Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The structure and biosynthesis of heinamides A1–A3 and B1–B5, antifungal members of the laxaphycin lipopeptide family†
Lassi Matti Petteri
Heinilä
 a,
David Peter
Fewer
a,
David Peter
Fewer
 a,
Jouni Kalevi
Jokela
a,
Jouni Kalevi
Jokela
 a,
Matti
Wahlsten
a,
Matti
Wahlsten
 a,
Xiaodan
Ouyang
a,
Xiaodan
Ouyang
 a,
Perttu
Permi
bc,
Anna
Jortikka
a,
Perttu
Permi
bc,
Anna
Jortikka
 a and
Kaarina
Sivonen
a and
Kaarina
Sivonen
 *a
*a
aDepartment of Microbiology, Faculty of Agriculture and Forestry, University of Helsinki, Helsinki, Finland. E-mail: kaarina.sivonen@helsinki.fi
bDepartment of Chemistry, University of Jyväskylä, Jyväskylä, Finland
cDepartment of Biological and Environmental Science, Nanoscience Center, University of Jyväskylä, Jyväskylä, Finland
First published on 4th June 2021
Abstract
Laxaphycins are a family of cyclic lipopeptides with synergistic antifungal and antiproliferative activities. They are produced by multiple cyanobacterial genera and comprise two sets of structurally unrelated 11- and 12-residue macrocyclic lipopeptides. Here, we report the discovery of new antifungal laxaphycins from Nostoc sp. UHCC 0702, which we name heinamides, through antimicrobial bioactivity screening. We characterized the chemical structures of eight heinamide structural variants A1–A3 and B1–B5. These variants contain the rare non-proteinogenic amino acids 3-hydroxy-4-methylproline, 4-hydroxyproline, 3-hydroxy-D-leucine, dehydrobutyrine, 5-hydroxyl β-amino octanoic acid, and O-carbamoyl-homoserine. We obtained an 8.6-Mb complete genome sequence from Nostoc sp. UHCC 0702 and identified the 93 kb heinamide biosynthetic gene cluster. The structurally distinct heinamides A1–A3 and B1–B5 variants are synthesized using an unusual branching biosynthetic pathway. The heinamide biosynthetic pathway also encodes several enzymes that supply non-proteinogenic amino acids to the heinamide synthetase. Through heterologous expression, we showed that (2S,4R)-4-hydroxy-L-proline is supplied through the action of a novel enzyme LxaN, which hydroxylates L-proline. 11- and 12-residue heinamides have the characteristic synergistic activity of laxaphycins against Aspergillus flavus FBCC 2467. Structural and genetic information of heinamides may prove useful in future discovery of natural products and drug development.
Introduction
Cyanobacteria produce a wide range of bioactive natural products with unusual structures and potent bioactivity.1,2 Knowledge of natural product chemical structures and biosynthetic mechanisms can facilitate their use in the pharmaceutical industry as new active compounds3–5 and provide insights into their ecological role.6 Laxaphycins are cyanobacterial natural products with two distinct macrocycles, 11- and 12-residue types, which act in synergy to produce antiproliferative and antifungal activities.7,8 Laxaphycins include characteristic non-proteinogenic amino acids, including 3-hydroxy-D-leucine (OHLeu), 3-hydroxy-D-asparagine (OHAsn), dehydrobutyrine (Dhb), and (2S,4R)-4-hydroxyproline ((2S,4R)-4-OHPro).9,10 Altogether 17 variants of 11-residue laxaphycins and 23 variants of 12-residue laxaphycins have been described from a broad range of cyanobacteria (Table S1†). Although 11- and 12-residue types are both found in most producer strains, there are reports of strains for which only one type of the compound is reported.11,12In an earlier study, we described the biosynthetic pathway of scytocyclamides, which belong to the laxaphycin peptide family.10 This pathway includes a shared initiating fatty-acyl AMP ligase (FAAL) and a polyketide synthase (PKS) module that branches with two non-ribosomal peptide synthetase (NRPS) pathways to produce the two distinct 11- and 12-residue compounds.10 Scytocyclamides are produced by Scytonema hofmannii PCC 7110.
The aim of this study was to identify and describe new antifungal compounds from cyanobacteria. Members of the genera Candida and Aspergillus can cause invasive infections in humans, typically in immunocompromised patients.13,14 Only a few chemical families of antimicrobials are currently used to treat fungal infections, and fungal resistance to these compounds is growing.13–15 Cyanobacteria are known producers of antifungal compounds such as laxaphycins, hassallidins, nostofungicidine, and cryptophycins, which could be used as antifungal drug leads.7,16–19
Here, we identified novel members of the laxaphycin family of natural products, heinamides, through bioactivity-guided fractionation of Nostoc sp. UHCC 0702 extracts. Heinamides have antifungal activity that inhibit the growth of Aspergillus flavus FBCC 2467 with synergistic effect between 11- and 12-residue type heinamides. We describe the chemical structures of heinamides A1–A3 and B1–B5 and identified the heinamide biosynthetic pathway. While the biosynthetic pathway is generally similar to the previously described scytocyclamide pathway, the differences provide a broader view of the laxaphycin biosynthesis pathways. The heinamide biosynthetic pathway encodes enzymes for the production of the unusual amino acids (2S,4R)-4-OHPro and 3-hydroxy-4-methylproline (OHMePro), which appear in heinamide structures. The action of the proline hydroxylase LxaN from Nostoc sp. UHCC 0702 was shown through heterologous expression. A homolog of LxaN was found also in the genome of S. hofmannii PCC 7110, a producer of scytocyclamides, which also contain (2S,4R)-4-OHPro.
Experimental
Strains
Cyanobacterial strains used in this study were Nostoc sp. UHCC 0702, isolated from the Finnish freshwater lake Villähteen Kukkanen (60°57′19.4′′N 25°53′02.4′′E) in 19.8.2013, and S. hofmannii PCC 7110, which originated from a limestone cave in Bermuda.Antimicrobial screening
Nostoc sp. UHCC 0702 was grown in 250 mL of Z8 medium at 20–21 °C with photon irradiation of 15 μmol m−2 s−1. Erlenmeyer flasks of 500 mL were used with 250 mL of medium, with constant sterilized air bubbling for 3–5 weeks. The biomass was collected by decanting excess media and centrifugation at 8000g for 5 min. The biomass was frozen at −80 °C and freeze-dried with a CHRIST BETA 2–8 LSC plus freeze drier with a LYO CUBE 4–8 chamber. A total of 100 mg of freeze-dried Nostoc sp. UHCC 0702 biomass was extracted with 1 mL of methanol and glass beads (0.5 mm glass beads, Scientific Industries Inc, USA) using a FastPrep cell disrupter at 6.5 m s−1 two times for 25 s with a resting time of 5 min between runs. The samples were centrifuged at room temperature at 8000g for 5 min. The supernatant was collected and extraction of the biomass was repeated with 1 mL of methanol. The 2 mL combined supernatant was stored at −20 °C.Antimicrobial activity screening was performed using 17 strains of fungi and bacteria (Table S2†). A total of 50 μL of cell extract, 50 μl of methanol (negative control) and 10 μL nystatin (5 mg mL−1, Nystatin, Streptomyces noursei, EMD Millipore Corp, Germany) or 10 μL ampicillin (50 mg mL−1 in 70% ethanol, Ampicillin sodium salt, Sigma, Israel) were placed directly on the agar surface prior to inoculation with an indicator strain. Nystatin was used as a positive control for fungal assays while ampicillin was used as a positive control for bacterial assays. Solvents of the extract and controls were allowed to evaporate, leaving the solids diffused in the agar. Inoculant was prepared by growing fungi for 2–14 days on potato dextrose agar (PDA) media at 28 °C and bacteria for 2 days on brain heart infusion (BHI) agar at 37 °C. Inoculant cell mass was transferred with a cotton swab from the agar to 3 mL of sterile 5 M NaCl solution, or sterile water in the case of A. flavus. Solution was spread on the assay plate with a fresh cotton swab. Fungal plates were incubated at 28 °C and bacterial plates at 37 °C for 2 days and examined for the presence of inhibition zones.
Disc-diffusion assays were performed with purified heinamide for more quantitative analysis of bioactivity. Paper discs (Blank monodiscs, Abtek biologicals Ltd, UK) were prepared with methanol solutions of the peptides, methanol as negative control, and nystatin as positive control. A. flavus inoculum was prepared as previously and spread on the plate. Disks were placed on agar, the plates were incubated at 28 °C for 2 days, and examined for the presence of inhibition zones.
Purification of heinamides
Mass cultures were grown in modified Z8 medium without added nitrogen at 20–21 °C with photon irradiation of 15 μmol m−2 s−1. Five-liter Erlenmeyer flasks were used with 2.7 L of medium, with constant sterilized air bubbling for 3–5 weeks. Cells were collected by decanting excess media and centrifugation at 8000g for 5 min. The cells were frozen at −80 °C and freeze-dried as described earlier.A total of 30 mL of methanol was used per 1 g of dry cells. Cells were homogenized with a Heidolph Silentcrusher M at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 s. The solution was centrifuged 10
000 rpm for 30 s. The solution was centrifuged 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min and supernatant was collected. The extraction was repeated with 30 mL of methanol using the cell pellet. Chomatorex (Fuji-Davison Chemical Ltd, Aichi, Japan) chromatography silica ODS powder (10 mL) was added to the supernatant pool and the mixture was dried with a rotary evaporator Büchi Rotavapor R-200 at 30 °C. Solid phase extraction (SPE) was performed with Phenomenex SPE strata SI-1 silica 5 g per 20 mL column, preconditioned with 20 mL isopropanol and 20 mL of heptane. Silica ODS powder with the dry extract was added on top of the column and extracted with heptane, ethyl acetate, acetone, acetonitrile, and methanol, 40 mL each, with every fraction collected individually. Fractions were dried with a nitrogen gas flow and re-dissolved in 1 mL of methanol for bioactivity assays.
000g for 5 min and supernatant was collected. The extraction was repeated with 30 mL of methanol using the cell pellet. Chomatorex (Fuji-Davison Chemical Ltd, Aichi, Japan) chromatography silica ODS powder (10 mL) was added to the supernatant pool and the mixture was dried with a rotary evaporator Büchi Rotavapor R-200 at 30 °C. Solid phase extraction (SPE) was performed with Phenomenex SPE strata SI-1 silica 5 g per 20 mL column, preconditioned with 20 mL isopropanol and 20 mL of heptane. Silica ODS powder with the dry extract was added on top of the column and extracted with heptane, ethyl acetate, acetone, acetonitrile, and methanol, 40 mL each, with every fraction collected individually. Fractions were dried with a nitrogen gas flow and re-dissolved in 1 mL of methanol for bioactivity assays.
The active methanol fraction was further fractionated with an Agilent 1100 Series liquid chromatograph with Phenomenex Luna C18(2) (150 × 10 mm, 100 Å) column. Sample was injected in 100 μL batches and eluted with acetonitrile/isopropanol 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (solvent B) and 0.1% HCOOH (solvent A) with initial isocratic stage of 40% solvent B in A for 15 min, followed by a linear gradient of solvent B from 40% to 100% in 10 min with a flow rate of 3 mL min−1. Four heinamide fractions were collected, dried with nitrogen flow, and weighed. Fraction 1 contained heinamides B1 and B2 (1
1 (solvent B) and 0.1% HCOOH (solvent A) with initial isocratic stage of 40% solvent B in A for 15 min, followed by a linear gradient of solvent B from 40% to 100% in 10 min with a flow rate of 3 mL min−1. Four heinamide fractions were collected, dried with nitrogen flow, and weighed. Fraction 1 contained heinamides B1 and B2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), fraction 2 contained heinamide B1, fraction 3 contained heinamides A1, B3, and B4 (7
1), fraction 2 contained heinamide B1, fraction 3 contained heinamides A1, B3, and B4 (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and fraction 4 contained heinamide A2. To further separate the products, fraction 3 was treated with an additional HPLC run with isocratic conditions of 41% solvent B in solvent A for 30 min with flow rate of 3 mL min−1 using the same column. Fraction 3 was thus separated to fractions 3a and 3b containing heinamides B3 + B4 and A1, respectively. Heinamides A3 and B5 were not purified due to low production levels.
1) and fraction 4 contained heinamide A2. To further separate the products, fraction 3 was treated with an additional HPLC run with isocratic conditions of 41% solvent B in solvent A for 30 min with flow rate of 3 mL min−1 using the same column. Fraction 3 was thus separated to fractions 3a and 3b containing heinamides B3 + B4 and A1, respectively. Heinamides A3 and B5 were not purified due to low production levels.
Amino acid analysis
To elucidate the stereochemistry of the eight heinamide structural variants, amino acid analysis was performed with the Marfey method as described before20 with the isolated heinamides A1–A2 and B1–B4. The reagents 1-fluoro-2,4-dinitrophenyl-5-L-alanine amide (FDAA), 1-fluoro-2,4-dinitrophenyl-5-L-leucine amide (L-FDLA), and fluoro-2,4-dinitrophenyl-5-D-leucine amide (D-FDLA) were used with reference amino acids L-Ser, D-Ser, L-Hse, D-Hse, L-Glu, D-Glu, L-Thr, D-Thr, L-Val, D-Val, D-Pro, L-Pro, L-Tyr, D-Tyr, D-allo-Thr, L-Leu, D-Leu, L-Ile, D-Ile, D-allo-Ile, L-allo-Ile, D-Phe, L-Phe, (2S,4S)-4-hydroxy-Pro, (2R,4S)-4-hydroxy-Pro, (2S,4R)-4-hydroxy-Pro, L-Ser, D-Ser, (Sigma, Switzerland), N-methyl-L-Ile (hydrochloride, ABCR, Germany), L-allo-Thr (ICN Biomedicals, USA), (2R,4R)-4-hydroxy-Pro (Aldrich, USA), and (3S)- and (3R)-3-amino octanoic acid (ABCR, Germany).Amino acid feeding experiment
Nostoc sp. UHCC 0702 and S. hofmannii PCC 7110 were cultivated in the presence of different Pro variants and hypothesized OHMePro synthesis intermediates to investigate if they are incorporated in the laxaphycin structures. Nostoc sp. UHCC 0702 was grown on Z8 media and S. hofmannii PCC 7110 was grown on Z8 media without added nitrogen. The media were modified by adding (2S,4S)-4-methyl-proline, (2R,4R)-4-methyl-proline, (2S,4R)-4-methyl-proline, (ABCR, Germany), (2S,4S)-4-hydroxyproline (Sigma, USA), (2S,4R)-4-hydroxyproline (Sigma, Japan), or L-Leu (Sigma, Switzerland) to concentrations of 10 μM. Racemic OHLeu was used at 40 μM concentration. Unaltered medium was used in control cultivation. Three replicates of each cultivation were grown and analyzed. The bacteria were grown in 100 mL Erlenmeyer flasks in 40 mL of medium for 17 days at 20–21 °C with photon irradiation of 15 μmol m−2 s−1. Cells were collected by centrifugation at 8000g for 5 min. Cells were frozen to −80 °C and freeze-dried. Freeze-dried biomass was weighed and extracted with 0.5 mL methanol and glass beads (0.5 mm glass beads, Scientific Industries Inc, USA) using a FastPrep cell disrupter two times for 25 s at a speed of 6.5 m s−1. Samples were centrifuged at room temperature at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min and supernatant was collected for LC-MS analysis.
000g for 5 min and supernatant was collected for LC-MS analysis.
Stable isotope labeling
Nostoc sp. UHCC 0702 was labelled with 15N in a nitrogen-free atmosphere in Z8 growth medium containing 15N-urea as the sole source of nitrogen. The medium was bubbled with nitrogen-free argon with 20.9% O2 and 0.45% CO2 (quality 5.7; AGA Gas Ab, Sweden). The strain was grown for 21 days at 20 °C under photon irradiation of 15 μmol m−2 s−1. The biomass was collected and freeze dried. A total of 100 mg of the dried cells were extracted with methanol as described earlier and analyzed with LC-MS.LC-MS
Freeze-dried bacterial cells were extracted with methanol as described earlier. Extracts and isolated heinamide fractions dissolved in methanol were analyzed with UPLC-QTOF (Acquity I-Class UPLC-SynaptG2-Si, Waters Corp., Milford, MA, USA) equipped with a Kinetex® C8 column (2.1 × 50 or 100 mm, 1.7 μm, 100 Å, Phenomenex, Torrance, CA, USA) injected with 0.5 or 1 μL of sample and eluted at 40 °C with 0.1% HCOOH in water (solvent A) and acetonitrile/isopropanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, +0.1% HCOOH, solvent B) at a flow rate of 0.3 mL min−1. Different solvent gradients were used (Table S3†).
1, +0.1% HCOOH, solvent B) at a flow rate of 0.3 mL min−1. Different solvent gradients were used (Table S3†).
QTOF was calibrated using sodium formate and Ultramark® 1621, which gave a calibrated mass range from m/z 91 to 1921. Leucine Enkephalin was used at 10 s intervals as a lock mass reference compound. Mass spectral data were accumulated in positive electrospray ionization resolution mode. In MS/MS mode, Trap Collision Energy Ramp proceeded from 40.0 eV to 70.0 eV.
NMR spectroscopy
NMR spectra were obtained using a Bruker Avance III HD 800 MHz NMR spectrometer equipped with the TCI 1H, 13C, 15N triple resonance cryoprobe. Data were collected at 30 °C in DMSO-d6. In addition to 1H and broadband-decoupled 13C experiments, we employed two-dimensional TOCSY (TOtal Correlation SpectroscopY), DQF-COSY (Double Quantum Filtered COrrelation SpectroscopY), and EASY-ROESY (Efficient Adiabatic SYmmetrized Rotating-frame Overhauser Effect SpectroscopY)21 experiments, and 13C HSQC, 15N HSQC, edited 13C HSQC (heteronuclear Single Quantum Coherence), and 13C HMBC (Heteronuclear Multiple Bond Correlation) experiments for the assignment of 1H, 13C, and 15N chemical shifts. The 2D TOCSY was acquired with isotropic mixing time of 60 ms (10 kHz RF field). The mixing time for 2D ROESY experiment was 200 ms (RF field strength 4 kHz). For observing long-range H–C connectivities, the 13C HMBC experiment was measured using nJCH transfer time optimized for 8 Hz couplings. NMR experiment parameters are presented in Table S4.†Genome sequencing and gene cluster analysis
Cultures were grown in 250 mL of modified Z8 medium lacking a source of combined nitrogen at 20–21 °C with photon irradiation of 15 μmol m−2 s−1. Half-liter Erlenmeyer flasks were used with 250 mL of medium, with constant sterilized air bubbling for 3 weeks. The fresh Nostoc sp. UHC 0702 cells were disrupted by bead beating in GOS buffer and DNA was extracted with the phenol–chloroform method as previously described.20 Sequencing of the extracted DNA was performed with PacBio RSII and MGI DBNSeq-G400 sequencing and assembled with HGAP3 (smrtportal 2.3.0).The Nostoc sp. UHCC 0702 complete genome data was analyzed with AntiSMASH 5.022 and AntiSMASH 4.123 to identify the lxa biosynthetic gene cluster. BLASTp and CDD database searches were used to assign a predicted function to the proteins encoded in the lxa biosynthetic gene clusters from the Nostoc sp. UHCC 0702 and S. hofmannii PCC 7110 genomes. The condensation domain of LxaC13 was analyzed with Natural Product Domain Seeker (NaPDoS)24 and compared with other known Dhb-related condensation domains LxaC13, HasO2, NdaA1, and PuwF2, which are involved in the biosynthesis of scytocyclamides,10 hassalladins,25 nodularins,20 and puwainaphycins,26 respectively.
Heterologous expression of lxaN gene
The function of LxaN, a putative proline 4-hydroxylase, was tested through heterologous expression in Escherichia coli BL21(DE3). LxaN was expected to hydroxylate available Pro in the E. coli cells. Plasmid constructs with the lxaN gene were prepared and transformed to E. coli, and amino acid analysis was performed to detect 4-OHPro in the transformed cells.Genomic DNA was extracted from Nostoc sp. UHCC 0702 as previously described. The lxaN gene was amplified by PCR with primers LxaN-F (5′-gtggtggtgctcgagtgcggccgcaTTAAATAAGAACTTTGTCCAATAG-3′) and LxaN-R (5′-ggacagcaaatgggtcgcggatccgATGTCCTATACCAATCAAAC-3′). The vector pET28a (+) was digested with restriction enzymes EcoRI and HindIII (Promega, USA). The lxaN gene was inserted into pET28a (+) using the NEBuilder® HiFi DNA Assembly Cloning Kit (New England BioLabs, USA) to create plasmid pET28a-LxaN. To allow inducible expression, the lxaN gene was placed behind a T7 promoter and lac operator. The plasmid pET28a-LxaN was transformed into E. coli BL21 (DE3). A negative control was prepared by transforming an empty pET28a plasmid into E. coli BL21(DE3). Clones were selected by using LB agar plates containing 50 μg mL−1 kanamycin, 1 g 100 mL−1 arabinose, and 1 g 100 mL−1 glucose. Three transformants and a negative control were transferred to liquid LB medium with 50 μg mL−1 kanamycin and 1 g 100 mL−1 glucose and incubated at 37 °C overnight with shaking (170 rpm). Aliquots of these primary cultures were used to inoculate 10 mL of fresh LB medium supplemented with selective antibiotics, 1 g 100 mL−1 arabinose, and 1 g 100 mL−1 glucose. The cultures were incubated at 37 °C with shaking at 170 rpm until OD600 of 0.6 was achieved. The cultures were induced with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and incubated at 18 °C for 48 h with shaking (200 rpm).
Biomass was collected from the growth culture by centrifugation and the supernatant was discarded. The biomass was frozen at −80 °C in screw-cap tubes. To extract biomass, 200 μL of 0.5 mm glass beads (Scientific Industries Inc, USA) and 1 mL 100% methanol were added into the tube. The mixture was homogenized by using Fastprep-24 twice for 20 seconds at a speed of 6 m s−1. Cell debris was pelleted by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400g for 5 min. Supernatant was transferred into new tubes and dried with nitrogen gas flow. Amino acid analysis was performed with Marfey's method as previously described with UPLC-QTOF mass spectrometry,20 using reference amino acids (2S,4S)-4-hydroxyproline, (2S,4R)-4-hydroxyproline, (2R,4S)-4-hydroxyproline (Sigma, Switzerland), and (2R,4R)-4-hydroxyproline (Aldrich, USA).
400g for 5 min. Supernatant was transferred into new tubes and dried with nitrogen gas flow. Amino acid analysis was performed with Marfey's method as previously described with UPLC-QTOF mass spectrometry,20 using reference amino acids (2S,4S)-4-hydroxyproline, (2S,4R)-4-hydroxyproline, (2R,4S)-4-hydroxyproline (Sigma, Switzerland), and (2R,4R)-4-hydroxyproline (Aldrich, USA).
Results
Antimicrobial screening
A methanol extract of Nostoc sp. UHCC 0702 inhibited the growth of A. flavus FBCC 2467. The other tested fungal and bacterial strains were not affected by the extract.Structure of heinamides
Eight peptides belonging to the laxaphycin family were identified from Nostoc sp. UHCC 0702 and named heinamides A1–A3 and B1–B5 (Fig. 1). Heinamides A1–A3 are 11-residue laxaphycins and heinamides B1–B5 are 12-residue laxaphycins. The 11-residue heinamides follow the amino acid sequence Aoa1-Ser2-Dhb3-((2S,4R)-4-OHPro/Pro)4-Ser5-Tyr6-Leu7-Ile8-Phe9-((2S,4R)-4-OHPro/Pro)10-Gly11; the difference between variants was the hydroxylation of Pro4,10. The 12-residue heinamides have the amino acid sequence (Aoa/5-OH-Aoa)1-Ile2-OHLeu3-(O-carbamoyl-Hse)4-Leu5-Gln6-(N-Me-Ile)7-OHHse8-Val9-(OHMePro/Pro/4-MePro)10-Tyr11-Thr12, where the differences between variants is in the hydroxylation of the octanoic acid1 and hydroxylation and methylation of the Pro10. The stereochemistry of the compounds is derived from stereospecific amino acid analysis (Table S11†) and NMR. | ||
| Fig. 1 Chemical structures of heinamides and relative intensities of their MS signals (%), purified from Nostoc sp. UHCC 0702. | ||
The methanol extract from Nostoc sp. UHCC 0702 was analyzed with UPLC-QTOF to determine the initial heinamide structures. Cultivation on 15N-containing medium and comparison of the mass data of 15N-labeled and unlabeled compounds showed the presence of 11 nitrogen atoms in 11-residue heinamides A1–A3 and 14 nitrogen atoms in 12-residue heinamides B1–B5 (Table S5, Fig. S2†). The product ion spectra of protonated heinamides consisted of many evenly intense ions, which is typical for some cyclic peptides (Fig. S3–S6†). The best continuous data for amino acid sequencing started as expected from the amino acids next to the Pro N-terminus (red markings in Fig. S3 and S4†). The structure of heinamides A3, B4 and B5 is proposed solely on the basis of analyses of their LC MS/MS data. The structures assigned by LC-MS were consistent with those assigned using NMR. 1H, 13C, DQF-COSY, TOCSY, EASY-ROESY, 13C HSQC, edited 13C HSQC, 15N HSQC, and 13C HMBC NMR spectra were obtained from the purified heinamides A2 and B2 and heinamide mixtures A1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B3
B3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B4 (7
B4 (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and B1
1) and B1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B2 (1
B2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). NMR spectra are presented in Fig. S7–S10† and numerical data with COSY, ROESY, and HMBC correlations in Tables S6–S10.† COSY, HMBC, and ROESY uninterrupted correlation chain specified the subunit sequences in both 11-residue (HA A1) and 12-residue (HA B2) heinamides (Fig. S11 and S12†). Heinamide structures were in good agreement with previously described laxaphycin common structural features (Table S1†).
1). NMR spectra are presented in Fig. S7–S10† and numerical data with COSY, ROESY, and HMBC correlations in Tables S6–S10.† COSY, HMBC, and ROESY uninterrupted correlation chain specified the subunit sequences in both 11-residue (HA A1) and 12-residue (HA B2) heinamides (Fig. S11 and S12†). Heinamide structures were in good agreement with previously described laxaphycin common structural features (Table S1†).
In heinamides A1–A3, the only new element was (2S,4R)-4-OHPro or L-Pro in position 10. Two different ppm value sets for 4-OHPro were present for HA A1. In amino acid analysis (2S,4R)-4-OHPro was the only 4-OHPro enantiomer present of the four possible alternatives (Tables S6 and S11†). More new elements were found in 12-residue heinamides B1–B5. The hydroxy group in the β-amino octanoic acid1 has not been described earlier in laxaphycins, but COSY, HSQC, and HMBC correlations show the presence of a methine (δH = 3.46, δC = 66.2 ppm) group in position 5, which is most probably bonded to oxygen in HA B1 (Fig. S9F, I and N and S12†). In many 12-residue laxaphycins, Leu5 is hydroxylated but not in heinamides according to NMR (Fig. S9E, L and M†). In previously described 12-residue laxaphycins, the amino acid in position 8 has almost exclusively been 3-OHAsn. However, the amino acid is 3-hydroxy-homoserine in heinamides B1–B5 (Tables S8–S10, Fig. S9 and S10†). Lastly, NMR data showed another Hse is in position 4 in 12-residue heinamides. Furthermore, COSY, HSQC, and HMBC correlations showed that this subunit was actually O-carbamoylated (Fig. S9, especially frame O, Tables S8–S10†). Isobaric 3- or 4-OHGln were ruled out as the C3 and C4 were methylenes according the edited 13C-HSQC (Fig. S9,† frame I). Product ion spectra of protonated heinamides B1–B5 show the loss of the carbamoyl group as carbamic acid (61.02 Da) until the loss of the O-carbamoyl-Hse4 subunit itself (Fig. S4–S6†). No 12-residue heinamide without an O-carbamoyl group in Hse was found.
Antifungal activity
After fractionation of the extract and identification of the products, disc diffusion assays were performed with the purified compounds. Inhibition of fungal growth was observed with peptides of a single laxaphycin type as a hazy inhibition zone. Synergy was observed between 11- and 12-residue compounds as a clear inhibition zone (Fig. 2).Predicted heinamide biosynthetic pathway from Nostoc sp. UHCC 0702
We sequenced the complete genome of Nostoc sp. UHCC 0702 (GenBank accession number CP071065). The genome is a single 8587![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 404 bp circular chromosome without extrachromosomal elements. Sequence coverage with PacBio RSII was 90x and with MGI DBNSeq-G400 46x. Bioinformatics analysis with AntiSMASH identified 13 putative NRPS/PKS pathways. Comparison with the S. hofmannii PCC 7110 lxa biosynthetic gene cluster and chemical structures of the identified heinamides confirmed the presence of a lxa biosynthetic gene cluster in Nostoc sp. UHCC 0702 (Fig. 3).
404 bp circular chromosome without extrachromosomal elements. Sequence coverage with PacBio RSII was 90x and with MGI DBNSeq-G400 46x. Bioinformatics analysis with AntiSMASH identified 13 putative NRPS/PKS pathways. Comparison with the S. hofmannii PCC 7110 lxa biosynthetic gene cluster and chemical structures of the identified heinamides confirmed the presence of a lxa biosynthetic gene cluster in Nostoc sp. UHCC 0702 (Fig. 3).
The cluster is 93 kb long and encodes 13 open reading frames annotated as ORF1–2, lxaI1, lxaJ1, lxaK1, lxaA2, ORF3, lxaB, lxaC1, lxaE, lxaF, lxaG, and lxaM (Fig. 3, Table 1). lxaC1 is an NRPS with adenylation domain binding pockets matching the amino acid sequence of 11-residue heinamides (Table S12†). lxaC1 is flanked with PKSs lxaB and lxaE followed by cupin-like domains lxaFGM (Fig. 3, Table 1). The Nostoc sp. UHCC 0702 genes lxaI1, lxaJ1, and lxaK1 adenylation domain binding pockets match the amino acid sequence of 12-residue heinamides (Fig. 3, Table S12†). The position of epimerase domains in the lxa biosynthetic enzymes match the positions of D-amino acids in the elucidated structures (Fig. 1 and 3). Laxaphycin biosynthesis is predicted to be initiated by an activating enzyme lxaA with FAAL and ACP domains (Fig. 3). However, such an enzyme was not encoded in the Nostoc sp. UHCC 0702 lxa biosynthetic gene cluster. Instead, an ACP in the cluster with 41% sequence identity in BLASTp to the ACP domain of LxaA was identified and designated LxaA2 (Fig. 3, Table 1).
| Nostoc sp. UHCC 0702 | Top blastp hit | ||||||
|---|---|---|---|---|---|---|---|
| Protein | Accession no. | Length (aa) | Proposed function | Organism | Function | Accession no. | Sequence identity (%) |
| a blastx. | |||||||
| LxaN | QSJ20407.1 | 271 | Pro hydroxylation | Nostoc sp. PA-18-2419 | Phytanoyl-CoA dioxygenase | WP_138506010.1 | 93 |
| LxaO | QSJ20406.1 | 265 | OHMePro synthesis | Nostoc calcicola | Hypothetical protein | WP_073641216.1 | 87 |
| LxaP | QSJ20405.1 | 368 | OHMePro synthesis | Nostoc sp. ‘Peltigera membranacea cyanobiont’ 210A | Zinc-binding dehydrogenase | WP_094347233.1 | 97 |
| LxaQ | QSJ20404.1 | 273 | OHMePro synthesis | Nostoc sp. ‘Peltigera membranacea cyanobiont’ 210A | Pyrroline-5-carboxylate reductase | WP_094347234.1 | 95 |
| LxaR | QSJ20403.1 | 259 | OHMePro hydroxylation | Nostoc calciola | 2OG-Fe(II) oxygenase | WP_084177703.1 | 83 |
| LxaH | QSJ20216.1 | 727 | ABC transporter | Nostoc sp. NIES-4103 | ABC transporter ATP-binding protein/permease | WP_096559342.1 | 78 |
| ORF1 | QSJ20141.1 | 108 | Unknown | Nodularia sp. NIES-3585 | Polyketide synthase | WP_089089655.1 | 75 |
| ORF2 | JYQ62_16360a | 105 | Unknown | Scytonema hofmannii PCC 7110 | Polyketide synthase | WP_017742671.1 | 67a |
| LxaI1 | QST87270.1 | 7826 | NRPS | Scytonema sp. UIC 10036 | Non-ribosomal peptide synthase/polyketide synthase | WP_155746081.1 | 63 |
| LxaJ1 | QSJ20140.1 | 1608 | NRPS | Scytonema hofmannii PCC 7110 | Non-ribosomal peptide synthetase | WP_066612897.1 | 70 |
| LxK1 | QSJ20139.1 | 5036 | NRPS | Scytonema hofmannii PCC 7110 | Non-ribosomal peptide synthetase | WP_066612897.1 | 72 |
| LxaA2 | QSJ20138.1 | 89 | ACP | Microcystis aeruginosa | Acyl carrier protein | WP_185240878.1 | 63 |
| ORF3 | QSJ20137.1 | 47 | Unknown | Bradyrhizobium genosp. B | Cytochrome P450 | WP_195799373.1 | 45 |
| LxaB | QSJ20136.1 | 475 | PKS | Scytonema hofmannii PCC 7110 | Polyketide synthase | WP_017742671.1 | 89 |
| LxaC1 | QSJ20135.1 | 12487 | NRPS | Nostoc flagelliforme CCNUN1 | Non-ribosomal peptide synthetase | WP_100898072.1 | 66 |
| LxaE | QSJ20134.1 | 1196 | PKS | Nodularia sp. NIES-3585 | Aminotransferase class III-fold pyridoxal phosphate-dependent enzyme | WP_089093801.1 | 81 |
| LxaF | QSJ20133.1 | 295 | Cupin-like domain | Scytonema hofmannii PCC 7110 | Cupin-like domain-containing protein | WP_017742676.1 | 69 |
| LxaG | QSJ20132.1 | 306 | Cupin-like domain | Scytonema hofmannii PCC 7110 | Cupin-like domain-containing protein | WP_017742677.1 | 79 |
| LxaM | QSJ20131.1 | 295 | Cupin-like domain | Scytonema hofmannii PCC 7110 | Cupin-like domain-containing protein | WP_017742677.1 | 68 |
| LxaA1 | QSJ18887.1 | 580 | FAAL | Anabaena azotica | AMP-binding protein | WP_190474150.1 | 79 |
The lxa biosynthetic gene cluster was missing the essential FAAL domain for the initiation of the pathway and the genes to synthesize the modified amino acids (2S,4R)-4-OHPro, OHMePro, O-carbamoyl homoserine, and an ABC-transporter. The LxaA FAAL domain of S. hofmannii PCC 7110 was used as BLASTp query to search for the initiating domain, and a FAAL with the highest identity was annotated LxaA1 (Fig. 3, Table 1). This gene was located 1.6 Mb upstream from the lxa biosynthetic gene cluster (Fig. 3). In S. hofmannii PCC 7110, the LxaA protein includes two domains, a FAAL and an ACP domain (Fig. S16†). Together LxaA1 and LxaA2 act as LxaA (Fig. 3).
We predict that the cupin-like domain proteins LxaF, LxaG, and LxaM hydroxylate Leu3, Hse8, and Aoa (Fig. 3). ORF1–3 were found in the gene cluster with no predicted function. Eleven-residue heinamides contain the non-proteinogenic amino acid Dhb3, the dehydration product of Thr. We discovered that the condensation domain LxaC13 groups in the modified AA clade of condensation domains that act in the dehydration of Thr and Ser from the previous module to Dhb or Dha (Fig. S13†). Three putative carbamoyltransferases were found in the genome, but none were assigned to the gene cluster. With the lack of known carbamoyltrasferases acting on amino acids, no reliable prediction could be made for the enzyme responsible for the O-cabamoylation of HSe.
A set of genes encoding enzymes homologous to genes producing (2S,4S)-4-methylproline in cyanobacterial metabolites were identified 389 kb downstream from the lxa biosynthetic gene cluster (Fig. 3, Table 1). These enzymes are a L-Leu 5-hydroxylase (LxaO), a zinc-binding dehydrogenase (LxaP), and a pyrroline-5-carboxylate reductase (LxaQ) (Fig. 3, Table 1). Flanking lxaOPQ were two genes lxaN and lxaR that encode α-ketoglutarate-dependent oxygenases (Fig. 3, Table 1). LxaN was discovered to have a homolog also in the S. hofmannii PCC 7110 genome 11 kb downstream of the scytocyclamide gene cluster (WP_017742662.1). LxaN enzymes from Nostoc sp. UHCC 0702 and S. hofmannii PCC 7110 share 93% amino acid sequence identity. LxaN belongs to the pfam05721 class of oxygenases. We predicted that LxaN hydroxylates L-Pro to (2S,4R)-4-OHPro found in heinamide and scytocyclamide structures and that LxaR acts in OHMePro production hydroxylating the 3-carbon (Scheme 1). LxaR belongs to the pfam13640 class of α-ketoglutarate-FeII dependent oxygenases known to hydroxylate amino acids.
LxaN of Nostoc sp. UHCC 0702 was heterologously expressed in E. coli to assess if it hydroxylates L-Pro to (2S,4R)-4-OHPro. Amino acid analysis performed with Marfey's method showed that a derivatized extract from E. coli with the LxaN construct matched the retention time of derivatized (2S,4R)-4-OHPro (Fig. 4). A control E. coli strain without added LxaN in transformed vector did not produce a signal in LC-MS with the corresponding mass. This confirms that hydroxylation in (2S,4R)-4-OHPro was produced by LxaN.
Proline diversity, biosynthesis, and adenylation domain substrate specificity
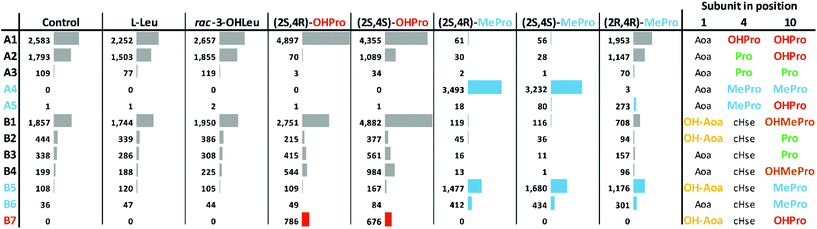
To obtain more information on laxaphycin biosynthesis, Nostoc sp. UHCC 0702 and S. hofmannii PCC 7110 were grown on modified media with added amino acids to determine if the selected amino acids are incorporated into the laxaphycin structures. The supplied amino acids were (2S,4S)-4-methyl-proline, (2R,4R)-4-methyl-proline, (2S,4R)-4-methyl-proline, (2S,4S)-4-hydroxyproline, (2S,4R)-4-hydroxyproline, racemic OHLeu, and L-Leu. Changes in product structures and proportions were measured with UPLC-QTOF (Fig. 5). Ten new laxaphycin variants were found from the cells grown on the amino acid supplemented media (Fig. 5 and Fig. S14†). Extracted ion chromatograms and product ion spectra of the most abundantly produced heinamides A4 and B5 are presented in Fig. S5, S6, and S15.† Heinamides A4, A5, B6 and scytocyclamides A3 and B5–B8 contain 4-MePro instead of (2S,4R)-4-OHPro, OHMePro, or Pro, which are normally present in these peptides (Fig. 1 and 5 and Fig. S14†). Addition of (2S)-4-MePro isomers practically prevented the formation of the original heinamide variants. Heinamide B5 is the only 4-Mepro-containing variant that was found among the original heinamide variants (Fig. 5). The added 4-MePro stereoisomers were not hydroxylated but incorporated as such to the structure in (2S,4S)-4-MePro and (2S,4R)-4-MePro.Likewise, new heinamide B7 and scytocyclamide B4 peptides were formed that contained OHPro stereoisomers instead of OHMePro or Pro, but only in small amounts (Fig. S14†). This indicates that adenylation domains that normally recognize OHMePro and Pro can recognize 4-MePro much more effectively than OHPro. Adding 4-hydroxyprolines did not result in new 11-residue variants. Feeding with (2S,4R)-4-OHPro (the stereoisomer naturally present in these laxaphycins) prevented the formation of Pro-containing heinamides A2 and A3 and scytocyclamide A2 so that only (2S,4R)-4-OHPro-containing variants heinamide A1 and scytocyclamide A were present. Feeding with L-Leu or a racemic mixture of OHLeu did not have any effect on the synthesis of heinamides or scytocyclamides. We tentatively hypothesized that OHLeu or Leu would be intermediates in the biosynthesis of OHMePro. However, supplementing the bacteria with these amino acids did not change OHMePro levels.
Discussion
Twenty-four laxaphycin structures have been described in the literature with antifungal, antiproliferative, and antibacterial activities7–12,27–38 (Table S1†). Here we report eight new laxaphycin variants, which we call heinamides. The 12-residue heinamides have unique substructures compared to known laxaphycins: OHMePro10, OH-Aoa (5-hydroxyl β-amino octanoic acid)1, and O-carbamoyl-homoserine4 (Table S1†). OHMePro is an unusual amino acid in peptides. In cyanobacteria, it has been described in nostopeptins where the hydroxyl group is esterified by the C-terminal carboxyl group to form a macrocyclic peptide structure.39 OHMePro is more common in fungal natural products, such as the clinically used antifungals echinocandins.40 The O-carbamoyl-homoserine appears to be a very unusual structure, as we did not find other O-carbamoyl homoserine-containing peptides or macrocyclic peptides containing O-carbamoylated amino acids in the literature. In banyaside A, the O-carbamoyl group in glucose showed 156.5 ppm carbonyl and 6.45 ppm NH2 proton signals,41 which are practically identical with O-carbamoyl-homoserine4O-carbamoyl signals. The adenylation domain binding pocket AA residue sequences of Hse4 (LxaI13) and cHse8 (LxaJ11) are identical (DLKNFGSDVK) (Table S12†) and we predict that the substrate for both is Hse, and the carbamoylation occurs at a later stage. While 3-aminooctanoic acid is common in laxaphycins, the hydroxylated amino octanoic acid of major heinamide variants is a novelty. Heinamides also include other non-proteinogenic amino acids characteristic to laxaphycins, such as E-Dhb3 and (2S,4R)-4-OHPro4,10 in 11-residue variants and OHLeu3 and NMe-Ile7 in 12-residue variants. The 3-OHHse8 of 12-residue heinamides is structurally identical to 4-hydroxy threonine8 of lobocyclamides B and C.31 In the light of genetic information on the adenylation domain substrate specifcity we decided to refer to it as Hse. We suggest that the reported 4-OHThr8 in lobocyclamides also originates from Hse which is 3-hydroxylated. This observation means that hydroxylation in the position-8 amino acid is C-3 specific, as this is the case in all 24 variants of 12-residue laxaphycins described. Heinamides share the strong synergistic antifungal activity of laxaphycins between 11- and 12-residue variants (Fig. 2).The general organization of laxaphycin biosynthesis for scytocyclamides was described recently.10 The heinamide biosynthetic gene clusters of Nostoc sp. UHCC 0702 and S. hofmannii PCC 7110 have a very similar NRPS domain organization (Fig. S16†). However, there are several differences in the organization of the genes between the strains (Fig. S16†). In S. hofmannii PCC 7110, the 11-residue laxaphycin NRPSs lxaC and lxaD are located before the 12-residue genes lxaI-L. In Nostoc sp. UHCC 0702, the order is reversed (Fig. S16†). There is also a difference in how the modules are organized at the gene level. In 11-residue heinamide NRPSs, all modules are in a single gene lxaC1, whereas in S. hofmannii PCC 7110 they are in two genes lxaC and lxaD. In 12-residue laxaphycin NRPSs, the modules are organized differently in the three ORFs as shown in Fig. 3. A rearrangement of genes appears to have occurred with probable gene fusions or fissions with the NRPS genes altering their length. Both lxa biosynthetic gene clusters include the PKS genes lxaB and lxaE, which are predicted to elongate an initiating hexanoic acid to octanoic acid (Fig. 3). The hexanoic acid was predicted to be loaded by LxaA.10 LxaA is a two-domain enzyme with a FAAL and an ACP domain. The heinamide biosynthetic gene cluster does not encode a direct LxaA homolog and the biosynthetic gene cluster lacks genes that encode FAAL (Fig. 3). However, a homolog to the ACP part of the LxaA protein is found in the heinamide biosynthetic gene cluster and annotated as LxaA2 (Fig. 3). A matching homolog to the FAAL was found in the genome and is predicted to initiate biosynthesis of heinamides. This homolog was annotated as LxaA1 (Fig. 3, Table 1). A similar situation has been described in biosynthesis of the lipopeptides puwainaphycins, where some gene clusters have a PuwI enzyme with both FAAL and ACP domains initiating biosynthesis and some have separate FAAL (PuwC) and ACP (PuwD) genes responsible for initiation.42
The amino acid feeding experiment demonstrated that the adenylation domains recognizing Pro are not highly specific and accepted all tested 4-MePro stereoisomers (Fig. 5 and Fig. S14†). The presence of heinamide B5 suggests that (2S,4R)-4-MePro was present in the cells without feeding but only in minuscule amounts. The fed 4-MePros were incorporated in the peptides in non-hydroxylated form (Fig. 5), which suggests that none of the 4-MePro is an intermediate in biosynthesis of OHMePro.
The branching of the biosynthetic pathway of lxa biosynthetic gene cluster in heinamide and scytocyclamide biosynthesis is unusual.10 This kind of branching in PKS-NRPS biosynthesis has also been reported with the cyanobacterial natural products vatiamides, where a single PKS cassette has three separate NRPS partner pathways and multiple products.43 This kind of genetic organization could be even more widespread in cyanobacteria and should be considered when identifying new biosynthetic gene clusters. The location of heinamide genes lxaA1HNOPQR in relation to the gene cluster is also unusual. Typically, all genes participating in the synthesis of a NRPS/PKS product are located directly in the gene cluster.44 However, in this case, the biosynthesis of heinamides could not be explained by genes encoded in the lxa biosynthetic gene cluster alone. Examples exist of crosstalk between gene clusters located in different parts of genome.45 The co-localization of the five OHMePro and (2S,4R)-4-OHPro tailoring genes lxaNOPQR suggests that they have a common function and target in the cell, which assisted in functional prediction. LxaN is also found in the S. hofmannii PCC 7110 genome, where it is also separated from the core gene cluster. The predictions made here are the most probable explanations for the biosynthesis of the elucidated structures, even when the scattering of the genes through the genome seems unconventional.
The predicted L-Pro hydroxylase activity of LxaN was confirmed through heterologous expression of the lxaN gene in E. coli (Scheme 1, Fig. 4). The enzyme is encoded by both the Nostoc sp. UHCC 0702 and S. hofmannii PCC 7110 genomes. Hydroxyprolines are important in the pharmaceutical industry and are produced through enzymatic reactions.46–49 (2S,4R)-4-OHPro is used in the production of carbapenem antibiotics50,51 and the anti-inflammatory agent oxaceprol.52 In human physiology, (2S,4R)-4-OHPro has a role in increasing collagen stability and has been shown to facilitate collagen biosynthesis in rats53 and is used in skin care products as Pro-(2S,4R)-4-OHPro dipeptides.54 (2S,4R)-4-OHPro can also be used in production of polymer materials based on polythioesters.55 Hydroxylated and methylated proline have been previously described in natural products and specific enzymes have been shown to produce these derivatives.46,56,57
3-Hydroxy-4-methylproline (OHMePro) appears in several antimicrobial compounds, such as echinocandins, and is proposed to potentiate their activity.58In vitro synthesis strategies have been developed for the production of OHMePro.58 In heinamides, OHMePro is present in variants B1 and B4, which together are the dominant 12-residue heinamides produced by this strain (Fig. 1). We propose that the OHMePro is synthesized by a group of four enzymes lxaO-R (Scheme 1). Three of these genes, lxaO-Q, are homologs to known cyanobacterial methylproline synthetic enzymes59 as in nostopeptolides,60 nostocyclopeptides,61 spumigins,62 and pseudoaeruginosins.63 The fourth gene is a putative 2OG-Fe(II)-dependent oxygenase lxaR, which 3-hydroxylates one of the methylproline intermediates but not the completed methylproline (Scheme 1). lxaR is related to the prolyl and lysyl hydroxylase family of AlkB, EGL-9, and leprecan. This class of enzymes is characterized as hydroxylating oxygenases, with prolyl 3- and 4-hydroxylases.64,65 4-MePro in heinamide B5 may be a byproduct, where LxaOPQ have acted without lxaR, leaking some 4-MePro to the cell. The biosynthesis of OHMePro in echinocandin variants pneumocandins has been described, where a α-ketoglutarate-FeII dependent enzyme GloF acts hydroxylating OHMePro, using MePro as substrate.66–68 The methylproline substrate is synthesized using Leu as starting point like in cyanobacterial MePro biosyntheses.67 GloF also hydoxylates Pro to 4-OHPro and 3-OHPro.66,68 No homolog to GloF could be found in Nostoc sp. UHCC 0702 genome. As we showed in the amino acid feeding experiment MePro is not used as substrate in OHMePro biosynthesis for laxaphycins, which shows that laxaphycin and pneumocandin OHMePros have different biosynthetic origins. We compared the OHMePro recognizing adenylation domain of LxaK12 to OHMePro adenylation domains in described echinocandin (BGC accession numbers KE145356 and JX421684).69,70 Both fungal adenylation domains had substrate binding pocket sequence identical to each other (DNTMITAMSK) with only 30% identity to the Lxa binding pocket (DVQFIAHAAK). A low similarity is however expected, as fungal adenylation domains differ from bacterial domains and the same training data is generally not used in the prediction of bacterial and fungal adenylation domain substrate specificities.71,72
The heinamide biosynthetic gene cluster contains three cupin domains (Fig. 3). We predict that two cupins hydroxylate the hydroxylated amino acids in positions 3, 5, and 8 in 12-residue heinamides and scytocyclamides. We predict that the third cupin domain unique for the heinamide cluster hydroxylates Aoa1, which is not hydroxylated in scytocyclamides.10 The adenylation domain binding pocket sequences for 3-OH-D-Leu and D-Leu are identical (DAWFLGNVVK) (Table S12†). This suggests that both domains identify the same substrate, and they both are predicted to recognize Leu. Both modules also have an epimerase domain, so stereospecificity does not explain the difference in hydroxylation. From this we conclude that the hydroxylation happens at a later stage. It is possible that the hydroxylation occurs as Leu is bound to the PCP or after the peptide is released. In the reported 12-residue laxaphycins Leu3 is always hydroxylated and Leu5 has hydroxylated and non-hydroxylated variants, with the exception of lyngbyacyclamides and heinamides (Table S1†). We propose, that a single cupin enzyme hydroxylates both leucins on the released peptide, with poorer affinity on Leu5. We also suggest that Leu5 in heinamides is not hydroxylated because structurally divergent cHse4 blocks the enzyme from interacting with the Leu5 residue. Although a previous article on laxaphycins predicted that the hydroxylation is performed by cytochrome p450 enzymes,36 we did not find these enzymes in the genomes of S. hofmannii PCC7110 or Nostoc sp. UHCC 0702. Thr dehydration to Dhb by the condensation domain of the modified AA clade has been shown experimentally in the biosynthesis of albopeptide.73 The condensation domain LxaC13 following the Thr incorporation module was found to belong this clade, as shown in phylogenetic studies10,74 (Fig. S13†).
Conclusions
In this study we characterized heinamides, which are new laxaphycin variants with several new modifications that possess antifungal activity, which is synergistic between the 11- and 12-residue compounds. These new laxaphycins broaden the known chemical space of bioactive natural products and provide new insight to potentially useful building blocks in antimicrobial molecules. We also describe the heinamide biosynthetic pathway, propose pathways for the amino acid modifications, and show experimentally the function of the proline hydroxylase LxaN. The synthesis of (2S,4R)-4-OHPro and OHMePro are of interest because their biosynthetic origins have been described in other organisms such as fungi but not in cyanobacteria.Author contributions
LH, KS, and DF designed the study. AJ, XO, MW, PP, and LH performed the experiments. LH, JJ, and DF analyzed and interpreted the data. LH, DF, JJ, and KS wrote the manuscript, which was corrected, revised, and approved by all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by a grant awarded to KS from the Jane and Aatos Erkko Foundation and LH was funded during the writing process by the Doctoral School of Environmental, Food and Biological Sciences of University of Helsinki. The authors thank Lyudmila Saari for maintaining the cyanobacterial strains Nostoc sp. UHCC 0702 and Scytonema hofmannii PCC 7110. S. hofmannii PCC 7110 was obtained from The Pasteur Culture Collection of Cyanobacteria.References
- E. Dittmann, M. Gugger, K. Sivonen and D. P. Fewer, Trends Microbiol., 2015, 23, 642–652 CrossRef CAS PubMed.
- J. Demay, C. Bernard, A. Reinhardt and B. Marie, Mar. Drugs, 2019, 17, 320 CrossRef CAS PubMed.
- T. Rodrigues, D. Reker, P. Schneider and G. Schneider, Nat. Chem., 2016, 8, 531–541 CrossRef CAS PubMed.
- E. Kim, B. S. Moore and Y. J. Yoon, Nat. Chem. Biol., 2015, 11, 649–659 CrossRef CAS PubMed.
- M. H. Medema, P. Cimermancic, A. Sali, E. Takano and M. A. Fischbach, PLoS Comput. Biol., 2014, 10, e1004016 CrossRef PubMed.
- L. Bohlin, U. Göransson, C. Alsmark, C. Wedén and A. Backlund, Phytochem. Rev., 2010, 9, 279–301 CrossRef CAS PubMed.
- W. P. Frankmölle, L. K. Larsen, F. R. Caplan, G. M. L. Patterson, G. Knubel, I. A. Levine and R. E. Moore, J. Antibiot., 1992, 45, 1451–1457 CrossRef PubMed.
- W. J. Cai, S. Matthew, Q. Y. Chen, V. J. Paul and H. Luesch, Bioorg. Med. Chem., 2018, 26, 2310–2319 CrossRef CAS PubMed.
- S. W. Luo, H. S. Kang, A. Krunic, W. L. Chen, J. L. Yang, J. L. Woodard, J. R. Fuchs, S. H. Cho, S. G. Franzblau, S. M. Swanson and J. Orjala, Bioorg. Med. Chem., 2015, 23, 3153–3162 CrossRef CAS PubMed.
- L. M. P. Heinilä, D. P. Fewer, J. K. Jokela, M. Wahlsten, A. Jortikka and K. Sivonen, Front. Microbiol., 2020, 11, 578878 CrossRef PubMed.
- P. Sullivan, A. Krunic, J. E. Burdette and J. Orjala, J. Antibiot., 2020, 73, 526–533 CrossRef CAS PubMed.
- N. Maru, O. Ohno and D. Uemura, Tetrahedron Lett., 2010, 51, 6384–6387 CrossRef CAS.
- G. Buda De Cesare, S. A. Cristy, D. A. Garsin and M. C. Lorenz, mBio, 2020, 11, e02123-20 CrossRef PubMed.
- J. A. Hendrickson, C. Hu, S. L. Aitken and N. Beyda, Curr. Infect. Dis. Rep., 2019, 21, 47 CrossRef PubMed.
- W. H. O. WHO, Antifungal drug resistance: The example of invasive Candidiasis, in Antimicrobial Resistance: Global Report on Surveillance, WHO Press, Geneva, Switzerland, 2014 Search PubMed.
- S.-i. Kajiyama, H. Kanzaki, K. Kawazu and A. Kobayashi, Tetrahedron Lett., 1998, 39, 3737–3740 CrossRef CAS.
- T. Neuhof, P. Schmieder, K. Preussel, R. Dieckmann, H. Pham, F. Bartl and H. von Döhren, J. Nat. Prod., 2005, 68, 695–700 CrossRef CAS PubMed.
- G. Trimurtulu, I. Ohtani, G. M. L. Patterson, R. E. Moore, T. H. Corbett, F. A. Valeriote and L. Demchik, J. Am. Chem. Soc., 1994, 116, 4729–4737 CrossRef CAS.
- T. K. Shishido, A. Humisto, J. Jokela, L. W. Liu, M. Wahlsten, A. Tamrakar, D. P. Fewer, P. Permi, A. P. D. Andreote, M. F. Fiore and K. Sivonen, Mar. Drugs, 2015, 13, 2124–2140 CrossRef CAS PubMed.
- J. Jokela, L. M. P. Heinilä, T. K. Shishido, M. Wahlsten, D. P. Fewer, M. F. Fiore, H. Wang, E. Haapaniemi, P. Permi and K. Sivonen, Front. Microbiol., 2017, 8, 1963 CrossRef PubMed.
- C. M. Thiele, K. Petzold and J. Schleucher, Chem. – Eur. J., 2009, 15, 585–588 CrossRef CAS PubMed.
- K. Blin, S. Shaw, K. Steinke, R. Villebro, N. Ziemert, S. Y. Lee, M. H. Medema and T. Weber, Nucleic Acids Res., 2019, 47, W81–W87 CrossRef CAS PubMed.
- K. Blin, T. Wolf, M. G. Chevrette, X. W. Lu, C. J. Schwalen, S. A. Kautsar, H. G. S. Duran, E. Santos, H. U. Kim, M. Nave, J. S. Dickschat, D. A. Mitchell, E. Shelest, R. Breitling, E. Takano, S. Y. Lee, T. Weber and M. H. Medema, Nucleic Acids Res., 2017, 45, W36–W41 CrossRef CAS PubMed.
- N. Ziemert, S. Podell, K. Penn, J. H. Badger, E. Allen and P. R. Jensen, PLoS One, 2012, 7, e34064 CrossRef CAS PubMed.
- J. Vestola, T. K. Shishido, J. Jokela, D. P. Fewer, O. Aitio, P. Permi, M. Wahlsten, H. Wang, L. Rouhiainen and K. Sivonen, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E1909 CrossRef CAS PubMed.
- J. Mareš, J. Hajek, P. Urajova, J. Kopecky and P. Hrouzek, PLoS One, 2014, 9, e111904 CrossRef PubMed.
- W. P. Frankmölle, G. Knubel, R. E. Moore and G. M. L. Patterson, J. Antibiot., 1992, 45, 1458–1466 CrossRef PubMed.
- L. Bornancin, E. Alonso, R. Alvarino, N. Inguimbert, I. Bonnard, L. M. Botana and B. Banaigs, Bioorg. Med. Chem., 2019, 27, 1966–1980 CrossRef CAS PubMed.
- I. Bonnard, M. Rolland, C. Francisco and B. Banaigs, Lett. Pept. Sci., 1997, 4, 289–292 CAS.
- W. H. Gerwick, Z. D. Jiang, S. K. Agarwal and B. T. Farmer, Tetrahedron, 1992, 48, 2313–2324 CrossRef CAS.
- J. B. MacMillan, M. A. Ernst-Russell, J. S. de Ropp and T. F. Molinski, J. Org. Chem., 2002, 67, 8210–8215 CrossRef CAS PubMed.
- J. C. Grewe, dissertation/doctoral thesis, Albert-Ludwigs-Universität Freiburg im Breisgau, 2005.
- I. Bonnard, M. Rolland, J. M. Salmon, E. Debiton, C. Barthomeuf and B. Banaigs, J. Med. Chem., 2007, 50, 1266–1279 CrossRef CAS PubMed.
- F. Boyaud, Z. Mahiout, C. Lenoir, S. Tang, J. Wdzieczak-Bakala, A. Witczak, I. Bonnard, B. Banaigs, T. Ye and N. Inguimbert, Org. Lett., 2013, 15, 3898–3901 CrossRef CAS PubMed.
- M. Gaillard, S. Das, M. Djibo, D. Raviglione, C. Roumestand, B. Legrand and N. Inguimbert, Tetrahedron Lett., 2018, 59, 3713–3718 CrossRef CAS.
- L. Bornancin, F. Boyaud, Z. Mahiout, I. Bonnard, S. C. Mills, B. Banaigs and N. Inguimbert, Mar. Drugs, 2015, 13, 7285–7300 CrossRef CAS PubMed.
- R. Alvarino, E. Alonso, L. Bornancin, I. Bonnard, N. Inguimbert, B. Banaigs and L. M. Botana, Mar. Drugs, 2020, 18, 364 CrossRef CAS PubMed.
- S. W. Luo, A. Krunic, H. S. Kang, W. L. Chen, J. L. Woodard, J. R. Fuchs, S. M. Swanson and J. Orjala, J. Nat. Prod., 2014, 77, 1871–1880 CrossRef CAS PubMed.
- T. Okino, S. Qi, H. Matsuda, M. Murakami and K. Yamaguchi, J. Nat. Prod., 1997, 60, 158–161 CrossRef CAS.
- Q. Yue, L. Chen, X. Zhang, K. Li, J. Sun, X. Liu, Z. An and G. F. Bills, Eukaryotic Cell, 2015, 14, 698–718 CrossRef CAS PubMed.
- A. Pluotno and S. Carmeli, Tetrahedron, 2005, 61, 575–583 CrossRef CAS.
- J. Mareš, J. Hájek, P. Urajová, A. Kust, J. Jokela, K. Saurav, T. Galica, K. Čapková, A. Mattila, E. Haapaniemi, P. Permi, I. Mysterud, O. M. Skulberg, J. Karlsen, D. P. Fewer, K. Sivonen, H. H. Tønnesen and P. Hrouzek, Appl. Environ. Microbiol., 2019, 85, e02675-18 CrossRef PubMed.
- N. A. Moss, G. Seiler, T. F. Leão, G. Castro-Falcón, L. Gerwick, C. C. Hughes and W. H. Gerwick, Angew. Chem., Int. Ed., 2019, 58, 9027–9031 CrossRef CAS PubMed.
- B. Baral, A. Akhgari and M. Metsä-Ketelä, Synth. Syst. Biotechnol., 2018, 3, 163–178 CrossRef PubMed.
- O. Lazos, M. Tosin, A. L. Slusarczyk, S. Boakes, J. Cortés, P. J. Sidebottom and P. F. Leadlay, Chem. Biol., 2010, 17, 160–173 CrossRef CAS PubMed.
- H.-L. Zhang, C. Zhang, C.-H. Pei, M.-N. Han, Z.-D. Xu, C.-H. Li and W. Li, Lett. Appl. Microbiol., 2018, 66, 400–408 CrossRef CAS PubMed.
- T. M. H. Bach and H. Takagi, Appl. Microbiol. Biotechnol., 2013, 97, 6623–6634 CrossRef CAS PubMed.
- P. Remuzon, Tetrahedron, 1996, 52, 13803–13835 CrossRef CAS.
- R. Hara and K. Kino, Appl. Microbiol. Biotechnol., 2020, 104, 4771–4779 CrossRef CAS PubMed.
- J. M. Williams, K. M. J. Brands, R. T. Skerlj, R. B. Jobson, G. Marchesini, K. M. Conrad, B. Pipik, K. A. Savary, F.-R. Tsay, P. G. Houghton, D. R. Sidler, U.-H. Dolling, L. M. DiMichele and T. J. Novak, J. Org. Chem., 2005, 70, 7479–7487 CrossRef CAS PubMed.
- J. D. Armstrong, J. L. Keller, J. Lynch, T. Liu, F. W. Hartner, N. Ohtake, S. Okada, Y. Imai, O. Okamoto, R. Ushijima, S. Nakagawa and R. P. Volante, Tetrahedron Lett., 1997, 38, 3203–3206 CrossRef CAS.
- M. Ionac, M. J. Parnham, M. Plauchithiu and K. Brune, Pharmacol. Res., 1996, 33, 367–373 CrossRef CAS PubMed.
- M. Aoki, K. Suto, M. Komatsu, A. Kamimura, K. Morishita, M. Yamasaki and T. Takao, Biosci. Biotechnol. Biochem., 2012, 76, 1242–1244 CrossRef CAS PubMed.
- H. Ohara, S. Ichikawa, H. Matsumoto, M. Akiyama, N. Fujimoto, T. Kobayashi and S. Tajima, J. Dermatol., 2010, 37, 330–338 CrossRef CAS PubMed.
- J. Yuan, W. Xiong, X. Zhou, Y. Zhang, D. Shi, Z. Li and H. Lu, J. Am. Chem. Soc., 2019, 141, 4928–4935 CrossRef CAS PubMed.
- H. Luesch, D. Hoffmann, J. M. Hevel, J. E. Becker, T. Golakoti and R. E. Moore, J. Org. Chem., 2003, 68, 83–91 CrossRef CAS PubMed.
- P. Lukat, Y. Katsuyama, S. Wenzel, T. Binz, C. Koenig, W. Blankenfeldt, M. Broenstrup and R. Mueller, Chem. Sci., 2017, 8, 7521–7527 RSC.
- D. K. Mohapatra, D. Mondal, M. S. Chorghade and M. K. Gurjar, Tetrahedron Lett., 2006, 47, 9215–9219 CrossRef CAS.
- M. Hibi, T. Kawashima, P. M. Sokolov, S. V. Smirnov, T. Kodera, M. Sugiyama, S. Shimizu, K. Yokozeki and J. Ogawa, Appl. Microbiol. Biotechnol., 2013, 97, 2467–2472 CrossRef CAS PubMed.
- D. Hoffmann, J. M. Hevel, R. E. Moore and B. S. Moore, Gene, 2003, 311, 171–180 CrossRef CAS PubMed.
- J. E. Becker, R. E. Moore and B. S. Moore, Gene, 2004, 325, 35–42 CrossRef CAS PubMed.
- D. P. Fewer, J. Jokela, L. Rouhiainen, M. Wahlsten, K. Koskenniemi, L. J. Stal and K. Sivonen, Mol. Microbiol., 2009, 73, 924–937 CrossRef CAS PubMed.
- L. Liu, A. Budnjo, J. Jokela, B. E. Haug, D. P. Fewer, M. Wahlsten, L. Rouhiainen, P. Permi, T. Fossen and K. Sivonen, ACS Chem. Biol., 2015, 10, 725–733 CrossRef CAS PubMed.
- W. G. Kaelin, Annu. Rev. Biochem., 2005, 74, 115–128 CrossRef CAS PubMed.
- J. A. Vranka, L. Y. Sakai and H. P. Bächinger, J. Biol. Chem., 2004, 279, 23615–23621 CrossRef CAS PubMed.
- S. Houwaart, L. Youssar and W. Huttel, ChemBioChem, 2014, 15, 2365–2369 CrossRef CAS PubMed.
- L. Chen, Q. Yue, Y. Li, X. Niu, M. Xiang, W. Wang, G. F. Bills, X. Liu and Z. An, Appl. Environ. Microbiol., 2015, 81, 1550–1558 CrossRef PubMed.
- Y. Li, L. Chen, Q. Yue, X. Liu, Z. An and G. F. Bills, ACS Chem. Biol., 2015, 10, 1702–1710 CrossRef CAS PubMed.
- L. Chen, Q. Yue, X. Zhang, M. Xiang, C. Wang, S. Li, Y. Che, F. J. Ortiz-López, G. F. Bills, X. Liu and Z. An, BMC Genomics, 2013, 14, 339 CrossRef CAS PubMed.
- R. A. Cacho, W. Jiang, Y.-H. Chooi, C. T. Walsh and Y. Tang, J. Am. Chem. Soc., 2012, 134, 16781–16790 CrossRef CAS.
- M. Röttig, M. H. Medema, K. Blin, T. Weber, C. Rausch and O. Kohlbacher, Nucleic Acids Res., 2011, 39, W362–W367 CrossRef.
- D. Baranašić, J. Zucko, J. Diminic, R. Gacesa, P. F. Long, J. Cullum, D. Hranueli and A. Starcevic, J. Ind. Microbiol. Biotechnol., 2014, 41, 461–467 CrossRef PubMed.
- S. Wang, Q. Fang, Z. Lu, Y. Gao, L. Trembleau, R. Ebel, J. H. Andersen, C. Philips, S. Law and H. Deng, Angew. Chem., Int. Ed., 2021, 60, 3229–3237 CrossRef CAS PubMed.
- D. Tillett, E. Dittmann, M. Erhard, H. von Dohren, T. Borner and B. A. Neilan, Chem. Biol., 2000, 7, 753–764 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ob00772f |
| This journal is © The Royal Society of Chemistry 2021 |