), 52.7 (C-2C), 51.3 (C-2E), 39.2 (C(CH3)3), 38.1 (CH2(Lev)), 29.7 (CH3(Lev)), 28.1 (CH2(Lev)), 27.1 (C(CH3)3); HR MS: m/z: calcd for C86H100F9N3O33Na: 1896.5988; found: 1896.5979 [M + Na]+.
1): δ 208.7, 179.3, 179.2, 172.7, 166.0, 165.9 (6 × CO), 158.4 (m, 3 × COCF3), 155.7–114.7 (Ar), 116.3 (q, JC,F = 287.5 Hz, COCF3), 115.9 (q, JC,F = 287.5 Hz, COCF3), 115.8 (q, JC,F = 287.5 Hz, COCF3), 101.3 (C-1B, C-1D), 100.5, 100.1 (C-1C, C-1E), 99.8 (C-1A), 80.7, 80.6 (C-3B, C-3D), 79.0 (C-3C), 78.2 (C-3A), 76.9, 76.8, 76.7 (2 × CH2(Bn), C-5B or D, C-5A or C or E), 75.2, 75.1 (C-5A or C or E, C-4B or D), 74.3 (C-4B or D), 73.8, 73.7 (C-5B or D, C-5A or C or E), 73.2 (C-3E), 73.0 (C-2B, C-2D), 68.2 (C-4A, C-4C), 66.3 (C-4E), 62.5, 62.2, 61.8, 61.6 (C–6A, C–6B, C-6C, C-6D, C-6E), 55.8 (Me (OMP)), 52.9 (C–2A), 52.7 (C-2C), 51.3 (C-2E), 39.2 (C(CH3)3), 38.1 (CH2(Lev)), 29.7 (CH3(Lev)), 28.1 (CH2(Lev)), 27.1 (C(CH3)3); HR MS: m/z: calcd for C86H100F9N3O33Na: 1896.5988; found: 1896.5979 [M + Na]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The residue was then purified by silica gel column choromatography (EtOAc–MeOH–H2O 20
1). The residue was then purified by silica gel column choromatography (EtOAc–MeOH–H2O 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 → EtOAc–MeOH–H2O 16
3 → EtOAc–MeOH–H2O 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) and finally eluted from a Dowex 50WX2-Na+ column (MeOH) to obtain 6 as sodium salt (55 mg, 78%, white amorphous solid). TLC (EtOAc–MeOH–H2O 16
3) and finally eluted from a Dowex 50WX2-Na+ column (MeOH) to obtain 6 as sodium salt (55 mg, 78%, white amorphous solid). TLC (EtOAc–MeOH–H2O 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) Rf 0.29; 1H-NMR (400 MHz, CD3OD): δ 7.95 (m, 4H, Ar), 7.62 (m, 2H, Ar), 7.45 (m, 4H, Ar), 7.16–6.97 (m, 10H, Ar), 6.96 (m, 2H, Ar), 6.80 (m, 2H, Ar), 5.23 (m, 2H, H-2B, H-2D), 5.05 (dd, 1H, J2,3 = 11.3 Hz, J3,4 = 3.2 Hz, H-3E), 4.99, 4.95 (2d, 2H, CH2(Bn)), 4.89 (m, 2H, H-4E, H-4A or C), 4.87 (d, 1H, J3,4 = 3.0 Hz, H-4A or C), 4.80–4.73 (m, 4H, H-1A, H-1B, H-1D, H-1E), 4.64 (d, 1H, J1,2 = 8.2 Hz, H-1C), 4.55, 4.52 (2d, 2H, CH2(Bn)), 4.44–4.29 (m, 7H, H-6aA, H-6aB, H-6aC, H-6aD, H-6aE, H-2E, H-6bA or C or E), 4.27–4.08 (m, 6H, H-2A, H-6bB, H-6bD, H-2C, H-6bA or C or E), 4.07–3.97 (m, 6H, H-4B, H-4D, H-3A, H-3C, H-5A or C or E), 3.92–3.83 (m, 3H, H-3B, H-3D, H-5A or C or E), 3.78–3.69 (m, 5H, H-5B, H-5D, Me (OMP)), 2.80 (m, 2H, CH2(Lev)), 2.59 (m, 2H, CH2(Lev)), 2.13 (s, 3H, CH3(Lev)), 1.26 (2s, 18H, C(CH3)3); 13C-NMR (100 MHz, CD3OD): δ 209.7, 180.1, 180.0, 173.9, 167.6 (6 × CO), 159.4 (q, JC,F = 37.1 Hz, COCF3), 159.3 (q, JC,F = 37.3 Hz, COCF3), 159.2 (q, JC,F = 37.0 Hz, COCF3), 156.9–115.5 (Ar), 117.4 (q, JC,F = 286.8 Hz, COCF3), 117.2 (q, JC,F = 287.3 Hz, 2 × COCF3), 102.6, 102.5 (C-1B, C-1D), 102.3 (C-1A), 101.2 (C-1E), 101.0 (C-1C), 81.1, 80.8 (C-3B, C-3D), 77.8, 77.2 (C-4B, C-4D), 76.9 (C-3A or C), 76.8 (C-4A, C-4C), 76.3 (C-3A or C), 76.1, 75.5 (2 × CH2(Bn)), 75.0, 74.9 (C-5B, C-5D), 74.8, 74.7, 74.6, 74.5 (C-2B, C-2D, C-5A or C or E), 74.0 (C-5A or C or E), 72.4 (C-4E), 71.9 (C-3E), 68.9, 68.3, 67.8 (C-6A, C-6C, C-6E), 64.4, 64.2 (C-6B, C-6D), 56.0 (Me (OMP)), 54.0 (C-2C), 53.3 (C-2A), 52.5 (C-2E), 40.0 (C(CH3)3), 38.5 (CH2(Lev)), 29.6 (CH3(Lev)), 29.2 (CH2(Lev)), 27.8 (C(CH3)3); 19F NMR (376 MHz, CD3OD): δ −76.5 (s, 3F), −76.9 (s, 3F), −77.1 (s, 3F); ESI MS: m/z: calcd for C86H94F9N3O51S6Na42−: 1219.6; found: 1219.2 [M + 4Na]2−.
3) Rf 0.29; 1H-NMR (400 MHz, CD3OD): δ 7.95 (m, 4H, Ar), 7.62 (m, 2H, Ar), 7.45 (m, 4H, Ar), 7.16–6.97 (m, 10H, Ar), 6.96 (m, 2H, Ar), 6.80 (m, 2H, Ar), 5.23 (m, 2H, H-2B, H-2D), 5.05 (dd, 1H, J2,3 = 11.3 Hz, J3,4 = 3.2 Hz, H-3E), 4.99, 4.95 (2d, 2H, CH2(Bn)), 4.89 (m, 2H, H-4E, H-4A or C), 4.87 (d, 1H, J3,4 = 3.0 Hz, H-4A or C), 4.80–4.73 (m, 4H, H-1A, H-1B, H-1D, H-1E), 4.64 (d, 1H, J1,2 = 8.2 Hz, H-1C), 4.55, 4.52 (2d, 2H, CH2(Bn)), 4.44–4.29 (m, 7H, H-6aA, H-6aB, H-6aC, H-6aD, H-6aE, H-2E, H-6bA or C or E), 4.27–4.08 (m, 6H, H-2A, H-6bB, H-6bD, H-2C, H-6bA or C or E), 4.07–3.97 (m, 6H, H-4B, H-4D, H-3A, H-3C, H-5A or C or E), 3.92–3.83 (m, 3H, H-3B, H-3D, H-5A or C or E), 3.78–3.69 (m, 5H, H-5B, H-5D, Me (OMP)), 2.80 (m, 2H, CH2(Lev)), 2.59 (m, 2H, CH2(Lev)), 2.13 (s, 3H, CH3(Lev)), 1.26 (2s, 18H, C(CH3)3); 13C-NMR (100 MHz, CD3OD): δ 209.7, 180.1, 180.0, 173.9, 167.6 (6 × CO), 159.4 (q, JC,F = 37.1 Hz, COCF3), 159.3 (q, JC,F = 37.3 Hz, COCF3), 159.2 (q, JC,F = 37.0 Hz, COCF3), 156.9–115.5 (Ar), 117.4 (q, JC,F = 286.8 Hz, COCF3), 117.2 (q, JC,F = 287.3 Hz, 2 × COCF3), 102.6, 102.5 (C-1B, C-1D), 102.3 (C-1A), 101.2 (C-1E), 101.0 (C-1C), 81.1, 80.8 (C-3B, C-3D), 77.8, 77.2 (C-4B, C-4D), 76.9 (C-3A or C), 76.8 (C-4A, C-4C), 76.3 (C-3A or C), 76.1, 75.5 (2 × CH2(Bn)), 75.0, 74.9 (C-5B, C-5D), 74.8, 74.7, 74.6, 74.5 (C-2B, C-2D, C-5A or C or E), 74.0 (C-5A or C or E), 72.4 (C-4E), 71.9 (C-3E), 68.9, 68.3, 67.8 (C-6A, C-6C, C-6E), 64.4, 64.2 (C-6B, C-6D), 56.0 (Me (OMP)), 54.0 (C-2C), 53.3 (C-2A), 52.5 (C-2E), 40.0 (C(CH3)3), 38.5 (CH2(Lev)), 29.6 (CH3(Lev)), 29.2 (CH2(Lev)), 27.8 (C(CH3)3); 19F NMR (376 MHz, CD3OD): δ −76.5 (s, 3F), −76.9 (s, 3F), −77.1 (s, 3F); ESI MS: m/z: calcd for C86H94F9N3O51S6Na42−: 1219.6; found: 1219.2 [M + 4Na]2−.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to obtain 4. This compound was then dissolved in H2O (2 mL) and Amberlite IR-120 H+ resin was added (pH 3.0). The mixture was immediately filtered, treated with 0.04 M NaOH (pH 7.1) and eluted from a column of Dowex 50WX4-Na+ (H2O) to obtain 4 as sodium salt (14 mg, 89%) after lyophilization. TLC (EtOAc–MeOH–H2O 10
1) to obtain 4. This compound was then dissolved in H2O (2 mL) and Amberlite IR-120 H+ resin was added (pH 3.0). The mixture was immediately filtered, treated with 0.04 M NaOH (pH 7.1) and eluted from a column of Dowex 50WX4-Na+ (H2O) to obtain 4 as sodium salt (14 mg, 89%) after lyophilization. TLC (EtOAc–MeOH–H2O 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) Rf 0.14; 1H-NMR (400 MHz, D2O): δ 7.58–7.38 (m, 10H, Ar), 7.12 (m, 2H, Ar), 7.00 (m, 2H, Ar), 5.08 (d, 1H, J1,2 = 8.6 Hz, H-1A), 5.01–4.97 (m, 3H, H-4A, CH2(Bn)), 4.93 (d, 1H, J3,4 = 2.7 Hz, H-4C), 4.88–4.82 (m, 2H, CH2(Bn)), 4.76–4.73 (m, 2H, H-4E, H-1C), 4.64 (d, 1H, J1,2 = 8.3 Hz, H-1E), 4.55, 4.52 (2d, 2H, J1,2 = 7.8 Hz, H-1B, H-1D), 4.39–4.19 (m, 6H, H-2A, H-6aA, H-6aC, H-6aE, H-6bA or C or E, H-5A or C or E), 4.16–3.88 (m, 13H, H-3A, H-2C, H-6bA or C or E, H-2E, H-3C, H-5A or C or E, H-3E, H-4B, H-4D, H-6aB, H-6aD), 3.83 (s, 3H, Me (OMP)), 3.76–3.65 (m, 4H, H-6bB, H-6bD, H-3B, H-3D), 3.60–3.46 (m, 4H, H-2B, H-2D, H-5B, H-5D), 2.07, 2.06, 2.05 (3s, 9H, NHAc); 13C-NMR (100 MHz, D2O; selected data from HSQC experiment): δ 104.0 (C-1B, C-1D), 100.6 (C-1E), 100.4 (C-1A), 100.1 (C-1C), 81.4 (C-3B, C-3D), 76.5 (C-3C), 76.4 (C-3A), 76.2 (C-4C), 76.0 (C-4A), 75.2 (C-4E), 75.0 (C-4B, C-4D), 74.7 (C-5B, C-5D), 73.4 (2 × CH2(Bn)), 72.4 (C-5A or C or E), 72.1 (C-2B, C-2D), 71.8 (C-5A or C or E), 69.6 (C-3E), 67.5, 67.1, 66.8 (C-6A, C-6C, C-6E), 60.0 (C-6B, C-6D), 55.6 (Me (OMP)), 52.9 (C-2E), 51.8 (C-2C), 51.3 (C–2A); ESI MS: m/z: calcd for C57H73N3O45S6Na42−: 901.6; found: 901.1 [M + 4Na]2−.
3) Rf 0.14; 1H-NMR (400 MHz, D2O): δ 7.58–7.38 (m, 10H, Ar), 7.12 (m, 2H, Ar), 7.00 (m, 2H, Ar), 5.08 (d, 1H, J1,2 = 8.6 Hz, H-1A), 5.01–4.97 (m, 3H, H-4A, CH2(Bn)), 4.93 (d, 1H, J3,4 = 2.7 Hz, H-4C), 4.88–4.82 (m, 2H, CH2(Bn)), 4.76–4.73 (m, 2H, H-4E, H-1C), 4.64 (d, 1H, J1,2 = 8.3 Hz, H-1E), 4.55, 4.52 (2d, 2H, J1,2 = 7.8 Hz, H-1B, H-1D), 4.39–4.19 (m, 6H, H-2A, H-6aA, H-6aC, H-6aE, H-6bA or C or E, H-5A or C or E), 4.16–3.88 (m, 13H, H-3A, H-2C, H-6bA or C or E, H-2E, H-3C, H-5A or C or E, H-3E, H-4B, H-4D, H-6aB, H-6aD), 3.83 (s, 3H, Me (OMP)), 3.76–3.65 (m, 4H, H-6bB, H-6bD, H-3B, H-3D), 3.60–3.46 (m, 4H, H-2B, H-2D, H-5B, H-5D), 2.07, 2.06, 2.05 (3s, 9H, NHAc); 13C-NMR (100 MHz, D2O; selected data from HSQC experiment): δ 104.0 (C-1B, C-1D), 100.6 (C-1E), 100.4 (C-1A), 100.1 (C-1C), 81.4 (C-3B, C-3D), 76.5 (C-3C), 76.4 (C-3A), 76.2 (C-4C), 76.0 (C-4A), 75.2 (C-4E), 75.0 (C-4B, C-4D), 74.7 (C-5B, C-5D), 73.4 (2 × CH2(Bn)), 72.4 (C-5A or C or E), 72.1 (C-2B, C-2D), 71.8 (C-5A or C or E), 69.6 (C-3E), 67.5, 67.1, 66.8 (C-6A, C-6C, C-6E), 60.0 (C-6B, C-6D), 55.6 (Me (OMP)), 52.9 (C-2E), 51.8 (C-2C), 51.3 (C–2A); ESI MS: m/z: calcd for C57H73N3O45S6Na42−: 901.6; found: 901.1 [M + 4Na]2−.
FP assays
FP measurements were performed in 384-well microplates (black polystyrene, non-treated, Corning). FP was recorded using a TRIAD multimode microplate reader (from Dynex), with excitation and emission wavelengths of 485 and 535 nm, respectively. The fluorescent probe (a fluorescein labelled heparin-like hexasaccharide previously prepared in our lab)35 was dissolved in PBS buffer (10 mM, pH 7.4). Recombinant human midkine (Peprotech) was dissolved in PBS buffer (10 mM, pH 7.4) containing 1% BSA (bovine serum albumin). Compounds 1–4 were dissolved in PBS buffer (10 mM, pH 7.4). 1 mM stock solutions of compounds 5 and 6 were prepared in PBS/DMSO 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) and serial dilutions were then performed in PBS buffer (10 mM, pH 7.4).
1 (v/v) and serial dilutions were then performed in PBS buffer (10 mM, pH 7.4).
For the determination of the IC50 values, we recorded the FP from wells containing 20 μL of a 125 nM midkine solution and 10 μL of a 40 nM probe solution in the presence of 10 μL of inhibitor solution, with concentrations ranging from 2 mM to 0.1 μM. The microplate was shaked in the dark for 5 min, before reading. The total sample volume in each well was 40 μL and the final buffer composition was PBS + 0.5% BSA. The final concentrations of fluorescent probe and midkine in each well were 10 nM and 63 nM, respectively, while the final inhibitor concentration ranged from 500 μM to 25 nM. The average polarization values of three replicates were plotted against the logarithm of inhibitor concentration. Two control samples were included in the competition experiment. The first one only contained fluorescent probe and afforded the expected minimum polarization value for 100% inhibition; the second one contained midkine and probe, in the absence of inhibitor, and gave the maximum polarization value corresponding to 0% inhibition. Blank wells contained 20 μL of protein solution and 20 μL of a 125 μM inhibitor solution and their measurements were subtracted from all values. The resulting curve was fitted to the equation for a one-site competition: y = A2 + (A1 − A2)/[1 + 10^(x − log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IC50)] where A1 and A2 are the maximal and minimal values of polarization, respectively, and IC50 is the inhibitor concentration that results in 50% inhibition. At least two independent experiments were carried out for each IC50 calculation.
IC50)] where A1 and A2 are the maximal and minimal values of polarization, respectively, and IC50 is the inhibitor concentration that results in 50% inhibition. At least two independent experiments were carried out for each IC50 calculation.
Nuclear magnetic resonance
Experiments were performed in a Bruker Avance III 700 MHz fitted with a QCI 5 mm cryoprobe for 1H, 13C, 15N, 31P with cold preamplifiers in 1H, 13C and 2H channels or in 600 MHz Avance III fitted with a QCI 5 mm cryoprobe for 1H, 13C, 15N, 19F with cold preamplifiers in 1H, 19F, 13C and 2H channels, at 298 K. STD-NMR samples were prepared in 300 μL of 99.9% D2O buffer containing 10 mM phosphate pD 7.5 and 150 mM NaCl. Ligand and midkine concentrations were 1.0 mM and 20 μM, respectively. NMR pulse sequences from the manufacturer library were used for the assignment.STD NMR experiments were carried out with watergate solvent suppression and a 10 ms spin-lock filter after the 90° pulse to reduce residual signals from the protein. For selective protein saturation, cascades of 50 ms Gaussian shaped pulses at a field strength of circa 50 Hz were employed, with a delay of 1 ms between successive pulses.45 The on-resonance and off-resonance frequencies were set to 1.7 ppm and 40 ppm, respectively. Blank experiments were performed to assure the absence of direct saturation of the ligand proton signals. The relaxation delay was properly adjusted so that the experiment time length was kept constant (5.0 s). STD NMR experiments were performed with 512–256 scans. Saturation times were of 1, 1.5, 2.0, 3.0, 4.0 and 5.0 s.
To avoid relaxation interference in STD, STD0 were calculated from the STD initial slopes.48,49 To do so, the evolution of the STD with the saturation time (tsat) was fitted to the equation STD (t) = a(1 − exp(−bt)), where the parameter a represents the asymptotic maximum of the STD build-up curve (STDmax), b is a rate constant related to the relaxation properties of a given proton that measures the speed of the STD build-up (ksat), and t is the saturation time (tsat). Thus, the STD0 values were obtained as the product of the ab coefficients.
Molecular dynamics (MD)
All starting structures were constructed with MAESTRO suite (Schrödinger, LLC, New York, NY, 2019). Partial charges were obtained from GLYCAM50 or derived from the molecular electrostatic potential, MEP, using the RESP method.51 The procedure has been developed under GLYCAM06 force fields available in the R.E.D web server. The partial charges for SO3 groups were established according to the protocol of GLYCAM.The prep and frcmod files were obtained by antechamber and parmchk modules in AmberTools using each residue of each sugar and the set of partial charges. By means of AMBER 12 version, the topology and coordinates initials were built with tLEAP module, where both tetrasacharides and pentasacharides were neutralized with sodium ions and immersed in a TIP3P water box as explicit solvent.52 This procedure was carried out using GLYCAM06 as force files and parm99 parameters.
To achieve system equilibrium, we followed a protocol that consisted in several steps. Firstly, an initial energy minimization of the solute in the water box was performed followed by a minimization of the entire system, including sodium ions. Next, the system was heated from 0 K to 300 K at a constant volume, and then equilibrated at a constant pressure (1 bar). At this point, the final step was the production dynamics simulations, with an overall length from 200 to 500 ns. The coordinates of the trajectories were saved each picosecond. We run the first steps of simulations with sander.MPI and the final production with pmemd.MPI extension, belonging to AMBER program.
For those mimetics that presented some distortions in their conformations, we carried out time-average distance restrained MD (MD-tar) simulations. This procedure has a shorter length (8–50 ns). NOE-derived distances between H1–H3, H1–H5 and H3–H5 protons were employed as restraints. The trajectories of all simulations were analyzed with cpptraj module of Amber12. All protocols have been described in detail in the ESI.†
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Spanish Ministry of Economy and Competitiveness and the Spanish Ministry of Science and Innovation (CTQ2015-70134-P, PGC2018-099497-B100), Junta de Andalucía (P12-BIO-1938) and the European Union (ERDF) for financial support. 700 MHz NMR experiments have been performed in the NMR service of the US (CITIUS).Notes and references
- S. Mizumoto, S. Yamada and K. Sugahara, Curr. Opin. Struct. Biol., 2015, 34, 35–42 CrossRef PubMed.
- K. Sugahara, T. Mikami, T. Uyama, S. Mizuguchi, K. Nomura and H. Kitagawa, Curr. Opin. Struct. Biol., 2003, 13, 612–620 CrossRef CAS PubMed.
- C. I. Gama, S. E. Tully, N. Sotogaku, P. M. Clark, M. Rawat, N. Vaidehi, W. A. Goddard, A. Nishi and L. C. Hsieh-Wilson, Nat. Chem. Biol., 2006, 2, 467–473 CrossRef CAS PubMed.
- C. J. Rogers, P. M. Clark, S. E. Tully, R. Abrol, K. C. Garcia, W. A. Goddard III and L. C. Hsieh-Wilson, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 9747–9752 CrossRef CAS PubMed.
- T. Muramatsu, Curr. Pharm. Des., 2011, 17, 410–423 CrossRef CAS.
- K. Kadomatsu, S. Kishida and S. Tsubota, J. Biochem., 2013, 153, 511–521 CrossRef CAS.
- E. Bedini and M. Parrilli, Carbohydr. Res., 2012, 356, 75–85 CrossRef CAS PubMed.
- M. Mende, C. Bednarek, M. Wawryszyn, P. Sauter, M. B. Biskup, U. Schepers and S. Braese, Chem. Rev., 2016, 116, 8193–8255 CrossRef CAS.
- X. Zhang, H. Liu, W. Yao, X. Meng and Z. Li, J. Org. Chem., 2019, 84, 7418–7425 CrossRef CAS.
- S. Jadhav, V. Gulumkar, P. Deshpande, E. T. Coffey, H. Lonnberg and P. Virta, Bioconjugate Chem., 2018, 29, 2382–2393 CrossRef CAS.
- S. Yang, Q. Liu, G. Y. Zhang, X. X. Zhang, Z. H. Zhao and P. S. Lei, J. Org. Chem., 2018, 83, 5897–5908 CrossRef CAS.
- J.-C. Jacquinet and C. Lopin-Bon, Carbohydr. Res., 2015, 402, 35–43 CrossRef CAS PubMed.
- J.-i. Tamura, N. Tsutsumishita-Nakai, Y. Nakao, M. Kawano, S. Kato, N. Takeda, S. Nadanaka and H. Kitagawa, Bioorg. Med. Chem. Lett., 2012, 22, 1371–1374 CrossRef CAS.
- C. Solera, G. Macchione, S. Maza, M. M. Kayser, F. Corzana, J. L. de Paz and P. M. Nieto, Chem. – Eur. J., 2016, 22, 2356–2369 CrossRef CAS.
- S. Ramadan, T. Li, W. Yang, J. Zhang, Z. Rashidijahanabad, Z. Tan, N. Parameswaran and X. Huang, ACS Cent. Sci., 2020, 6, 913–920 CrossRef CAS PubMed.
- N. S. Gandhi and R. L. Mancera, Drug Discovery Today, 2010, 15, 1058–1069 CrossRef CAS.
- P. Dominguez-Rodriguez, C. Vives, M. Thepaut, F. Fieschi, P. M. Nieto, J. L. de Paz and J. Rojo, Biomacromolecules, 2020, 21, 2726–2734 CrossRef CAS.
- S. Yang, H. Zhang, Q. Liu, S. Sun, P. Lei, Z. Zhao, L. Wu and Y. Wang, Future Med. Chem., 2019, 11, 1403–1415 CrossRef CAS PubMed.
- X. Zhang, W. Yao, X. J. Xu, H. F. Sun, J. H. Zhao, X. B. Meng, M. Y. Wu and Z. J. Li, Chem. – Eur. J., 2018, 24, 1694–1700 CrossRef CAS PubMed.
- S.-G. Lee, J. M. Brown, C. J. Rogers, J. B. Matson, C. Krishnamurthy, M. Rawat and L. C. Hsieh-Wilson, Chem. Sci., 2010, 1, 322–325 RSC.
- C.-J. Yeh, C.-C. Ku, W.-C. Lin, C.-Y. Fan, M. M. L. Zulueta, Y. Manabe, K. Fukase, Y.-K. Li and S.-C. Hung, ChemBioChem, 2019, 20, 237–240 CrossRef CAS PubMed.
- J. Revuelta, R. Fuentes, L. Lagartera, M. Jose Hernaiz, A. Bastida, E. Garcia-Junceda and A. Fernandez-Mayoralas, Chem. Commun., 2018, 54, 13455–13458 RSC.
- S. Kohling, J. Blaszkiewicz, G. Ruiz-Gomez, M. I. Fernandez-Bachiller, K. Lemmnitzer, N. Panitz, A. G. Beck-Sickinger, J. Schiller, M. T. Pisabarro and J. Rademann, Chem. Sci., 2019, 10, 866–878 RSC.
- K. D. Johnstone, T. Karoli, L. Liu, K. Dredge, E. Copeman, C. P. Li, K. Davis, E. Hammond, I. Bytheway, E. Kostewicz, F. C. K. Chiu, D. M. Shackleford, S. A. Charman, W. N. Charman, J. Harenberg, T. J. Gonda and V. Ferro, J. Med. Chem., 2010, 53, 1686–1699 CrossRef CAS.
- S. Maza, J. L. de Paz and P. M. Nieto, Molecules, 2019, 24, 1591, DOI:10.3390/molecules24081591.
- E. Lisztes, E. Mezo, F. Demeter, L. Horvath, S. Bosze, B. I. Toth, A. Borbas and M. Herczeg, ChemMedChem, 2021, 16, 1467–1476 CrossRef CAS PubMed.
- T. C. Lim, S. Cai, R. G. Huber, P. J. Bond, P. X. S. Chia, S. L. Khou, S. Gao, S. S. Lee and S.-G. Lee, Chem. Sci., 2018, 9, 7940–7947 RSC.
- R. N. Gangji, N. V. Sankaranarayanan, J. Elste, R. A. Al-Horani, D. K. Afosah, R. Joshi, V. Tiwari and U. R. Desai, ACS Med. Chem. Lett., 2018, 9, 797–802 CrossRef CAS.
- D. K. Afosah, R. A. Al-Horani, N. V. Sankaranarayanan and U. R. Desai, J. Med. Chem., 2017, 60, 641–657 CrossRef CAS PubMed.
- G. Despras, C. Bernard, A. Perrot, L. Cattiaux, A. Prochiantz, H. Lortat-Jacob and J.-M. Mallet, Chem. – Eur. J., 2013, 19, 530–539 CrossRef.
- O. P. Dhamale, C. Zong, K. Al-Mafraji and G.-J. Boons, Org. Biomol. Chem., 2014, 12, 2087–2098 RSC.
- D. J. Sheppard, S. A. Cameron, P. C. Tyler and R. Schworer, Org. Biomol. Chem., 2020, 18, 4728–4733 RSC.
- S. Maza, N. Gandia-Aguado, J. L. de Paz and P. M. Nieto, Bioorg. Med. Chem., 2018, 26, 1076–1085 CrossRef CAS PubMed.
- G. Macchione, S. Maza, M. M. Kayser, J. L. de Paz and P. M. Nieto, Eur. J. Org. Chem., 2014, 3868–3884 CrossRef CAS.
- S. Maza, M. Mar Kayser, G. Macchione, J. Lopez-Prados, J. Angulo, J. L. de Paz and P. M. Nieto, Org. Biomol. Chem., 2013, 11, 3510–3525 RSC.
- Q. Zhang, E. R. van Rijssel, M. T. C. Walvoort, H. S. Overkleeft, G. A. van der Marel and J. D. C. Codee, Angew. Chem., Int. Ed., 2015, 54, 7670–7673 CrossRef CAS.
- H. Lucas, J. E. M. Basten, T. G. Vandinther, D. G. Meuleman, S. F. Vanaelst and C. A. A. Vanboeckel, Tetrahedron, 1990, 46, 8207–8228 CrossRef CAS.
- J. L. de Paz and P. M. Nieto, Org. Biomol. Chem., 2016, 14, 3506–3509 RSC.
- B. Kuhnast, A. El Hadri, R. Boisgard, F. Hinnen, S. Richard, A. Caravano, V. Nancy-Portebois, M. Petitou, B. Tavitian and F. Dolle, Org. Biomol. Chem., 2016, 14, 1915–1920 RSC.
- R. Karuturi, R. A. Al-Horani, S. C. Mehta, D. Gailani and U. R. Desai, J. Med. Chem., 2013, 56, 2415–2428 CrossRef CAS PubMed.
- V. Ferro, L. Liu, K. D. Johnstone, N. Wimmer, T. Karoli, P. Handley, J. Rowley, K. Dredge, C. P. Li, E. Hammond, K. Davis, L. Sarimaa, J. Harenberg and I. Bytheway, J. Med. Chem., 2012, 55, 3804–3813 CrossRef CAS.
- J. O. Duus, C. H. Gotfredsen and K. Bock, Chem. Rev., 2000, 100, 4589–4614 CrossRef CAS PubMed.
- S. Macura and R. R. Ernst, Mol. Phys., 2002, 100, 135–147 CrossRef.
- A. E. Torda, R. M. Scheek and W. F. Vangunsteren, Chem. Phys. Lett., 1989, 157, 289–294 CrossRef CAS.
- B. Meyer and T. Peters, Angew. Chem., Int. Ed., 2003, 42, 864–890 CrossRef CAS.
- F. Ni, Prog. Nucl. Magn. Reson. Spectrosc., 1994, 26, 517–606 CrossRef CAS.
- P. M. Nieto, Front. Mol. Biosci., 2018, 5, 33, DOI:10.3389/fmolb.2018.00033.
- J. Angulo and P. M. Nieto, Eur. Biophys. J., 2011, 40, 1357–1369 CrossRef CAS.
- M. Mayer and T. L. James, J. Am. Chem. Soc., 2004, 126, 4453–4460 CrossRef CAS.
- K. N. Kirschner, A. B. Yongye, S. M. Tschampel, J. Gonzalez-Outeirino, C. R. Daniels, B. L. Foley and R. J. Woods, J. Comput. Chem., 2008, 29, 622–655 CrossRef CAS PubMed.
- F.-Y. Dupradeau, C. Cezard, R. Lelong, E. Stanislawiak, J. Pecher, J. C. Delepine and P. Cieplak, Nucleic Acids Res., 2008, 36, D360–D367 CrossRef CAS.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ob00882j |
| This journal is © The Royal Society of Chemistry 2021 |