Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Constrained beta-amino acid-containing miniproteins†
Magda
Drewniak-Świtalska
,
Barbara
Barycza
,
Ewa
Rudzińska-Szostak
 ,
Paweł
Morawiak
,
Paweł
Morawiak
 and
Łukasz
Berlicki
and
Łukasz
Berlicki
 *
*
Department of Bioorganic Chemistry, Wrocław University of Science and Technology, Wybrzeże Wyspiańskiego 27, 50-370 Wrocław, Poland. E-mail: lukasz.berlicki@pwr.edu.pl
First published on 17th March 2021
Abstract
The construction of β-amino acid-containing peptides that fold to tertiary structures in solution remains challenging. Two model miniproteins, namely, Trp-cage and FSD, were scanned using a constrained β-amino acid in order to evaluate its impact on the folding process. Relationships between forces stabilizing the miniprotein structure and conformational stability of analogues were found. The possibility of a significant increase of the conformational stability of the studied miniproteins by substitution with the β-amino acid at the terminus of a helix is shown. On the basis of these results, β-amino acid containing-peptide analogs with helical fragments substantially altered by the incorporation of several constrained β-amino acids were designed, synthesized and evaluated with respect to their structure and stability. The smallest known β-amino acid-containing peptide with a well-defined tertiary structure is described.
Introduction
The construction of β-amino acid containing peptides that fold into well-defined structures in solution has attracted much attention in recent years.1 Numerous studies have indicated the possibility of the formation of secondary structures with various geometries and properties. In particular, conformationally constrained, cycloalkane-based β-amino acids are of interest due to their significant structuring effect combined with good synthetic availability.2 Folding propensities are related to the applied sequence pattern and the stereochemistry of neighboring amino acid residues. The theory of the so-called ‘stereochemical patterning’ indicates that the combination of chosen types of amino acid residues with a tendency for specific φ and ψ dihedral angles, in a chosen sequence pattern results in the formation of a predictable secondary structure.3 If the signs of these angles flanking amide bonds match, a helical structure is formed. Therefore, secondary structure formation is considered to be well predictable, and several examples with various sequence patterns, including αβ, ααβ, and ααββ, have been described.4However, building β-amino acid-containing peptides adopting higher order structures remains challenging.5 The number of studies is very limited and related to the modification of only three small proteins that form hydrophobic cores, as well as a few examples of metal-binding or disulfide rich miniproteins.6 Villin headpiece subdomain (VHP),7 the B domain of protein G (GB1)8 and betabellin-14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 are examples of miniproteins with a hydrophobic core, which is considered to be the major driving force for the folding of native globular proteins. Gellman and coworkers tested a single substitution of four positions of VHP, indicating the possibility of the formation of a native-like structure. For four trans-(1S,2S)-2-aminocyclopentanecarboxylic acid (trans-ACPC) substituted VHP structures, it was proven that the impact of this mutation is generally destabilizing and position-specific as ΔTm values (difference between substituted and native miniproteins) were in a range of −15 to 0.3 °C. In the case of betabellin-14, two positions have been tested by substitution with six various β-amino acids. It has been shown that the incorporation of a β3-open chain residue leads to hydrophobic core disruption, while cyclopentane-based β-amino acid (cis-ACPC) substitution induced conformational mismatch between interacting backbones. Only modification by cyclohexane-based β-amino acid resulted in full conservation of the tertiary structure. Most extensive studies have been performed on the GB1 model, wherein several modifications using β-amino acids were introduced. Substitution of the helical fragment of GB1 by β3-open chain or trans-ACPC residues results in a significant decrease of conformational stability, as indicated by lowering of the Tm value by 11–26 °C. It was concluded that these modifications are enthalpically unfavorable and entropically favorable. Interestingly, in the above-mentioned studies, no β-amino acid-modified miniprotein has shown increased conformational stability compared to its native counterpart.
9 are examples of miniproteins with a hydrophobic core, which is considered to be the major driving force for the folding of native globular proteins. Gellman and coworkers tested a single substitution of four positions of VHP, indicating the possibility of the formation of a native-like structure. For four trans-(1S,2S)-2-aminocyclopentanecarboxylic acid (trans-ACPC) substituted VHP structures, it was proven that the impact of this mutation is generally destabilizing and position-specific as ΔTm values (difference between substituted and native miniproteins) were in a range of −15 to 0.3 °C. In the case of betabellin-14, two positions have been tested by substitution with six various β-amino acids. It has been shown that the incorporation of a β3-open chain residue leads to hydrophobic core disruption, while cyclopentane-based β-amino acid (cis-ACPC) substitution induced conformational mismatch between interacting backbones. Only modification by cyclohexane-based β-amino acid resulted in full conservation of the tertiary structure. Most extensive studies have been performed on the GB1 model, wherein several modifications using β-amino acids were introduced. Substitution of the helical fragment of GB1 by β3-open chain or trans-ACPC residues results in a significant decrease of conformational stability, as indicated by lowering of the Tm value by 11–26 °C. It was concluded that these modifications are enthalpically unfavorable and entropically favorable. Interestingly, in the above-mentioned studies, no β-amino acid-modified miniprotein has shown increased conformational stability compared to its native counterpart.
β-Amino acid-containing peptides exhibit great potential for various applications10 due to predictable and highly stable conformations, as well as the possibility of incorporation of a wide range of various functional groups in side chains. In particular, several types of biological activities11 have been evidenced for this class of compounds, including antimicrobial activity,12 inhibition of protein–protein interactions,13 agonism/antagonism of GPCR,14 and antiangiogenic activity.15 Moreover, catalytic activity has also been shown.16
Taking into account the possible high potential of the extended, protein-like structures of β-amino acid-containing peptides, further studies towards a better understanding of folding processes are of high importance. In this paper, we explore the methodology of building β-amino acid-containing peptide foldamers based on systematic scanning of helical fragments with constrained β-amino acids, namely, trans-ACPC. Two model miniproteins are used in order to possibly draw general conclusions.
Results and discussion
The miniproteins Trp-cage17 and FSD,18 incorporating helical and extended structures stabilized by hydrophobic interactions in the protein core, were selected (Fig. 1). Trp-cage consists of an α-helical fragment, a short 310-helix and a poly-proline helix. The van der Waals interactions of the tryptophan side chain with proline residues are crucial for miniprotein tertiary structure stabilization. The FSD miniprotein was designed de novo and it contains two extended fragments and an α-helix. π–π stacking is a major interaction between phenylalanine residues that form the hydrophobic core. Importantly, the small size of the chosen models (sequence lengths of 20 and 28 amino acid residues) makes them relatively sensitive to substitutions; therefore, the influence of β-residues could be evaluated well. Moreover, differences in miniprotein construction including the size and positioning of the helical fragment (N- versus C-terminal for Trp-cage and FSD, respectively) as well as different types of interactions stabilizing the hydrophobic core could allow us to draw general conclusions regarding the effect of β-amino acid substitution.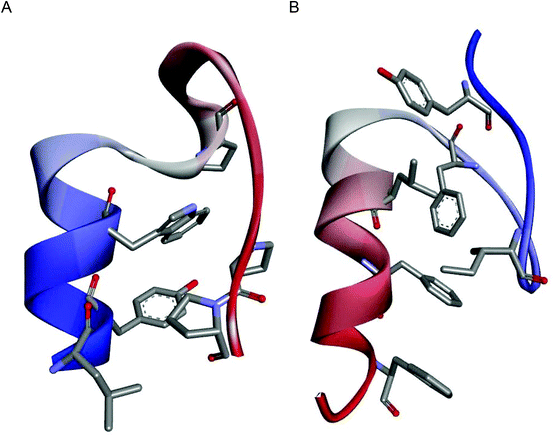 | ||
| Fig. 1 NMR structures of the model miniproteins: Trp-cage (PDB id 1L2Y)17 (A) and FSD (PDB id 2FSV)18 (B). The main chains are shown as ribbons colored from blue (N-terminus) to red (C-terminus), and the side chains forming the hydrophobic cores are shown as sticks colored by atoms. | ||
The scan of the α-helical fragment of Trp-cage with trans-ACPC (peptides 2–9, Tables S1 and S2†) was performed in order to investigate the impact of single substitutions on the conformational stability (Table 1, Fig. 2). The CD spectrum of peptide 2 showed two minima which are characteristic of α-helix-like structures. In the case of the CD spectra of peptides 4–6 and 9, the major peak at 205 nm is accompanied by a less intense band at 220 nm or 215 nm for structures 4–6 and 9, respectively. Therefore, the helical conformation of peptides 4–6, which are substituted in the central part of the helix, is affected more than in the case of peptide 2, which is modified at the N-terminus. Their CD spectra are also analogous to those observed for trans-ACPC substituted helical peptides reported previously.19 The trans-ACPC substitution of the C-terminal part of the Trp-cage helix (peptide 9), which is in the central part of the whole sequence, appears to destabilize the structure, as the major signal and the shoulder are of similar intensity, but shifted towards lower wavelengths. The CD spectra of peptides 3, 7, and 8 indicate unordered structures. The CD spectral analysis result is consistent with the observed temperature dependences, which were of sigmoidal shape allowing the measurement of melting points for peptides 2, 4–6 and 9 (Table 1 and Fig. S1†).7 Most of these peptides showed Tm values that were slightly lower than that of the parent peptide 1, with the exception of peptide 5, where the substitution of Ile4 resulted in a Tm = 46.6 °C. The observed increase of the Tm value for peptide 5 is a result of the combination of a few phenomena: (a) similar size and character (hydrophobic) of trans-ACPC and Ile side chains; (b) outside position in the three-dimensional structure of Trp-cage, and (c) position in the sequence close to the N-terminus, which does not affect the critical stabilizing interaction of the Trp6 residue. Therefore, the position of trans-ACPC substitution correlates well with the conformational stability of the peptide. Modification of residues forming the hydrophobic core (Leu2, Trp6 and Leu7) led to the loss of the tertiary structure. Mutations of other residues of the helix only slightly decreased (or kept constant) the conformational stability of Trp-cage. Interestingly, the addition of a trans-ACPC residue at the N-terminus of the Trp-cage miniprotein led to a significant increase in the Tm value (compound 10, Tm = 54.2 °C). The CD spectrum of peptide 10 (Fig. 2B) is very similar to the CD spectrum of the parent peptide 1, indicating that this mutation does not influence the conformation of the molecule. Therefore, the enhanced conformational stability of peptide 10 can be attributed to the structuration of the end of the helix by the conformationally constrained trans-ACPC residue, which also promotes helical folding. Moreover, the terminal position of this β-amino acid does not affect the interactions stabilizing the tertiary structure.
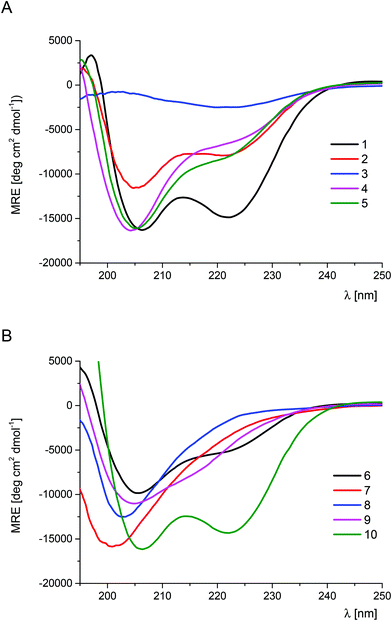 | ||
| Fig. 2 CD spectra of the Trp-cage (1) and its trans-ACPC mono-substituted analogs (peptides 1–5 – panel A, peptides 5–10 – panel B) measured in phosphate buffer solution at pH 7.0 (293 K). | ||
| No. | Sequence | T m [°C] |
|---|---|---|
| All peptides are C-amidated and N-acetylated; pentagons denote trans-ACPC.a Measurement of the Tm value was not possible due to peptide precipitation; n/a – the Tm value is not available due to the lack of sigmoidal transition in the observed CD dependence. | ||
| 1 | NLYIQWLKDGGPSSGRPPPS | 44.0 ± 1.3 |
| 2 | ⬟LYIQWLKDGGPSSGRPPPS | 35.6 ± 1.5 |
| 3 | N⬟YIQWLKDGGPSSGRPPPS | n/a |
| 4 | NL⬟IQWLKDGGPSSGRPPPS | 37.2 ± 4.4 |
| 5 | NLY⬟QWLKDGGPSSGRPPPS | 46.6 ± 1.1 |
| 6 | NLYI⬟WLKDGGPSSGRPPPS | 41.9 ± 1.3 |
| 7 | NLYIQ⬟LKDGGPSSGRPPPS | n/a |
| 8 | NLYIQW⬟KDGGPSSGRPPPS | n/a |
| 9 | NLYIQWL⬟DGGPSSGRPPPS | 39.3 ± 2.2 |
| 10 | ⬟NLYIQWLKDGGPSSGRPPPS | 54.2 ± 1.3 |
| 11 | ⬟LY⬟QWLKDGGPSSGRPPPS | 49.5 ± 1.9 |
| 12 | NLY⬟QWL⬟DGGPSSGRPPPS | 38.0 ± 2.3 |
| 13 | ⬟LY⬟QWL⬟DGGPSSGRPPPS | 47.1 ± 3.6 |
| 14 | ⬟LYI⬟WL⬟DGGPSSGRPPPS | |
| 15 | ⬟NLY⬟QWL⬟DGGPSSGRPPPS | 38.3 ± 4.2 |
| 16 | QQYTAKIKGRTFRNEKELRDFIEKFKGR | 55.7 ± 3.0 |
| 17 | QQYTAKIKGRTFRNEKELRDFIEKFKG⬟ | 61.3 ± 1.9 |
| 18 | QQYTAKIKGRTFRNEKELRDFIEKFK⬟R | 58.3 ± 3.3 |
| 19 | QQYTAKIKGRTFRNEKELRDFIEKF⬟GR | 66.3 ± 1.4 |
| 20 | QQYTAKIKGRTFRNEKELRDFIE⬟FKGR | 41.7 ± 2.9 |
| 21 | QQYTAKIKGRTFRNEKELRDFI⬟KFKGR | 51.9 ± 3.9 |
| 22 | QQYTAKIKGRTFRNEKELRDF⬟EKFKGR | n/a |
| 23 | QQYTAKIKGRTFRNEKELR⬟FIEKFKGR | n/a |
| 24 | QQYTAKIKGRTFRNEKEL⬟DFIEKFKGR | n/a |
| 25 | QQYTAKIKGRTFRNEKE⬟RDFIEKFKGR | n/a |
| 26 | QQYTAKIKGRTFRNEK⬟LRDFIEKFKGR | n/a |
| 27 | QQYTAKIKGRTFRNEKELRDFI⬟KF⬟GR | 50.2 ± 1.5 |
| 28 | QQYTAKIKGRTFRN⬟KEL⬟DF⬟EKFKGR | n/a |
| 29 | QQYTAKIKGRTFRNEKEL⬟DF⬟EKF⬟GR | n/a |
The results of the trans-ACPC scan of the FSD miniprotein (peptides 17–26) were considerably different (Table 1 and Fig. 3 and S3†). Only the C-terminal substitutions (peptides 17–21) led to conformationally stable miniproteins. Notably, peptides with mutations at the last three positions (17–19) showed CD spectra with a shape similar to that of the parent peptide 16, but their absolute MRE values were higher, which indicated a higher helical content. These peptides also exhibited significantly increased conformational stability in comparison with the reference structure 16. The most stable derivative 19 exceeded the Tm value of the original peptide 16 by approximately 10 °C (Tm = 55.7 °C versus 66.3 °C for 16 and 19, respectively). On the basis of these observations, it could be concluded that the substitution of FSD at positions 26–28 by trans-ACPC leads to the structuration of the C-terminal fragment, enhancing the stability of the helical fragment. These modifications do not interfere with the interactions between the secondary structures. Therefore, in both studied cases, it was possible to indicate the positions at the end of helical structures that led to significant conformational stabilization of the whole miniprotein.
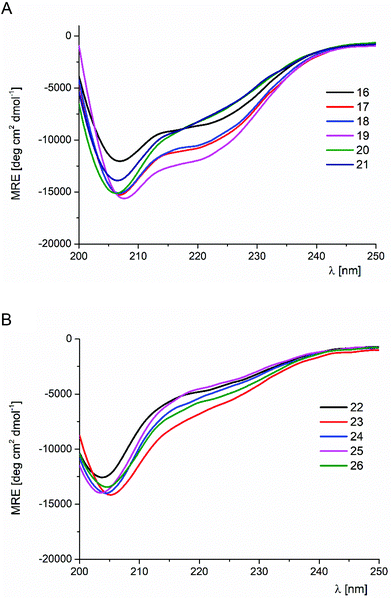 | ||
| Fig. 3 CD spectra of FSD (16) and its trans-ACPC mono-substituted analogs (peptides 16–21 – panel A, peptides 22–26 – panel B), measured in phosphate buffer solution at pH 7.0 (293 K). | ||
The CD spectra of peptides 20 and 21, with trans-ACPC substitutions at positions 23 and 24 which are close to the center of the helix, are slightly different from those of the parent peptide 16. The minimum at 206 nm is deeper, while the signal at 220 nm is less intense. This change of the CD spectral shape could be related to the conformational changes introduced by trans-ACPC substitution in the middle of the helix. These changes still allow the formation of a tertiary structure, although they are characterized by lower Tm values (42 and 52 °C for peptides 20 and 21, respectively).
Peptides 22–26 with trans-ACPC substitutions at positions 16–22 were found to be conformationally unstable. The CD spectra were characterized by the shift of the minimum towards lower wavelengths, while the CD signal temperature dependences did not show the characteristic sigmoidal shape.
The observed differences in trans-ACPC scan results between the two studied miniproteins are most probably the consequence of different packing of the hydrophobic core and interactions stabilizing this core. In the case of Trp-cage, the van der Waals interactions between the central Trp side chain and the surrounding lipophilic residues allow minimal changes to the structure. In the case of FSD, the face-to-edge π–π stacking between the side chains of Phe12 and Phe21 is the key miniprotein stabilizing interaction, and any substitution to the sequence between these residues (or close to Phe21) changes their relative positioning and destabilizes their interaction.
Subsequently, on the basis of the performed scan results, miniproteins with the conformation of helical fragments significantly altered by the incorporation of several trans-ACPC residues were designed. In the case of Trp-cage, positions 1, 3–5 and 8 can be substituted. The application of the ααβαααβ sequence pattern, which has already been shown to provide stable helical structures,20 resulted in peptides 13 and 14 with three β-amino acid moieties. Two sequences with two trans-ACPC residues, namely, compounds 11 and 12, were also synthesized. Moreover, one sequence (peptide 15), which explored the possibility of an N-terminal extension of the peptide, was prepared. The CD spectra of peptides 11–15 showed very similar shapes, although with variable intensities of the peaks (Fig. 4).
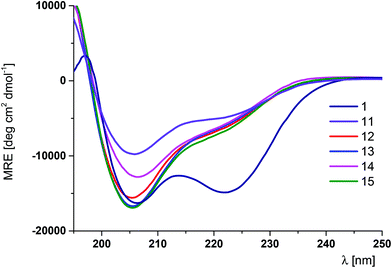 | ||
| Fig. 4 CD spectra of peptides 11–15 and reference peptide 1 dissolved in phosphate buffer solution at pH 7.0 (293 K). | ||
The presence of two bands at around 206 and 220 nm of different intensities in the CD spectra suggests the presence of α-helix-like structures. The majority of these peptides showed thermal denaturation curves typical of cooperatively folded proteins (Fig. S2†), with the exception of peptide 14, which precipitated during measurements. The melting temperature for peptide 13 was slightly higher than that observed for the parent peptide 1. Comparison of the Tm values of partially substituted peptides 11 and 12, which differ significantly, indicates that N-terminal substitutions are stabilizing while substitutions closer to the protein core are destabilizing, which, in total, combine to contribute to small differences observed in the Tm values for peptides 1 and 13.
Analogous trials for the construction of FSD analogs were undertaken. Consistent with the scan results, only peptides with substitutions in regions 24–28 in the sequence gave peptides that fold, namely, derivative 27. Peptides 28 and 29 with trans-ACPC incorporated at positions close to the miniprotein core were found to be conformationally unstable.
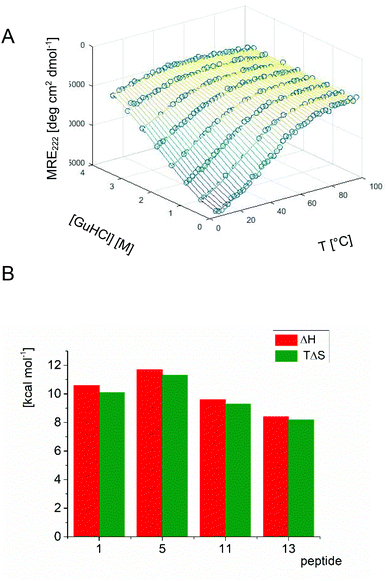
In order to gain insight into the folding process of trans-ACPC substituted analogs, a detailed analysis was performed for unsubstituted peptide 1, monosubstituted oligomer 5, disubstituted compound 11 and trisubstituted compound 13. Monitoring the CD signal (MRE at 222 nm) as a function of the denaturant (guanidine hydrochloride) and temperature provides a dependence that allows the determination of the set of thermodynamic parameters of unfolding (ΔG, ΔH, TΔS, Cp and m, Fig. 5 and S4†).8 The enthalpy of unfolding (ΔH) decreases with an increasing number of trans-ACPC substitutions, indicating that the intramolecular interactions are disturbed by these modifications. On the other hand, rigidification of the peptide structure by increasing the number of constrained β-amino acid residues causes a decrease in entropy. Therefore, the trans-ACPC residue preorganizes the structure of the peptide, which is in agreement with previously observed helix-promoting properties. However, the changes in the two abovementioned components of the free energy of folding balance each other out, and the values of ΔG° and equilibrium constant K (Fig. S4†) remain similar for the analyzed peptides indicating similar conformational stability.
The three-dimensional structure in solution of the most stable analog 13 has been analyzed in detail using NMR techniques. All 1H resonances of this miniprotein have been assigned using 2D TOCSY and NOESY spectra (Table S3, Fig. S10, S11 and S13†). Numerous nonsequential interproton contacts covering the whole sequence were found and they indicated a well-folded structure. In particular, regular i—(i + 3) contacts spanning residues 2 to 11 confirmed the presence of an α-helix-like structure (Table S4, Fig. S6†). In this region, contacts HA(i) − HN(i + 3) or HB(i) − HN(i + 3) (if the β-amino acid is at the i position) were detected with the exception of the interaction between trans-ACPC2 and trans-ACPC5, where HB(2)–HA(5) contact was observed. Long distance contacts were observed between the Trp6 side chain and Gly11, Pro12, Arg16 and Pro18, as well as between the Tyr3 side chain and Pro18 (Fig. S12†). Secondary chemical shifts for the Hα protons of peptide 13 (Fig. S5†) were found to be negative for nearly all the residues except Gly15, Arg17 and Pro18, indicating the formation of helical structures analogous to those found in Trp-cage, where α-helix, 310-helix and polyproline helix are present.21 Using NMR-derived constraints, the three-dimensional structure of peptide 13 was modeled using X-plor NIH software (Table S5†).22 A well-ordered structure with secondary elements arranged in a manner similar to that observed for the original Trp-cage was obtained (Fig. 6 and S7†). The hydrophobic core of miniprotein 13 contained the side chains of Trp7, Tyr4, and Pro19 and partially Pro13 and Leu3. The Trp6 side chain was sandwiched between Pro13 and Pro19. The major difference between peptide 13 and Trp-cage is observed in the construction of the helical part, and also the shift of N-terminal part in relation to the C-terminal part and the different conformation of the Tyr4 side chain could be noticed (Fig. S8†). The N-terminal part of the peptide forms a helical structure that is analogous to that reported by Gellman and coworkers for a peptide with the ααβαααβ sequence pattern.20 A regular network of hydrogen bonds NH(i)⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(i − 4) was observed for residues 2–9. The last turn of this helix was disturbed, and the Asp10-Gln7 hydrogen bond was very weak, while Gly12-Trp7 was additionally formed. In summary, although the three-dimensional structure of peptide 13 was well-defined, the observed small conformational changes, i.e., relative shifts of the secondary structures or some weaker hydrogen bonds, may contribute to the lower ΔH values measured for this peptide compared to the native Trp-cage.
C(i − 4) was observed for residues 2–9. The last turn of this helix was disturbed, and the Asp10-Gln7 hydrogen bond was very weak, while Gly12-Trp7 was additionally formed. In summary, although the three-dimensional structure of peptide 13 was well-defined, the observed small conformational changes, i.e., relative shifts of the secondary structures or some weaker hydrogen bonds, may contribute to the lower ΔH values measured for this peptide compared to the native Trp-cage.
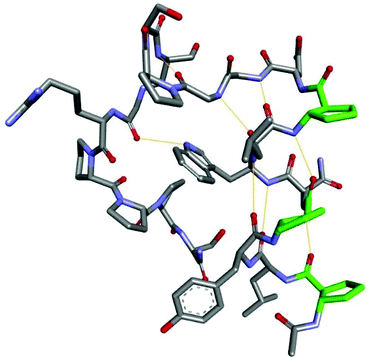 | ||
| Fig. 6 Average NMR-derived structure of peptide 13. Hydrogen bonds are shown as orange thin lines, and the carbon atoms of β-amino acid residues are shown in green. | ||
Conclusions
In summary, we performed a detailed analysis of the possibility of incorporation of constrained β-amino acid residues in the helical fragments of two miniproteins, namely, Trp-cage and FSD. Trans-ACPC scans revealed different results for the two studied miniproteins, indicating that the possibility of trans-ACPC substitution is related to the nature of the interactions that stabilize the hydrophobic core. The van der Waals interactions in the Trp-cage core were not significantly affected by the changes introduced by helix modifications, while the π–π interactions in the FSD core were damaged by the structural changes resulting from trans-ACPC incorporation into the core side of the helix. On the other hand, in both cases, it was possible to indicate the substitution, which led to considerably increased conformational stability of the miniproteins, i.e., the trans-ACPC residue added to the N-terminus of the Trp-cage helix (compound 10) or included in the C-terminal part of FSD (peptide 19).Finally, on the basis of the scan results, we were able to construct a miniprotein analogous to Trp-cage with a fully modified helical fragment according to the ααβαααβ sequence pattern (peptide 13) with conformational stability similar to that of its native counterpart. The detailed CD measurements in combination with structural NMR analysis provided insights into the contribution of the stabilization factors. The incorporation of trans-ACPC residues rigidified the structure and preorganized it for folding, decreasing the change in entropy. However, the trans-ACPC incorporation also caused structural disturbances that decreased intramolecular stabilizing interactions (ΔH). Notably, compound 13 is the smallest known β-amino acid-containing peptide that folds into a well-defined tertiary structure. We hope that the presented results will contribute to a better understanding of β-amino acid-containing peptide folding and provide a basis for the construction of more extended structures.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work was financially supported by the National Science Centre, Poland (grant no. 2016/21/B/ST5/00269).References
- (a) K. L. George and W. S. Horne, Acc. Chem. Res., 2018, 51, 1220–1228 CrossRef PubMed; (b) L. K. Pilsl and O. Reiser, Amino Acids, 2011, 41, 709–718 CrossRef PubMed; (c) L. M. Johnson and S. H. Gellman, Methods Enzymol., 2013, 523, 407–429 Search PubMed.
- (a) T. A. Martinek and F. Fülöp, Chem. Soc. Rev., 2012, 41, 687–702 RSC; (b) F. Fülöp, T. A. Martinek and G. K. Tóth, Chem. Soc. Rev., 2006, 35, 323–334 RSC.
- I. M. Mándity, E. Wéber, T. A. Martinek, G. Olajos, G. K. Tóth, E. Vass and F. Fülöp, Angew. Chem., Int. Ed., 2009, 48, 2171–2175 CrossRef PubMed.
- (a) Ł. Berlicki, L. Pilsl, E. Wéber, I. M. Mándity, C. Cabrele, T. A. Martinek, F. Fülöp and O. Reiser, Angew. Chem., Int. Ed., 2012, 51, 2208–2212 CrossRef; (b) M. Szefczyk, E. Węglarz-Tomczak, P. Fortuna, A. Krzysztoń, E. Rudzińska-Szostak and Ł. Berlicki, Angew. Chem., Int. Ed., 2017, 56, 2087–2091 CrossRef CAS; (c) S. H. Choi, I. A. Guzei, L. C. Spencer and S. H. Gellman, J. Am. Chem. Soc., 2009, 131, 2917–2924 CrossRef CAS; (d) S. H. Choi, I. A. Guzei, L. C. Spencer and S. H. Gellman, J. Am. Chem. Soc., 2008, 130, 6544–6570 CrossRef CAS PubMed; (e) M. A. Schmitt, S. H. Choi, I. A. Guzei and S. H. Gellman, J. Am. Chem. Soc., 2006, 128, 4538–4539 CrossRef CAS PubMed; (f) G. Srinivasulu, S. K. Kumar, G. V. M. Sharma and A. C. Kunwar, J. Org. Chem., 2006, 71, 8395–8400 CrossRef CAS; (g) E. Rudzińska-Szostak and Ł. Berlicki, Chem. – Eur. J., 2017, 23, 14980–14986 CrossRef.
- (a) W. S. Horne and T. N. Grossmann, Nat. Chem., 2020, 12, 331–337 CrossRef CAS; Z. E. Reinert and W. S. Horne, Org. Biomol. Chem., 2014, 12, 8796–8802 Search PubMed.
- (a) C. C. Cabalteja, D. S. Mihalko and W. S. Horne, ChemBioChem, 2019, 20, 103–110 CrossRef CAS; (b) K. L. George and W. S. Horne, J. Am. Chem. Soc., 2017, 139, 7931–7938 CrossRef CAS.
- D. F. Kreitler, D. E. Mortenson, K. T. Forest and S. H. Gellman, J. Am. Chem. Soc., 2016, 138, 6498–6505 CrossRef CAS.
- (a) Z. E. Reinert, G. A. Lengyel and W. S. Horne, J. Am. Chem. Soc., 2013, 135, 12528–12531 CrossRef CAS; (b) Z. E. Reinert and W. S. Horne, Chem. Sci., 2014, 5, 3325–3330 RSC; (c) N. A. Tavenor, Z. E. Reinert, G. A. Lengyel, B. D. Griffith and W. S. Horne, Chem. Commun., 2016, 52, 3789–3792 RSC.
- G. Olajos, A. Hetényi, E. Wéber, L. J. Németh, Z. Szakonyi, F. Fülöp and T. A. Martinek, Chem. – Eur. J., 2015, 21, 6173–6180 CrossRef CAS.
- (a) R. P. Cheng, S. H. Gellman and W. F. DeGrado, Chem. Rev., 2001, 101, 3219–3232 CrossRef CAS PubMed; (b) C. M. Goodman, S. Choi, S. Shandler and W. F. DeGrado, Nat. Chem. Biol., 2007, 3, 252–262 CrossRef CAS.
- (a) W. S. Horne, Expert Opin. Drug Discovery, 2011, 6, 1247–1262 CrossRef CAS; (b) C. Cabrele, T. A. Martinek, O. Reiser and Ł. Berlicki, J. Med. Chem., 2014, 57, 9718–9739 CrossRef CAS PubMed.
- (a) G. N. Tew, R. W. Scott, M. L. Klein and W. F. Degrado, Acc. Chem. Res., 2010, 43, 30–39 CrossRef CAS PubMed; (b) M. A. Schmitt, B. Weisblum and S. H. Gellman, J. Am. Chem. Soc., 2007, 129, 417–428 CrossRef CAS PubMed.
- (a) P. Wójcik and Ł. Berlicki, Bioorg. Med. Chem. Lett., 2016, 26, 707–713 CrossRef; (b) H. S. Haase, K. J. Peterson-Kaufman, S. K. Lan Levengood, J. W. Checco, W. Murphy and S. H. Gellman, J. Am. Chem. Soc., 2012, 134, 7652–7655 CrossRef CAS; (c) W. S. Horne, M. D. Boersma, M. A. Windsor and S. H. Gellman, Angew. Chem., Int. Ed., 2008, 47, 2853–2856 CrossRef CAS.
- (a) Ł. Berlicki, M. Kaske, R. Gutierrez-Abad, G. Bernhardt, O. Illa, R. M. Ortuño, C. Cabrele, A. Buschauer and O. Reiser, J. Med. Chem., 2013, 56, 8422–8431 CrossRef; (b) N. Koglin, C. Zorn, R. Beumer, C. Cabrele, C. Bubert, N. Sewald, O. Reiser and A. G. Beck-Sickinger, Angew. Chem., Int. Ed., 2003, 42, 202–205 CrossRef CAS.
- Z. Hegedüs, E. Wéber, É. Kriston-Pál, I. Makra, Á. Czibula, É. Monostori and T. A. Martinek, J. Am. Chem. Soc., 2013, 135, 16578–16584 CrossRef PubMed.
- (a) M. M. Muller, M. A. Windsor, W. C. Pomerantz, S. H. Gellman and D. Hilvert, Angew. Chem., Int. Ed., 2009, 48, 922–925 CrossRef CAS PubMed; (b) P. S. P. Wang, J. B. Nguyen and A. Schepartz, J. Am. Chem. Soc., 2014, 136, 6810–6813 CrossRef CAS; (c) M. Drewniak, E. Węglarz-Tomczak, K. Ożga, E. Rudzińska-Szostak, K. Macegoniuk, J. M. Tomczak, M. Bejger, W. Rypniewski and Ł. Berlicki, Bioorg. Chem., 2018, 81, 356–361 CrossRef CAS.
- J. W. Neidigh, R. M. Fesinmeyer and N. H. Andersen, Nat. Struct. Biol., 2002, 9, 425–430 CrossRef CAS.
- B. I. Dahiyat and S. L. Mayo, Science, 1997, 278, 82–87 CrossRef CAS.
- P. Fortuna, A. Twarda-Clapa, L. Skalniak, K. Ożga, T. A. Holak and Ł. Berlicki, Eur. J. Med. Chem., 2020, 208, 112814 CrossRef CAS.
- W. S. Horne, L. M. Johnson, T. J. Ketas, P. J. Klasse, M. Lu, J. P. Moore and S. H. Gellman, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 14751–14756 CrossRef CAS.
- D. S. Wishart, B. D. Sykes and F. M. Richards, Biochemistry, 1992, 31, 1647–1651 CrossRef CAS PubMed.
- C. D. Schwieters, J. J. Kuszewski and G. M. Clore, Prog. Nucl. Magn. Reson. Spectrosc., 2006, 48, 47–62 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ob00309g |
| This journal is © The Royal Society of Chemistry 2021 |