A buried glutamate in the cross-β core renders β-endorphin fibrils reversible†
Yuying
Liu
a,
Yu
Zhang
a,
Yunxiang
Sun
 *ab and
Feng
Ding
*ab and
Feng
Ding
 *b
*b
aSchool of Physical Science and Technology, Ningbo University, Ningbo 315211, China
bDepartment of Physics and Astronomy, Clemson University, Clemson, South Carolina 29634, USA. E-mail: sunyunxiang@nbu.edu.cn; fding@clemson.edu
First published on 8th November 2021
Abstract
Functional amyloids are abundant in living organisms from prokaryotes to eukaryotes playing diverse biological roles. In contrast to the irreversible aggregation of most known pathological amyloids, we postulate that naturally-occurring functional amyloids are reversible under evolutionary pressure to be able to modulate the fibrillization process, reuse the composite peptides, or perform their biological functions. β-Endorphin, an endogenous opioid peptide hormone, forms such kinds of reversible amyloid fibrils in secretory granules for efficient storage and returns to the functional state of monomers upon release into the blood. The environmental change between low pH in secretory granules and neutral pH in extracellular spaces is believed to drive the reversible fibrillization of β-endorphin. Here, we investigate the critical role of a buried glutamate, Glu8, in the pH-responsive disassembly of β-endorphin fibrils using all-atom molecular dynamics simulations along with structure-based pKa prediction. The fibril was stable at pH 5.5 or lower with all the buried Glu8 residues protonated and neutrally charged. After switching to neutral pH, the Glu8 residues of peptides at the outer layers of the ordered fibrils became deprotonated due to partial solvent exposure, causing sheet-to-coil conformational changes and subsequent exposure of adjacent Glu8 residues in the inner chains. Via iterative deprotonation of Glu8 and induced structural disruption, all Glu8 residues would be progressively deprotonated. Electrostatic repulsion between deprotonated Glu8 residues along with their high solvation tendency disrupted the hydrogen bonding between the β1 strands and increased the solvent exposure of those otherwise buried residues in the cross-β core. Overall, our computational study reveals that the strategic positioning of ionizable residues into the cross-β core is a potential approach for designing reversible amyloid fibrils as pH-responsive smart bio-nanomaterials.
Introduction
The self-assembly of proteins into nanosized fibrils has attracted extensive attention due to their association with numerous human degenerative diseases,1–5 physiological functions,6–9 and potential applications in biomedicine and bio-nanotechnology.10–12 Besides many commonly-known amyloid diseases13 caused by the pathological aggregation of amyloid peptides (e.g., Aβ in Alzheimer's disease,14 αS in Parkinson's disease,15 hIAPP in type-2 diabetes,16 and Tau in multiple neurodegenerative diseases17), functional amyloids (e.g., curli,18 suckerin,19–21 blood fibrin,22 and β-endorphin23,24) have been discovered in all kingdoms of life, including organisms ranging from bacteria to humans playing diverse functional roles.25 One recently discovered functional amyloid in human is formed by β-endorphin in the central and peripheral nervous systems.23,26 β-Endorphin is a 31-residue endogenous opioid neuropeptide responsible for the regulation of pain perception and stress response.27–29 The peptides synthesized in the cells are stored in secretory granules in a highly compact amyloid state for efficient storage until a signal triggers their release into the blood or extracellular space.27,30,31One possible feature separating disease-related amyloids1–5,32 from functional amyloids might be fibril stability or fibrillization reversibility, since functional amyloids likely experience the evolutionary pressure of reused or recycled peptides forming these aggregates but pathological amyloids do not. Most of the disease-related aggregations are irreversible and the corresponding fibrils are stable even under highly denaturing conditions,33–35 while peptide hormone fibrils (e.g., β-endorphin) are often required to dissociate into monomers when the cellular environment changes.6 For example, prior experiments showed that 31 out of 42 randomly selected peptide hormones could assemble into amyloid fibrils in the presence of heparin at granule-relevant pH 5.5.24,27 All of these pre-formed hormone fibrils would disaggregate into monomers at pH 6.0 or 7.4.27 Since the dissociation process of β-endorphin fibrils is much faster than the synthesis rate, the reversible aggregation enables rapid responses to the stimulus – i.e., releases enough amounts of the hormone/neuropeptide into the blood in a short period of time.23,30,36 pH-responsive fibrillization was also observed in other functional amyloids.37,38 For example, suckerin amyloids, formed by the squid ring teeth protein at neutral pH, could dissociate into unstructured forms in an acidic environment due to the protonation of histidine residues.21,39,40 Owing to their high biocompatibility, low immunogenicity, pH-responsiveness, and reversibility of fibrillization, functional amyloids have been explored for potential applications in biomedicine and bio-nanotechnology.41,42 Therefore, uncovering the molecular mechanism for the pH-responsive regulation of hormone/neuropeptide aggregation will not only help understand the secretory pathway of the peptide hormones6,43,44 but also assist in the future design of peptide-based stimuli-responsive smart materials.
The aggregation and disaggregation of β-endorphin strongly depend on the chemical environment (including pH, phosphate, and cofactors).6,23,24 The β-endorphin fibril structure at pH 5.5 determined by solid-state NMR (ssNMR) is similar to most known fibril structures,45 featuring a cross-β core with the β-strands of each chain aligned parallel in-register and perpendicular to the fibril axis.14,23,46 Each chain has three β-strands formed by residues 4–10 (β1), 14–19 (β2), and 21–24 (β3) (Fig. 1). Sidechains buried inside the fibril core partition into one hydrophobic (i.e., Phe4, Phe18, Ala21, Ile23, and Ala26) and one hydrophilic (i.e., Thr6, Glu8, Ser10, and Thr16) clusters. As the only residue with an ionizable sidechain in the core, Glu8 has been postulated to contribute to the pH-responsive reversibility of the β-endorphin fibril.27 The pH change during the exocytosis of β-endorphin from the acidic secretory granule at pH ∼5.5 to the physiological pH of ∼7.4 in the blood could induce the change of the Glu8 protonation state, which subsequently modulates the stability of the β-endorphin fibril. However, since the sidechain pKa value of the glutamate in solution is ∼4.5 which is out of the above pH ranges, the molecular mechanism for the reversible β-endorphin fibril remains to be uncovered.
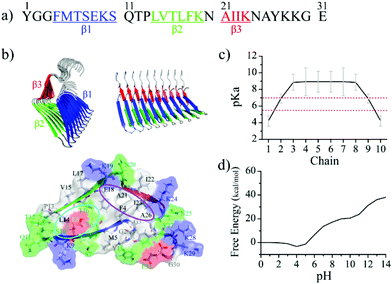 | ||
| Fig. 1 Experimentally determined β-endorphin fibril structure. (a) The sequence of β-endorphin used in our simulation. Three β-strands forming the fibrils are highlighted in blue, green, and red, respectively. (b) The reconstructed β-endorphin proto-fibril structure composed of 10 peptides based on the prior ssNMR determined fibril structure (PDB ID: 6TUB) is shown in both top and side views. Structural details of a single chain in β-endorphin proto-fibril are also shown, highlighting a hydrophobic cluster formed by residues F4, F18, A21, I23, and A26 (indicated by the purple circle), and also a hydrophilic cluster in the core consisted of residues T6, E8, S10, and T16 (the cyan circle). (c) The computationally predicted pKa values of Glu8 in each chain based on the experimental structure of the β-endorphin fibril. (d) The free energy of the β-endorphin fibril structure as a function of the pH values. | ||
Here, we applied all-atom molecular dynamics (MD) simulations with an explicit solvent to systemically investigate the role of Glu8 in the pH-dependent stability of a model β-endorphin fibril comprised of 10 peptides. The protonation states of 10 Glu8 residues were estimated according to the structure-based pKa calculations.47–49 The experimentally determined β-endorphin fibril structure27 was stable at secretory granule-relevant pH ∼5.5, where only the two exposed Glu8 residues located at two fibril ends were deprotonated with their pKa values lower than 5.0 and all the buried Glu8s of inner chains in the β-endorphin fibril remained protonated and neutrally charged with pKa above 5.5. Specifically, by using the structural ensemble sampled in MD simulations at pH 5.5 the Glu8 residues in the second outer layers were estimated to have pKa values below 7.0 due to partial solvent exposure while all other inner Glu residues have pKa values above 7.0. After switching to pH 7.4, the Glu8 residues in both the first and second outer layers were thus deprotonated, which induced a sheet-to-coil transition in the corresponding β1 strands and subsequently exposed adjacent Glu8 in the inner layers. Via iterative Glu8 deprotonation and induced structural disruption, all Glu8 residues would be progressively deprotonated. Due to electrostatic repulsions between the negatively charged Glu8 residues and the increased solvation, the cross-β core of the fibril was disrupted with the increased solvent accessible surface area of the originally buried hydrophobic residues. Although the complete fibril disaggregation and monomer release caused by the deprotonation of Glu8 were not observed in our simulations due to the high free energy barrier expected to overcome from the aggregated state to monomers, our computational study captured the early processes of pH-induced fibril disaggregation27,50 and revealed the molecular mechanism for the pH-dependent stability of the β-endorphin fibrils, which is important for the efficient storage and rapid release of the analgesic hormone. Moreover, our study suggests that the strategic positioning of ionizable residues into the cross-β core might be an attractive approach for designing reversible amyloid fibrils as pH-responsive smart bio-nanomaterials.
Materials and methods
The simulated molecular systems
The β-endorphin peptide consists of 31 residues (the primary sequence in Fig. 1a). Based on the prior solid-state NMR determined data (PDB: 6TUB),27 a 10-peptide fibril structure was constructed using our in-house code as the initial structure for simulation (Fig. 1b). The protonated Glu8 was buried in the fibril core (Fig. 1b). To probe whether the Glu8 plays a key role in the disaggregation of the β-endorphin fibril, two additional systems with E8Q and E8L mutations, where the negatively charged glutamate was replaced with either polar glutamine or hydrophobic leucine, were also simulated.Molecular dynamics simulations
All the MD simulations were performed in the isothermal–isobaric (NPT) ensemble using the Gromacs-2020.3 software51 with the AMBER99SB-ILDN force field.52 For each system, the 10-peptide β-endorphin fibril was placed in the center of a rectangular box of 10.0 × 10.0 × 10.0 nm3 and solvated with TIP3P water.53 Counterions, including Na+ and Cl−, were added to neutralize the system and mimic a physiological salt concentration (0.15 mM). The bond length of the peptides and water molecules was constrained respectively using the LINCS54 and SETTLE55 algorithms, allowing an integration time step of 2 fs. Nonbonded pair lists were updated every twenty integration steps. The protein, counterion and water groups were separately coupled to an external heat bath at 310 K with a coupling constant of 0.1 ps using a velocity rescaling coupling method.56 The pressure was kept around 1 bar applying the Parrinello–Rahman algorithm with a coupling time constant of 1.0 ps.57 Electrostatic interactions were analysed by the particle mesh Ewald (PME) method58 with a real space cutoff of 1.4 nm. The van der Waals interactions were calculated using a cutoff of 1.4 nm. Periodic boundary conditions were applied in all the simulations.Analysis methods
All data analyses were performed using our in-house-developed codes and the tools implemented in the Gromacs-2020.3 software package.51,59 Secondary structural analyses were performed using the dictionary secondary structure of protein (DSSP) method.60 The structural stability of the β-endorphin proto-filament was examined by the time evolution of backbone-root-mean-square-derivation (backbone-RMSD) corresponding to the optimized initial structure. pKa prediction was performed using a fast empirical approach, pKa predictors, PROPKA 3.0,49 which is based on better physical description of the desolvation and dielectric response of the protein.47,48Results and discussion
The fibril structure is favorable for β-endorphin under the acidic environment
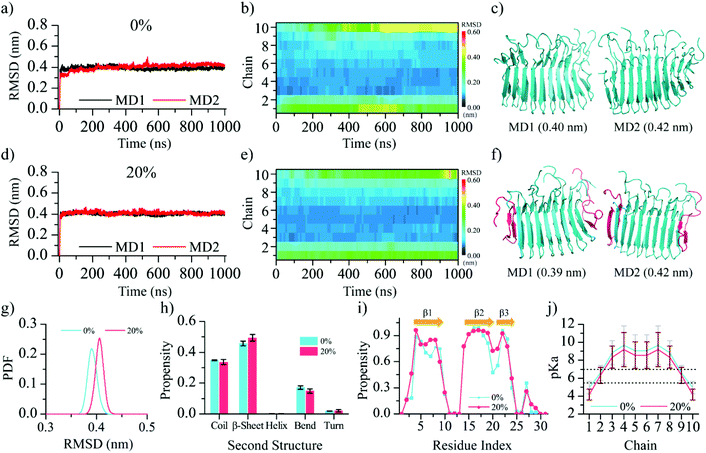
To investigate the effect of pH environmental changes on the protonation state of the Glu8 residues and subsequently the fibril stability, we first calculated the pKa values of Glu8 in the fibril (Fig. 1c) using PROPKA 3.0, a rapid empirical pKa prediction tool.47 Different from the solvent-exposed glutamate in solution, which has the side-chain pKa value around 4.35 as measured by the NMR experiment,61 the estimated pKa values of the buried Glu8 residues in the β-endorphin fibril were much higher than the secretory granule-relevant pH of 5.527,62 while the pKa values of the two exposed Glu8 residues located at the elongation ends were lower than 5. Hence, all buried Glu8 residues except the two exposed ones are expected to be protonated (i.e., uncharged) in the fibril conformation at the pH value of 5.5. Such pKa shifts of the ionizable groups when embedded in the hydrophobic protein cores have been well established by prior computational and experimental studies.63–68 The free energy of the β-endorphin fibril as a function of pH values was also predicted using PROPKA 3.047 (Fig. 1d). There is no obvious free energy change when the pH value ranges from 0 to 5 followed by a significant increase once the pH is larger than 6. Hence, the pKa analysis suggests that the experimentally determined fibril structure is favorable in the acidic environment with a pH value less than 6, but becomes unfavorable once it is released to the blood circulation with a pH value of ∼7.4.To evaluate the stability of the β-endorphin fibril under an acidic environment, two molecular systems – one with all Glu8 protonated corresponding to pH environments lower than 4.0 and the other with only two exposed Glu8 deprotonated (corresponding to 20% deprotonation) at pH around 5.5 were simulated. Here, the Glu8 residues from the two outer layer chains at two fibril elongation ends were deprotonated according to the pKa prediction (chains 1 & 10 in Fig. 1c.) For each molecular system, two 1000 ns independent simulations were performed (Fig. 2). The time evolution of backbone root-mean square deviation (RMSD) of the whole β-endorphin fibril was saturated around 0.40 nm with a small fluctuation (Fig. 2a & d) for both systems. The time evolution of the RMSD per chain was also analyzed (Fig. 2b & e). Except for two outer chains located at two fibril ends which had RMSD larger than 0.4 nm after reaching equilibrium, the RMSD of most inner chains in the fibril was less than 0.2 nm. Examination of the final representative snapshots from each trajectory revealed that the fibril structure was well retained without much structural destabilization (Fig. 2c & f). Deprotonation of the two exposed Glu8 only slightly increased the average RMSD of the β-endorphin fibril from 0.39 nm to 0.40 nm (Fig. 2g). The secondary structure analysis (Fig. 2h & i) revealed that the overall β-sheet content was ∼47%, and the three β-strands were well maintained without much destruction, which was further confirmed by the RMSD analysis of each β-strand (Fig. S1†).
Since pKa is known to be sensitive to structural changes, we recomputed the average Glu8 pKa values using the equilibrium conformations of the β-endorphin fibril during the last 50 ns for each of the two molecular systems (Fig. 2j). Both systems had similar pKa profiles and the pKa values of all buried Glu8 residues in the inner chains were still higher than 5.5, suggesting no further changes in the protonation states compared to the initial state. Overall, our simulation results confirmed that the experimentally determined β-endorphin fibril27 was stable under an acidic environment with a pH value of 5.5 or lower.
Progressive deprotonation of Glu8 at neutral pH triggers β-endorphin fibril disruption
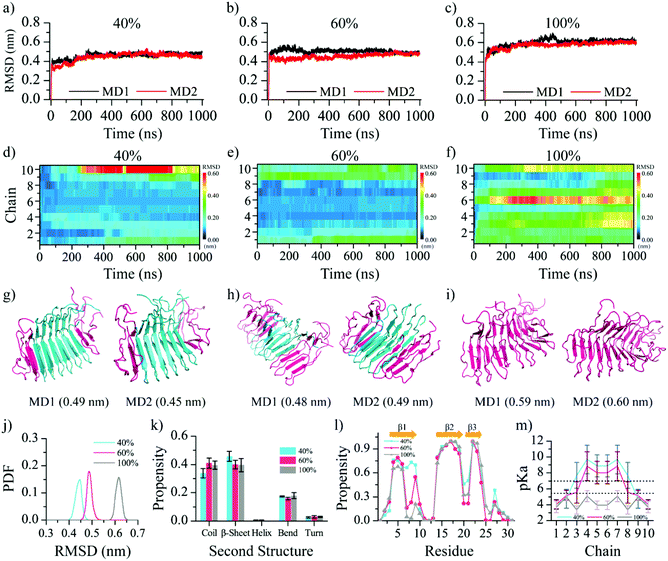
Using the equilibrium structural ensemble derived from MD simulations, the Glu8 residues in the second outer layer chains (chains 2 & 9, Fig. 2j) have reduced the pKa values below 7 compared to the predicted values using the static fibril structure (Fig. 1c). Therefore, upon entering the neutral pH environment, the β-endorphin fibril stable at pH 5.5 with the only outer layer Glu8 deprotonated is expected to have Glu8 residues in the second outer layer also deprotonated. We next simulated the β-endorphin fibril with the Glu8 residues in both first and second outer layers deprotonated (i.e., Glu8 in chains 1, 2, 9, and 10; denoted as 40% deprotonation). The time evolution of the fibril backbone RMSD (Fig. 3a) reached equilibrium around 0.44 nm, having a larger deviation from the initial state compared to the systems with 0% and 20% Glu8 deprotonated (Fig. 2g). The final structures in both independent simulations featured partially disrupted fibril structures (Fig. 3g), especially the first β-strands in the four outer layer peptides with the corresponding Glu8 deprotonated (Fig. S2a–c†). This observation resulted from the electrostatic repulsion interactions of Glu8 in adjacent peptides in the two outer layers. Indeed, the time evolution of the RMSD per chain showed that the chains with deprotonated Glu8 featured a higher RMSD than that of the systems with 0% and 20% outer Glu8 residues deprotonated (Fig. 3d and 2b & e).With structural changes accompanying the deprotonation of the Glu8 residues in the first two outer layer peptides, we re-calculated the pKa values of Glu8 using the structural ensemble from the equilibrated MD simulations. Compared to the initial pKa values above 8 for adjacent Glu8 residues in the 3rd outer layer (e.g., chains 3 & 8 in Fig. 2j), the pKa values decreased close to ∼7.0 (Fig. 3m), indicating the significantly increased probability of these two glutamates to be deprotonated at neutral pH. To test whether this observation led to an iterative deprotonation of adjacent Glu8 and subsequent structural disruption, we also performed MD simulations with the first three outer layer Glu8 residues protonated (i.e., chains 1–3 and 8–10, or 60% deprotonated). As expected, the β-endorphin fibril structure was further destabilized with the backbone RMSD saturated around ∼0.5 nm and increased β-sheet loss in the β1 region (Fig. 3b, j, l and Fig. S2†). For example, the average β-sheet probability decreased to ∼40% in β1 by its structural conversion into a random coil. The β-sheets formed by β2 and β3 were still retained (Fig. S2a–d†). Moreover, the recalculated pKa values of all Glu8 residues in chains 1–3 and 8–10 were ∼4.0–5.5 after reaching equilibrium, suggesting that the sampled structural ensemble with all these Glu8 residues protonated was the thermodynamically stable state at neutral pH (Fig. 3m). The pKa values of other inner Glu8 residues also significantly reduced. Together, our simulation results suggested an iterative deprotonation of the Glu8 residues and structural disruptions of the β-endorphin fibril at neutral pH until all Glu8s become deprotonated and negatively charged. The progressive deprotonation of the Glu8 residues and structural disruptions agree with the prior experimental study of the pH-triggered release of β-endorphin, where the disaggregation rate of the sonicated β-endorphin fibrils was much faster than that of the non-sonicated long fibril.27 Although a complete dissociation of the preformed fibril into monomers was not observed in our all-atom MD simulations, the observed cascades suggested a cooperative dissociation of the fibrils. Physiologically, the β-endorphin usually reached the maximal level in the blood within 20–60 min upon stimulation by physical exercise,69,70 which was much faster than the average duration of protein synthesis in eukaryotes (2–3 h).23 Further studies using either multiscale simulations or enhanced samplings are necessary to reach long timescales and overcome the large energy barriers associated with fibril dissociation.
Next, we also studied the fibril stability with all of the Glu8 residues deprotonated (i.e., 100% deprotonation). The back RMSD with respect to the initial ideal fibril structure reached ∼0.60 nm after 1000 ns in both independent MD simulations (Fig. 3c). The structural deviation was much larger than the simulations with the fractions of Glu8 deprotonated (Fig. 2g and 3j). Importantly, the slowly increasing trends of the overall RMSD in both simulations also suggested that the equilibrium was yet to be achieved due to a large energy barrier expected for the complete disaggregation of the fibril. Such a trend of the increasing fibril structural disruption was also obvious in the time evolution of the backbone RMSD per chain (Fig. 3f). Interestingly, the largest structural deviation was in the middle of the fibril, chain 6. The representative final snapshots further confirmed that the fibril structure was significantly destabilized (Fig. 3i). The calculation of the RMSD of each of the three β-strands revealed that β1 was significantly destroyed, although β2 and β3 still retained some of the β-sheet structures (Fig. S2†). The analysis of the secondary structures (Fig. 3k) showed ∼10% β-sheet content converted into random coils compared to fibril simulation in an acidic environment (Fig. 2h). The plot of the averaged β-sheet and random coil propensity per residue demonstrated that the β-sheet structure in the β1 region was significantly lost due to the deprotonation of Glu8 (Fig. 3l and Fig. S2†). Using the structures in the last 50 ns, the pKa value of all Glu8 decreased less than 5.0 due to the structural disruption of the β1 region, confirming that the state with fully deprotonated Glu8 was the thermodynamically stable state at neutral pH (Fig. 3m).
The deprotonated Glu8 residues disrupted the hydrophilic core of the β-endorphin fibril
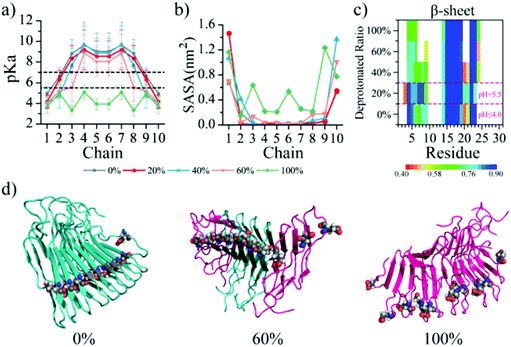
In the fibril core (Fig. 1b), interactions between the residues in the hydrophobic cluster are expected to stabilize the fibril while the hydrophobic cluster helps solvate the Glu8 and determine the reversibility of the fibril. To characterize the solvent accessibility of these residues, the averaged solvent accessible surface area (SASA) of each of these residues in simulations of different molecular systems was also analyzed (Fig. 4 and S3†). The pKa value of Glu8 residues correlated well with their SASA (Fig. 4a and b). Under acidic conditions (pH ≤ 5.5), all the inner Glu8 residues buried with the SASA equal to 0 were protonated, while the two outer Glu8 residues with the largest SASA were deprotonated. In the neutral pH environment, the partial solvent exposure of adjacent Glu8 with a small increase of the SASA from 0 resulted in a significant decrease of pKa from above 7.0 to lower values, and thus, deprotonation. Eventually, all the Glu8 residues became solvent exposed and correspondingly deprotonated due to the significant structural disruption of the β1 region (Fig. 4c) caused by the electrostatic repulsion of the charged Glu8s and the corresponding high solvation propensity. As a result, the solvent exposure of other buried residues in β1 – Phe4, Thr6, and Ser10 – increased with increasing Glu8 deprotonation ratio (Fig. S3a†). Although the β2 sheet was not destroyed at neutral pH in our simulations (Fig. 3l), the SASA of the residues Leu14, Thr16 and Phe18 in β2 also increased. For β3, the deprotonation of Glu8 only rendered Ala26 more exposed, but did not alter the SASA of Ala21 and Ile23 from the region (Fig. S3c†). Hence, the SASA analysis confirmed that the deprotonation of Glu8 destructed the cross-β core by exposing the buried residues in the hydrophilic and hydrophobic clusters.The E8Q and E8L mutants did not alter the structure but lost the pH-responsive reversibility of the β-endorphin fibrils
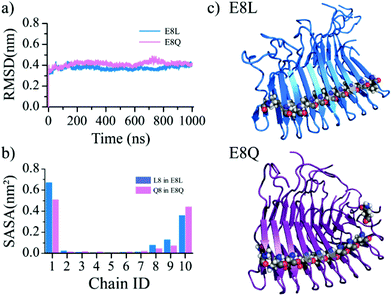
Prior experiments showed that the disassembly of the β-endorphin fibrils triggered by the pH change from 5.5 to 7.4 or higher was significantly inhibited when the Glu8 mutated to non-ionizable residues – polar Gln or hydrophobic Leu (denoted as E8Q and E8L, respectively).27 We also performed MD simulations for the two mutant fibrils (Fig. 5) to evaluate the effects of single-point mutations, E8Q and E8L, on the fibril stability. Both mutant fibril structures were well maintained during 1000 ns simulations with the RMSD fluctuating around ∼0.4 nm (Fig. 5a), which was similar to the wild-type simulation results at a pH value of 5.5 or lower (Fig. 2). The average SASA computed using the equilibrated conformations revealed that the mutated residues in the inner chains of the corresponding mutant fibrils remained buried (Fig. 5b and c). Since there are no ionizable residues in the fibril within the pH range of 5.5–7.4, the simulation results suggest that the mutant fibrils remain stable over the pH change from acidic 5.5 to neutral 7.4. In other words, the E8Q and E8L mutations did not alter the structure but lost the pH-responsive reversibility of the β-endorphin fibril. The results confirmed that the ionizable residues buried inside the fibril played an important role in the reversible aggregation of the β-endorphin.Positioning ionizable residues inside the cross-β core as a common strategy to render pH-responsive reversible fibrils
It has been well established that charged amino acids like glutamate in globular proteins were predominantly found on the solvent-exposed surfaces, and only occasionally buried in the hydrophobic core paring with the opposite charges (i.e., forming salt-bridges).71–74 With the expense of reduced protein stability, the unpaired glutamates in the protein cores are evolutionally selected to play important functional roles, such as catalysis and pH-dependent conformational changes. Similar to protein folding, the self-assembly of the amyloid fibrils is also driven by hydrophobic interactions and in the case of β-endorphin the strategic positioning of an unpaired glutamate in the core renders its fibrils pH-responsive and reversible by reducing the fibril stability. After entering the circulation with neutral pH 7.4, the β-endorphin fibrils formed at low pH for efficient storage release the peptide hormone as intrinsically disordered monomers containing partial helices75 (Fig. 6a left panel). Such environment-dependent reversible fibrillization is especially important for naturally-occurring functional amyloids so that the fibrillization process can be modulated by changing the environments such as pH and the composite proteins or peptides can be reused. For example, suckerin, a squid ring teeth protein, could self-assemble into well-ordered fibrils in the neutral pH environment and the pre-formed fibrils disaggregate under acidic conditions.39,76 Our previous computational study demonstrated that the protonation of the histidine residues buried in the cross-β core is important for destabilizing the fibril at low pH40 (Fig. 6a right panel). Another example is pre-melanosomal protein Pmel17.6,37,38 The aggregation of Pmel17 repeats only occurred under the conditions of pH lower than 5, and the preformed fibrils rapidly dissociated under neutral pH solution conditions.37,77 Although the Pmel17 fibril structure is still unknown, each Pmel17 repeat contained at least one glutamate (shown in Fig. 6b), and thus, the corresponding fibrils likely have unpaired glutamates buried in the cross-β core, also making the fibrils pH-responsive and reversible. Hence, positioning ionizable residues inside the cross-β core serves as a common strategy to render pH-responsive and reversible fibrils for naturally-occurring functional amyloids.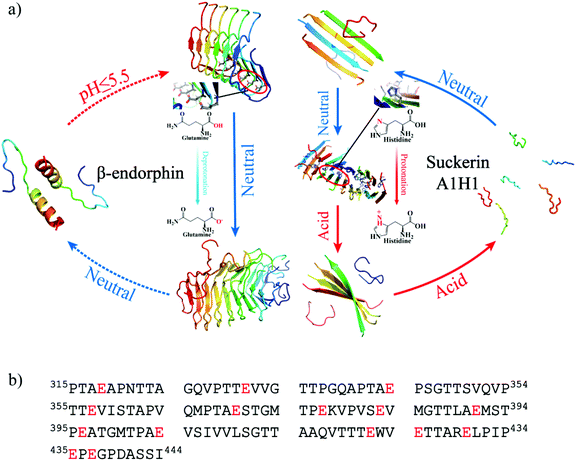 | ||
| Fig. 6 The schematic diagram illustrates that burying ionizable amino acids is a common strategy to render pH-responsive reversible fibrils for functional amyloids. (a) One example is the β-endorphin fibrils containing a buried glutamate, which is stable at low pH but disaggregates at neutral pH. The other example is the suckerin A1H1 peptide with histidines buried in the core, which could self-assemble into fibrils at neutral pH but disaggregate in the acidic environment.40 (b) The amino acid sequences of Pmel17 repeat, also forming pH-responsive functional amyloids. The ionizable glutamates are highlighted in red. | ||
The pathological aggregations are irreversible because the corresponding fibrils are highly stable and remain intact even under highly denaturing conditions. Whether and how the disease-related amyloid peptides could be rendered pH-sensitive by strategically introducing one or more ionizable residues to the beta-sheet core still need further computational and experimental exploration. Nevertheless, the pH-responsive and reversible properties of these functional amyloids endow them with great application prospects, such as controlled drug delivery and release,78 regulating the drug release rate by the pH change.39,79 Reversible fibrillization also enables promising applications in 3D printing and tissue regeneration. Therefore, introducing ionizable groups into the cross-β core could allow the design of reversible amyloid fibrils as pH-responsive smart bio-nanomaterials.
Conclusion
In summary, we investigated the structural stability of the β-endorphin fibril using atomistic MD simulations along with structure-based pKa predication. Our result showed that the experimentally determined amyloid fibril27 was stable under acidic conditions at a pH of 5.5 or lower. After switching to neutral pH, residues Glu8 from the outer layers along the fibrils would be deprotonated and cause sheet-to-coil conversion in the β1 region, which could make adjacent Glu8 residues in the inner layers partially exposed and subsequently deprotonated. Eventually, all Glu8 residues were progressively deprotonated and the fibril structures were significantly destabilized. Although a complete fibril disaggregation and monomer release were not observed in our simulation due to a high free energy barrier and long timescales, our computational study demonstrated the early processes for the pH-triggered fibril disassembly and uncovered the key role of buried Glu8 in the pH-responsive reversible fibrillization of β-endorphin. Introducing ionizable residues into the cross-β core might be, therefore, a potential approach for designing reversible amyloid fibrils as pH-responsive smart bio-nanomaterials for drug delivery, 3D printing and tissue regeneration.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported in part by the National Natural Science Foundation of China under grant no. 11904189 (Y. S.), K. C. Wong Magna Fund in Ningbo University, China (Y. S.), NSF CBET-1553945 (F. D.), and NIH R35GM119691 (F. D.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NSFC, NIH, and NSF.References
- P. C. Ke, E. H. Pilkington, Y. Sun, I. Javed, A. Kakinen, G. Peng, F. Ding and T. P. Davis, Mitigation of Amyloidosis with Nanomaterials, Adv. Mater., 2020, 32, e1901690 CrossRef PubMed.
- P. C. Ke, M. A. Sani, F. Ding, A. Kakinen, I. Javed, F. Separovic, T. P. Davis and R. Mezzenga, Implications of peptide assemblies in amyloid diseases, Chem. Soc. Rev., 2017, 46, 6492–6531 RSC.
- P. H. Nguyen, A. Ramamoorthy, B. R. Sahoo, J. Zheng, P. Faller, J. E. Straub, L. Dominguez, J. E. Shea, N. V. Dokholyan, A. De Simone, B. Ma, R. Nussinov, S. Najafi, S. T. Ngo, A. Loquet, M. Chiricotto, P. Ganguly, J. McCarty, M. S. Li, C. Hall, Y. Wang, Y. Miller, S. Melchionna, B. Habenstein, S. Timr, J. Chen, B. Hnath, B. Strodel, R. Kayed, S. Lesne, G. Wei, F. Sterpone, A. J. Doig and P. Derreumaux, Amyloid Oligomers: A Joint Experimental/Computational Perspective on Alzheimer's Disease, Parkinson's Disease, Type II Diabetes, and Amyotrophic Lateral Sclerosis, Chem. Rev., 2021, 121, 2545–2647 CrossRef CAS PubMed.
- J. Nasica-Labouze, P. H. Nguyen, F. Sterpone, O. Berthoumieu, N. V. Buchete, S. Cote, A. De Simone, A. J. Doig, P. Faller, A. Garcia, A. Laio, M. S. Li, S. Melchionna, N. Mousseau, Y. Mu, A. Paravastu, S. Pasquali, D. J. Rosenman, B. Strodel, B. Tarus, J. H. Viles, T. Zhang, C. Wang and P. Derreumaux, Amyloid beta Protein and Alzheimer's Disease: When Computer Simulations Complement Experimental Studies, Chem. Rev., 2015, 115, 3518–3563 CrossRef CAS PubMed.
- P. Chen, F. Ding, R. Cai, I. Javed, W. Yang, Z. Zhang, Y. Li, T. P. Davis, P. C. Ke and C. Chen, Amyloidosis Inhibition, a New Frontier of the Protein Corona, Nano Today, 2020, 35, 100937 CrossRef CAS PubMed.
- S. K. Maji, M. H. Perrin, M. R. Sawaya, S. Jessberger, K. Vadodaria, R. A. Rissman, P. S. Singru, K. P. Nilsson, R. Simon, D. Schubert, D. Eisenberg, J. Rivier, P. Sawchenko, W. Vale and R. Riek, Functional amyloids as natural storage of peptide hormones in pituitary secretory granules, Science, 2009, 325, 328–332 CrossRef CAS PubMed.
- L. P. Blanco, M. L. Evans, D. R. Smith, M. P. Badtke and M. R. Chapman, Diversity, biogenesis and function of microbial amyloids, Trends Microbiol., 2012, 20, 66–73 CrossRef CAS PubMed.
- D. M. Fowler, A. V. Koulov, W. E. Balch and J. W. Kelly, Functional amyloid–from bacteria to humans, Trends Biochem. Sci., 2007, 32, 217–224 CrossRef CAS PubMed.
- P. A. Guerette, S. Hoon, Y. Seow, M. Raida, A. Masic, F. T. Wong, V. H. Ho, K. W. Kong, M. C. Demirel, A. Pena-Francesch, S. Amini, G. Z. Tay, D. Ding and A. Miserez, Accelerating the design of biomimetic materials by integrating RNA-seq with proteomics and materials science, Nat. Biotechnol., 2013, 31, 908–915 CrossRef CAS PubMed.
- X. Zhao, F. Pan, H. Xu, M. Yaseen, H. Shan, C. A. Hauser, S. Zhang and J. R. Lu, Molecular self-assembly and applications of designer peptide amphiphiles, Chem. Soc. Rev., 2010, 39, 3480–3498 RSC.
- C. A. Hauser and S. Zhang, Designer self-assembling peptide nanofiber biological materials, Chem. Soc. Rev., 2010, 39, 2780–2790 RSC.
- G. Wei, Z. Su, N. P. Reynolds, P. Arosio, I. W. Hamley, E. Gazit and R. Mezzenga, Self-assembling peptide and protein amyloids: from structure to tailored function in nanotechnology, Chem. Soc. Rev., 2017, 46, 4661–4708 RSC.
- M. G. Iadanza, M. P. Jackson, E. W. Hewitt, N. A. Ranson and S. E. Radford, A new era for understanding amyloid structures and disease, Nat. Rev. Mol. Cell Biol., 2018, 19, 755–773 CrossRef CAS PubMed.
- Y. Xiao, B. Ma, D. McElheny, S. Parthasarathy, F. Long, M. Hoshi, R. Nussinov and Y. Ishii, Abeta(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer's disease, Nat. Struct. Mol. Biol., 2015, 22, 499–505 CrossRef CAS PubMed.
- D. R. Boyer, B. Li, C. Sun, W. Fan, M. R. Sawaya, L. Jiang and D. S. Eisenberg, Structures of fibrils formed by alpha-synuclein hereditary disease mutant H50Q reveal new polymorphs, Nat. Struct. Mol. Biol., 2019, 26, 1044–1052 CrossRef CAS PubMed.
- D. C. R. Camargo, D. Garg, K. Buday, A. Franko, A. R. Camargo, F. Schmidt, S. J. Cox, S. Suladze, M. Haslbeck, Y. G. Mideksa, G. Gemmecker, M. Aichler, G. Mettenleiter, M. Schulz, A. K. Walch, M. Hrabe de Angelis, M. J. Feige, C. A. Sierra, M. Conrad, K. Tripsianes, A. Ramamoorthy and B. Reif, hIAPP forms toxic oligomers in plasma, Chem. Commun., 2018, 54, 5426–5429 RSC.
- L. Martin, X. Latypova, C. M. Wilson, A. Magnaudeix, M. L. Perrin, C. Yardin and F. Terro, Tau protein kinases: involvement in Alzheimer's disease, Ageing Res. Rev., 2013, 12, 289–309 CrossRef CAS PubMed.
- Z. Abdali, M. Aminzare, X. Zhu, E. DeBenedictis, O. Xie, S. Keten and N. M. Dorval, Courchesne,Curli-Mediated Self-Assembly of a Fibrous Protein Scaffold for Hydroxyapatite Mineralization, ACS Synth. Biol., 2020, 9, 3334–3343 CrossRef CAS PubMed.
- D. Ding, P. A. Guerette, J. Fu, L. Zhang, S. A. Irvine and A. Miserez, From Soft Self-Healing Gels to Stiff Films in Suckerin-Based Materials Through Modulation of Crosslink Density and beta-Sheet Content, Adv. Mater., 2015, 27, 3953–3961 CrossRef CAS PubMed.
- K. Deepankumar, C. Lim, I. Polte, B. Zappone, C. Labate, M. P. De Santo, H. Mohanram, A. Palaniappan, D. S. Hwang and A. Miserez, Supramolecular β–Sheet Suckerin–Based Underwater Adhesives, Adv. Funct. Mater., 2020, 30(16), 1907534 CrossRef CAS.
- P. A. Guerette, S. Hoon, D. Ding, S. Amini, A. Masic, V. Ravi, B. Venkatesh, J. C. Weaver and A. Miserez, Nanoconfined beta-sheets mechanically reinforce the supra-biomolecular network of robust squid Sucker Ring Teeth, ACS Nano, 2014, 8, 7170–7179 CrossRef CAS PubMed.
- A. Undas and R. A. Ariens, Fibrin clot structure and function: a role in the pathophysiology of arterial and venous thromboembolic diseases, Arterioscler., Thromb., Vasc. Biol., 2011, 31, e88–e99 CrossRef CAS PubMed.
- N. Nespovitaya, J. Gath, K. Barylyuk, C. Seuring, B. H. Meier and R. Riek, Dynamic Assembly and Disassembly of Functional beta-Endorphin Amyloid Fibrils, J. Am. Chem. Soc., 2016, 138, 846–856 CrossRef CAS PubMed.
- N. Nespovitaya, P. Mahou, R. F. Laine, G. S. K. Schierle and C. F. Kaminski, Heparin acts as a structural component of beta-endorphin amyloid fibrils rather than a simple aggregation promoter, Chem. Commun., 2017, 53, 1273–1276 RSC.
- M. F. Gebbink, D. Claessen, B. Bouma, L. Dijkhuizen and H. A. Wosten, Amyloids–a functional coat for microorganisms, Nat. Rev. Microbiol., 2005, 3, 333–341 CrossRef CAS PubMed.
- A. Pilozzi, C. Carro and X. Huang, Roles of beta-Endorphin in Stress, Behavior, Neuroinflammation, and Brain Energy Metabolism, Int. J. Mol. Sci., 2020, 22(1), 338 CrossRef PubMed.
- C. Seuring, J. Verasdonck, J. Gath, D. Ghosh, N. Nespovitaya, M. A. Walti, S. K. Maji, R. Cadalbert, P. Guntert, B. H. Meier and R. Riek, The three-dimensional structure of human beta-endorphin amyloid fibrils, Nat. Struct. Mol. Biol., 2020, 27, 1178–1184 CrossRef CAS PubMed.
- J. F. Dalayeun, J. M. Nores and S. Bergal, Physiology of beta-endorphins. A close-up view and a review of the literature, Biomed. Pharmacother., 1993, 47, 311–320 CrossRef CAS.
- R. J. Bodnar, Endogenous opiates and behavior: 2013, Peptides, 2014, 62, 67–136 CrossRef CAS PubMed.
- R. B. Kelly, Pathways of protein secretion in eukaryotes, Science, 1985, 230, 25–32 CrossRef CAS PubMed.
- P. Arvan and D. Castle, Sorting and storage during secretory granule biogenesis: looking backward and looking forward, Biochem. J., 1998, 332(Pt 3), 593–610 CrossRef CAS PubMed.
- Y. Sun, A. Kakinen, Y. Xing, P. Faridi, A. Nandakumar, A. W. Purcell, T. P. Davis, P. C. Ke and F. Ding, Amyloid Self-Assembly of hIAPP8-20 via the Accumulation of Helical Oligomers, alpha-Helix to beta-Sheet Transition, and Formation of beta-Barrel Intermediates, Small, 2019, 15, e1805166 CrossRef PubMed.
- A. Kakinen, Y. Sun, I. Javed, A. Faridi, E. H. Pilkington, P. Faridi, A. W. Purcell, R. Zhou, F. Ding, S. Lin, P. C. Ke and T. P. Davis, Physical and Toxicological Profiles of Human IAPP Amyloids and Plaques, Sci. Bull., 2019, 64, 26–35 CrossRef CAS PubMed.
- Y. Jin, Y. Sun, J. Lei and G. Wei, Dihydrochalcone molecules destabilize Alzheimer's amyloid-beta protofibrils through binding to the protofibril cavity, Phys. Chem. Chem. Phys., 2018, 20, 17208–17217 RSC.
- Y. Zou, Z. Qian, Y. Chen, H. Qian, G. Wei and Q. Zhang, Norepinephrine Inhibits Alzheimer's Amyloid-beta Peptide Aggregation and Destabilizes Amyloid-beta Protofibrils: A Molecular Dynamics Simulation Study, ACS Chem. Neurosci., 2019, 10, 1585–1594 CrossRef CAS PubMed.
- R. B. Kelly, Protein transport. From organelle to organelle, Nature, 1987, 326, 14–15 CrossRef CAS PubMed.
- D. N. Dean and J. C. Lee, pH-Dependent fibril maturation of a Pmel17 repeat domain isoform revealed by tryptophan fluorescence, Biochim. Biophys. Acta, Proteins Proteomics, 2019, 1867, 961–969 CrossRef CAS PubMed.
- D. Dharmadana, N. P. Reynolds, C. E. Conn and C. Valéry, pH-Dependent Self-Assembly of Human Neuropeptide Hormone GnRH into Functional Amyloid Nanofibrils and Hexagonal Phases, ACS Appl. Bio Mater., 2019, 2, 3601–3606 CrossRef CAS.
- D. Ding, J. Pan, S. H. Lim, S. Amini, L. Kang and A. Miserez, Squid suckerin microneedle arrays for tunable drug release, J. Mater. Chem. B, 2017, 5, 8467–8478 RSC.
- Y. Sun and F. Ding, Thermo- and pH-responsive fibrillization of squid suckerin A1H1 peptide, Nanoscale, 2020, 12, 6307–6317 RSC.
- B. Chen, X. Y. He, X. Q. Yi, R. X. Zhuo and S. X. Cheng, Dual-peptide-functionalized albumin-based nanoparticles with pH-dependent self-assembly behavior for drug delivery, ACS Appl. Mater. Interfaces, 2015, 7, 15148–15153 CrossRef CAS PubMed.
- J. Wang, K. Liu, R. Xing and X. Yan, Peptide self-assembly: thermodynamics and kinetics, Chem. Soc. Rev., 2016, 45, 5589–5604 RSC.
- M. M. Glombik and H. H. Gerdes, Signal-mediated sorting of neuropeptides and prohormones: secretory granule biogenesis revisited, Biochimie, 2000, 82, 315–326 CrossRef CAS PubMed.
- B. Borgonovo, J. Ouwendijk and M. Solimena, Biogenesis of secretory granules, Curr. Opin. Cell Biol., 2006, 18, 365–370 CrossRef CAS PubMed.
- C. Seuring, J. Gath, J. Verasdonck, R. Cadalbert, J. Rivier, A. Bockmann, B. H. Meier and R. Riek, Solid-state NMR sequential assignment of the beta-endorphin peptide in its amyloid form, Biomol. NMR Assignments, 2016, 10, 259–268 CrossRef CAS PubMed.
- R. Nelson, M. R. Sawaya, M. Balbirnie, A. O. Madsen, C. Riekel, R. Grothe and D. Eisenberg, Structure of the cross-beta spine of amyloid-like fibrils, Nature, 2005, 435, 773–778 CrossRef CAS PubMed.
- H. Li, A. D. Robertson and J. H. Jensen, Very fast empirical prediction and rationalization of protein pKa values, Proteins, 2005, 61, 704–721 CrossRef CAS PubMed.
- M. H. Olsson, Improving the desolvation penalty in empirical protein pKa modeling, J. Mol. Model., 2012, 18, 1097–1106 CrossRef CAS PubMed.
- M. H. Olsson, C. R. Sondergaard, M. Rostkowski and J. H. Jensen, PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions, J. Chem. Theory Comput., 2011, 7, 525–537 CrossRef CAS PubMed.
- V. Colomer, G. A. Kicska and M. J. Rindler, Secretory granule content proteins and the luminal domains of granule membrane proteins aggregate in vitro at mildly acidic pH, J. Biol. Chem., 1996, 271, 48–55 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- K. Lindorff-Larsen, S. Piana, K. Palmo, P. Maragakis, J. L. Klepeis, R. O. Dror and D. E. Shaw, Improved side-chain torsion potentials for the Amber ff99SB protein force field, Proteins, 2010, 78, 1950–1958 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, Comparison of Simple Potential Functions for Simulating Liquid Water, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, LINCS: A linear constraint solver for molecular simulations, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- S. Miyamoto and P. A. Kollman, Settle - an Analytical Version of the Shake and Rattle Algorithm for Rigid Water Models, J. Comput. Chem., 1992, 13, 952–962 CrossRef CAS.
- G. Bussi, D. Donadio and M. Parrinello, Canonical sampling through velocity rescaling, J. Chem. Phys., 2007, 126, 014101 CrossRef PubMed.
- M. Parrinello and A. Rahman, Polymorphic Transitions in Single-Crystals - a New Molecular-Dynamics Method, J. Appl. Phys., 1981, 52, 7182–7190 CrossRef CAS.
- S. Grimme, J. Antony, S. Ehrlich and H. Krieg, A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu, J. Chem. Phys., 2010, 132, 154104 CrossRef PubMed.
- H. J. C. Berendsen, D. Vanderspoel and R. Vandrunen, Gromacs - a Message-Passing Parallel Molecular-Dynamics Implementation, Comput. Phys. Commun., 1995, 91, 43–56 CrossRef CAS.
- W. Kabsch and C. Sander, Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features, Biopolymers, 1983, 22, 2577–2637 CrossRef CAS PubMed.
- C. A. Castaneda, C. A. Fitch, A. Majumdar, V. Khangulov, J. L. Schlessman and B. E. Garcia-Moreno, Molecular determinants of the pKa values of Asp and Glu residues in staphylococcal nuclease, Proteins, 2009, 77, 570–588 CrossRef CAS PubMed.
- P. Paroutis, N. Touret and S. Grinstein, The pH of the secretory pathway: measurement, determinants, and regulation, Physiology, 2004, 19, 207–215 CrossRef CAS PubMed.
- M. J. Harms, C. A. Castaneda, J. L. Schlessman, G. R. Sue, D. G. Isom, B. R. Cannon and E. B. Garcia-Moreno, The pK(a) values of acidic and basic residues buried at the same internal location in a protein are governed by different factors, J. Mol. Biol., 2009, 389, 34–47 CrossRef CAS PubMed.
- J. A. Wallace, Y. Wang, C. Shi, K. J. Pastoor, B. L. Nguyen, K. Xia and J. K. Shen, Toward accurate prediction of pKa values for internal protein residues: the importance of conformational relaxation and desolvation energy, Proteins, 2011, 79, 3364–3373 CrossRef CAS PubMed.
- M. H. Olsson, Protein electrostatics and pKa blind predictions; contribution from empirical predictions of internal ionizable residues, Proteins, 2011, 79, 3333–3345 CrossRef CAS PubMed.
- C. T. Han, J. Song, T. Chan, C. Pruett and S. Han, Electrostatic Environment of Proteorhodopsin Affects the pKa of Its Buried Primary Proton Acceptor, Biophys. J., 2020, 118, 1838–1849 CrossRef CAS PubMed.
- Y. Huang, W. Chen, J. A. Wallace and J. Shen, All-Atom Continuous Constant pH Molecular Dynamics With Particle Mesh Ewald and Titratable Water, J. Chem. Theory Comput., 2016, 12, 5411–5421 CrossRef CAS PubMed.
- Y. Huang, W. Chen, D. L. Dotson, O. Beckstein and J. Shen, Mechanism of pH-dependent activation of the sodium-proton antiporter NhaA, Nat. Commun., 2016, 7, 12940 CrossRef CAS PubMed.
- P. M. Radosevich, J. A. Nash, D. B. Lacy, C. O'Donovan, P. E. Williams and N. N. Abumrad, Effects of low- and high-intensity exercise on plasma and cerebrospinal fluid levels of ir-beta-endorphin, ACTH, cortisol, norepinephrine and glucose in the conscious dog, Brain Res., 1989, 498, 89–98 CrossRef CAS PubMed.
- L. Schwarz and W. Kindermann, Beta-endorphin, adrenocorticotropic hormone, cortisol and catecholamines during aerobic and anaerobic exercise, Eur. J. Appl. Physiol. Occup. Physiol., 1990, 61, 165–171 CrossRef CAS PubMed.
- P. Kota, F. Ding, S. Ramachandran and N. V. Dokholyan, Gaia: automated quality assessment of protein structure models, Bioinformatics, 2011, 27, 2209–2215 CrossRef CAS PubMed.
- A. Honegger and A. Pluckthun, The influence of the buried glutamine or glutamate residue in position 6 on the structure of immunoglobulin variable domains, J. Mol. Biol., 2001, 309, 687–699 CrossRef CAS PubMed.
- K. Suzuki, T. Yamada and T. Tanaka, Role of the buried glutamate in the alpha-helical coiled coil domain of the macrophage scavenger receptor, Biochemistry, 1999, 38, 1751–1756 CrossRef CAS PubMed.
- D. G. Isom, C. A. Castaneda, B. R. Cannon, P. D. Velu and E. B. Garcia-Moreno, Charges in the hydrophobic interior of proteins, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 16096–16100 CrossRef CAS PubMed.
- G. Saviano, O. Crescenzi, D. Picone, P. Temussi and T. Tancredi, Solution structure of human beta-endorphin in helicogenic solvents: an NMR study, J. Pept. Sci., 1999, 5, 410–422 CrossRef CAS PubMed.
- D. Ding, P. A. Guerette, S. Hoon, K. W. Kong, T. Cornvik, M. Nilsson, A. Kumar, J. Lescar and A. Miserez, Biomimetic production of silk-like recombinant squid sucker ring teeth proteins, Biomacromolecules, 2014, 15, 3278–3289 CrossRef CAS PubMed.
- C. M. Pfefferkorn, R. P. McGlinchey and J. C. Lee, Effects of pH on aggregation kinetics of the repeat domain of a functional amyloid, Pmel17, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 21447–21452 CrossRef CAS PubMed.
- J. Zhu, H. Han, T. T. Ye, F. X. Li, X. L. Wang, J. Y. Yu and D. Q. Wu, Biodegradable and pH Sensitive Peptide Based Hydrogel as Controlled Release System for Antibacterial Wound Dressing Application, Molecules, 2018, 23(12), 3383 CrossRef PubMed.
- Y. Ping, D. Ding, R. Ramos, H. Mohanram, K. Deepankumar, J. Gao, G. Tang and A. Miserez, Supramolecular beta-Sheets Stabilized Protein Nanocarriers for Drug Delivery and Gene Transfection, ACS Nano, 2017, 11, 4528–4541 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S3. See DOI: 10.1039/d1nr05679d |
| This journal is © The Royal Society of Chemistry 2021 |