Open Access Article
Open Access ArticleAmphiphilic nanoparticles generate curvature in lipid membranes and shape liposome–liposome interfaces†
E.
Lavagna
a,
Z. P.
Güven
b,
D.
Bochicchio
a,
F.
Olgiati
b,
F.
Stellacci
 b and
G.
Rossi
b and
G.
Rossi
 *a
*a
aPhysics Department, University of Genoa, Via Dodecaneso 33, 16146 Genoa, Italy. E-mail: rossig@fisica.unige.it
bInstitute of Materials and Bioengineering Institute, École Polytechnique Fédérale de Lausanne, 1015 Lausanne, Switzerland
First published on 4th October 2021
Abstract
We show by molecular dynamics that amphiphilic Au nanoparticles (NP) with a diameter of 4 nm generate curvature in phosphatidylcholine lipid membranes. NPs generate negative curvature when they adsorb on the membrane surface but, as they get spontaneously and progressively embedded into the membrane core, the curvature turns positive. As membrane embedding is kinetically slow, both configurations can be observed by Cryo-EM. NP-induced curvature explains the peculiar structure of liposome–liposome interfaces in presence of NPs.
The spontaneous curvature of a pure lipid bilayer with symmetrical composition is zero. A non-zero curvature can arise from the asymmetry of lipid composition, or from different solution conditions across the membrane.1 In most biologically relevant cases, the presence of membrane inclusions (proteins) is fundamental to have stable, large curvature regions in the bilayer.2 Indeed, it is now well known that different families of membrane proteins have an important remodeling effect on the lipid membrane and influence its local curvature, either favoring a positive or negative curvature.
Membrane proteins adhering to the membrane surface can sense and modify the local curvature by different mechanisms. The family of BAR (Bin/amphiphysin/Rvs domain) proteins,3–5 which have a banana-shaped backbone and can vary in length, intrinsic curvature, and binding affinity to the membrane,4 can generate membrane curvature by scaffolding,4 that is, by leaving their imprint on the membrane surface. Another way to generate membrane curvature is by inserting a protein segment into the bilayer (wedging),6 a mechanism that has been shown to co-operate to stabilize hemispherical clathrin-coated structures.7 Oligomerization and crowding at large concentrations, resulting from curvature-mediated forces,8 can also cause or increase membrane bending.4,6,9–11
Synthetic molecules that stably interact with the lipid bilayer can have important remodeling effects, as well. Synthetic DNA origami scaffolds, for example, can reproduce the activity of membrane-sculpting proteins,12 an effect that is modulated by DNA curvature, membrane affinity, and surface density. Synthetic nanoparticles (NPs), such as carbon NPs, can induce local membrane curvature in a size-dependent fashion, an effect enhanced upon spontaneous aggregation.13,14 Overall, the effect of synthetic NPs on membrane curvature has yet to be understood from a fundamental perspective.15 NPs are often functionalized by shells of flexible, amphiphilic organic ligands, which make the NP capable of adopting different shapes; ligand flexibility allows to change the hydrophobicity of the NP accessible surface as a response to different environments. These features cause different and dynamic interactions with biological membranes, with effects on membrane curvature that cannot be easily interpreted based on the knowledge of NP size and composition only.
Gold nanoparticles (Au NPs) are very promising as diagnostic or therapeutic agents.16,17 It has been shown that they can favorably interact with lipid membranes and spontaneously penetrate them in the 2–5 nm core diameter range.18–20 Functionalized Au NPs can aggregate on liposome surfaces,21 destabilize the phase separation in multi-domain membranes,22,23 mediate the interaction between liposomes (populating liposome–liposome interfaces)21 and even affect membrane fusion in presence of calcium.24
Recently, experimental and computational studies shed some light on the molecular details of the interaction of small (2–4 nm diameter) amphiphilic Au NPs with lipid bilayers. It is now well established that Au NPs functionalized with a mixture of the negatively charged 11-mercapto-1-undecanesulfonate (MUS) and 1-octanethiol (OT) ligands can adsorb onto a lipid bilayer and spontaneously penetrate into its hydrophobic core.19,21 This spontaneous insertion (see ESI Fig. S1†) is a multi-step process that has been elucidated by molecular dynamics simulation studies using both atomistic and coarse-grained models.25–28 The first step is the one in which the MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT Au NP is simply adsorbed on the bilayer surface, with the MUS ligands in contact with the lipid heads. Then, as soon as a hydrophobic contact is established between the NP monolayer and the membrane core, the NP starts diving into the bilayer and eventually anchors one of its charged ligands to the headgroups of the distal leaflet (semi-snorkeling configuration). Snorkeling proceeds by anchoring more and more ligands to the distal leaflet: the NP eventually reaches the final, most stable state, in which the NP core is symmetrically immersed in the membrane hydrophobic core (snorkeling configuration). The energy barriers associated to these kinetic steps can be very large,29,30 meaning that the various metastable states can have long and experimentally relevant lifetimes. Indeed, cryo-EM images are able to capture several stages of this interaction, showing the NPs adsorbed, or partially embedded into the bilayer.21
OT Au NP is simply adsorbed on the bilayer surface, with the MUS ligands in contact with the lipid heads. Then, as soon as a hydrophobic contact is established between the NP monolayer and the membrane core, the NP starts diving into the bilayer and eventually anchors one of its charged ligands to the headgroups of the distal leaflet (semi-snorkeling configuration). Snorkeling proceeds by anchoring more and more ligands to the distal leaflet: the NP eventually reaches the final, most stable state, in which the NP core is symmetrically immersed in the membrane hydrophobic core (snorkeling configuration). The energy barriers associated to these kinetic steps can be very large,29,30 meaning that the various metastable states can have long and experimentally relevant lifetimes. Indeed, cryo-EM images are able to capture several stages of this interaction, showing the NPs adsorbed, or partially embedded into the bilayer.21
In this paper, we show that the interaction between MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT Au NPs and model DOPC membranes can severely affect the bilayer curvature, making it shift from negative, to positive and zero curvature as the NP–membrane interaction unrolls. NP-mediated curvature generation is enhanced by NPs aggregation, leading to stabilization of liposome–liposome interfaces with very different geometries.
OT Au NPs and model DOPC membranes can severely affect the bilayer curvature, making it shift from negative, to positive and zero curvature as the NP–membrane interaction unrolls. NP-mediated curvature generation is enhanced by NPs aggregation, leading to stabilization of liposome–liposome interfaces with very different geometries.
Cryo-EM results: NPs induce liposome clustering, affect bilayer curvature and shape liposome–liposome interfaces
We synthesized Au NPs with a core diameter between 2 and 5 nm and a mixed ligand shell composed of 11-mercaptoundecane-1-sulfonic acid and 1-octane thiol (MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
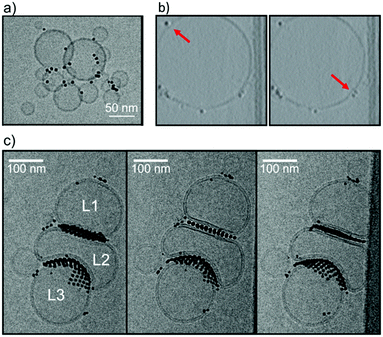
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT NPs) via one-phase method, as described previously31,32 and in the ESI.† Experimental cryo-EM images, whose setup is described in the ESI,† show that NPs induce liposome clustering in the 0–30% OT composition range (Fig. 1a and ESI Fig. S2†). The observation of liposome clustering is robust against the variation of the NP core diameter in the 2–5 nm range. When liposomes are small (<60 nm diameter), NPs gather at the liposome–liposome interface and interact with two or even three liposomes at a time, neither leading to membrane fusion nor to stalk formation.24 With larger liposomes (60 nm < diameter < 150 nm) Cryo-EM has provided evidence for both adsorbed (Fig. 1b) and snorkeled21 NPs. As for the latter, the resolution did not allow to distinguish between semi- and fully snorkeling configurations. The tomograms shown in Fig. 1b suggest that isolated NPs, adsorbed at the membrane surface of large liposomes, may induce a slight negative curvature.
OT NPs) via one-phase method, as described previously31,32 and in the ESI.† Experimental cryo-EM images, whose setup is described in the ESI,† show that NPs induce liposome clustering in the 0–30% OT composition range (Fig. 1a and ESI Fig. S2†). The observation of liposome clustering is robust against the variation of the NP core diameter in the 2–5 nm range. When liposomes are small (<60 nm diameter), NPs gather at the liposome–liposome interface and interact with two or even three liposomes at a time, neither leading to membrane fusion nor to stalk formation.24 With larger liposomes (60 nm < diameter < 150 nm) Cryo-EM has provided evidence for both adsorbed (Fig. 1b) and snorkeled21 NPs. As for the latter, the resolution did not allow to distinguish between semi- and fully snorkeling configurations. The tomograms shown in Fig. 1b suggest that isolated NPs, adsorbed at the membrane surface of large liposomes, may induce a slight negative curvature.
A characteristic feature of NPs in the 10–30% OT composition range interacting with large DOPC liposomes is NP aggregation on the membrane surface. NPs preferably form large bidimensional aggregates on the liposome surface such as those shown in Fig. 1c: the aggregates are ordered, as previously observed in supported model neuronal membranes.23 Like isolated NPs, also NPs within the bidimensional NP aggregates can be found adsorbed on the membrane surface, or snorkeled in the membrane. An example of an aggregate of embedded NPs is shown in the tomogram reconstruction in ESI Fig. S3.†
The effect of NP aggregates on the membrane curvature of large liposomes, though, is hard to decipher. In Fig. 1c we observe a cluster of 3 large liposomes (L1, L2 and L3, diameter > 100 nm), and extended bidimensional NP aggregates. NPs are found mainly at the interface between the liposomes, confirming their ability to induce liposome aggregation. The most interesting feature is that the curvature of the L2 bilayer is modified by the presence of the NPs in two different ways. At the L1–L2 interface both bilayers are flattened, while at the L2–L3 interface one liposome, L2, inverts its curvature to the advantage of L3, whose bilayer remains positively curved. Further examples of this NP-induced interface reshaping are shown in the ESI ( Fig. S4†). Quite strangely, NP aggregates can cause both curvature inversion and flattening when mediating the interaction between two liposomes.
In the following, we use Molecular Dynamics (MD) to explore the effect that isolated NPs and NP aggregates have on membrane curvature, and we provide a molecular level interpretation of the unexpected heterogeneity of liposome–liposome interfaces observed in the experiments. We used the MARTINI coarse grained force field to model NPs and DOPC liposomes. Model and simulation details are reported in the Computational methods section of the ESI.†
First, we solvated 2 DOPC small liposomes (27 nm diameter) and a variable number of MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT 2
OT 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 NPs in a box of water. On the microsecond time scale, we observed liposome–liposome interactions in each of the simulations containing NPs, while no liposome aggregation was observed in the control run without NPs. As soon as the first NP-mediated liposome–liposome contact was established, other NPs, previously adsorbed on the surface of the liposomes, gathered at the liposome–liposome contact point (ESI Fig. S5†). Overall, these simulations depict a scenario which fits well the experimental data related to small liposomes.
1 NPs in a box of water. On the microsecond time scale, we observed liposome–liposome interactions in each of the simulations containing NPs, while no liposome aggregation was observed in the control run without NPs. As soon as the first NP-mediated liposome–liposome contact was established, other NPs, previously adsorbed on the surface of the liposomes, gathered at the liposome–liposome contact point (ESI Fig. S5†). Overall, these simulations depict a scenario which fits well the experimental data related to small liposomes.
Effect of isolated NPs on membrane curvature, in equilibrium conditions
The unbiased simulation of a dispersion of large liposomes and NPs is cumbersome. We thus resorted to quantifying the curvature induced by NPs on a planar bilayer. We started with isolated NPs, and set up three systems with one 4 nm MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
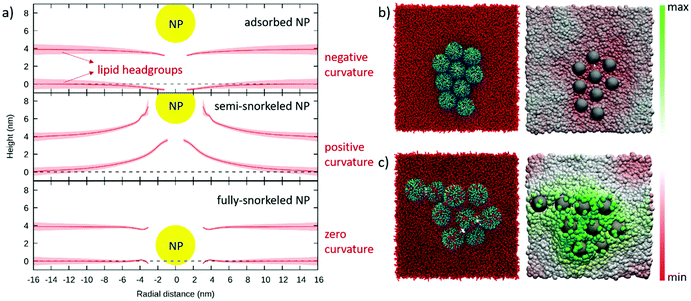
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT 30%OT NP (i) adsorbed on, (ii) partially snorkeled and (iii) fully snorkeled in a flat DOPC membrane (15 × 15 nm2). In Fig. 2a we show the average membrane profile as a function of the radial distance from the NP center obtained after 2 μs of unbiased sampling. In the adsorbed state, the membrane adopts a slightly negative local curvature, to increase the contacts between the hydrophilic ligand terminals and the polar lipid heads. The negative curvature induced by adsorbed NPs is visible also in the experimental image of Fig. 1b. In this configuration, the NP acts by scaffolding. The NP wrapping proceeds only as long as the enthalpic gain associated to favorable NP-lipid headgroups overcomes the cost of the elastic membrane deformation. As the NP diameter is very small, the bilayer does not completely wrap the NP and bends only slightly.
OT 30%OT NP (i) adsorbed on, (ii) partially snorkeled and (iii) fully snorkeled in a flat DOPC membrane (15 × 15 nm2). In Fig. 2a we show the average membrane profile as a function of the radial distance from the NP center obtained after 2 μs of unbiased sampling. In the adsorbed state, the membrane adopts a slightly negative local curvature, to increase the contacts between the hydrophilic ligand terminals and the polar lipid heads. The negative curvature induced by adsorbed NPs is visible also in the experimental image of Fig. 1b. In this configuration, the NP acts by scaffolding. The NP wrapping proceeds only as long as the enthalpic gain associated to favorable NP-lipid headgroups overcomes the cost of the elastic membrane deformation. As the NP diameter is very small, the bilayer does not completely wrap the NP and bends only slightly.
In the semi-snorkeled state, the NP induces a much more pronounced positive curvature in the membrane, deforming the membrane up to much longer radial distances (>10 nm, Fig. 2a). This curvature is caused by the asymmetric deformations that the NP imposes on the two leaflets. Since the anchoring of more and more ligands on the distal leaflet would shift the NP position towards the center of the bilayer, we expect these deformations to decrease with the number of anchored ligands. This is indeed the case: in the fully snorkeled state, in which the NP position coincides with the center of the bilayer, the curvature induced by the NP vanishes. Here, we observe only a small and short-range deformation of the membrane thickness, likely caused by the hydrophobic mismatch between the NP and the lipids.
Effect of NP aggregates on membrane curvature, in equilibrium conditions
Our simulations reproduce the spontaneous NP aggregation of adsorbed, semi- as well as fully snorkeled NPs (see Fig. S6†). What is, then, the effect of NP aggregates on membrane curvature?When NPs are simply adsorbed at the bilayer surface, the membrane adopts a slight negative curvature at the edges of the NP aggregate and flattens elsewhere (Fig. 2b). When the NPs are partially embedded, the aggregate induces a significant positive curvature on the membrane: we see the formation of a dome-like region in the bilayer (Fig. 2c). The curvature vanishes in the presence of the aggregate of symmetrically embedded NPs (Fig. S6†).
The presence of NP aggregates drives the membrane response to mechanical stress
MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT NPs can affect membrane curvature also in out-of-equilibrium conditions, such as under the action of a mechanical stress. In absence of NPs, when a flat membrane is subject to an in-plane, uniaxial mechanical compression, the membrane undergoes a buckling transition and assumes a roughly sinusoidal shape with period equal to the length of the box.33 Curvature-inducing NPs have been shown to induce spontaneous buckling in large flat membranes, especially when aggregating in linear patterns.13 MUS
OT NPs can affect membrane curvature also in out-of-equilibrium conditions, such as under the action of a mechanical stress. In absence of NPs, when a flat membrane is subject to an in-plane, uniaxial mechanical compression, the membrane undergoes a buckling transition and assumes a roughly sinusoidal shape with period equal to the length of the box.33 Curvature-inducing NPs have been shown to induce spontaneous buckling in large flat membranes, especially when aggregating in linear patterns.13 MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
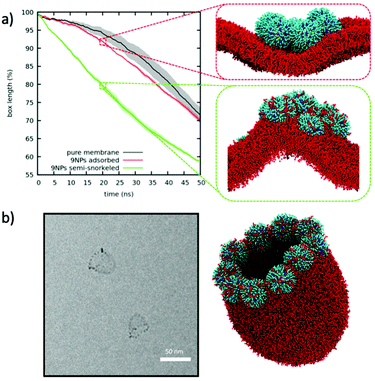
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT NPs can facilitate membrane buckling, as a result of the NP-induced local membrane curvature. Fig. 3a shows the time evolution of the lateral size of the box during the compression (Pxx = 4 bar, Pyy = Pzz = 1 bar) of a bare membrane, of a membrane with an adsorbed 9-NP aggregate, and of a membrane with a semi-snorkeled 9-NP aggregate. The bare membrane has the slowest kinetics. The slight negative curvature induced by the adsorbed aggregate facilitates bending, with the NP aggregate sitting on the concave side of the membrane. The strong positive curvature due to the presence of the semi-snorkeled aggregate has the largest effect on the kinetics of bending, quickly leading to halve the x box edge, with the NP aggregate sitting on the convex side of the buckle. Isolated NPs (adsorbed and semi-snorkeled) also affect the kinetics of buckling, as shown in Fig. S7.†
OT NPs can facilitate membrane buckling, as a result of the NP-induced local membrane curvature. Fig. 3a shows the time evolution of the lateral size of the box during the compression (Pxx = 4 bar, Pyy = Pzz = 1 bar) of a bare membrane, of a membrane with an adsorbed 9-NP aggregate, and of a membrane with a semi-snorkeled 9-NP aggregate. The bare membrane has the slowest kinetics. The slight negative curvature induced by the adsorbed aggregate facilitates bending, with the NP aggregate sitting on the concave side of the membrane. The strong positive curvature due to the presence of the semi-snorkeled aggregate has the largest effect on the kinetics of bending, quickly leading to halve the x box edge, with the NP aggregate sitting on the convex side of the buckle. Isolated NPs (adsorbed and semi-snorkeled) also affect the kinetics of buckling, as shown in Fig. S7.†
The strong positive curvature induced by semi-snorkeled NPs can also lead to the formation of metastable open liposome configurations, as the ones resembling jellyfish bells shown in Fig. 3b. Here, semi-snorkeled NPs seal the open liposome edge that, in absence of NPs, would spontaneously and quickly close up to form a liposome. It has not been possible to observe the spontaneous formation of this open liposome configuration in unbiased simulations but, once artificially set up, we could assess its stability. We did not observe any structural evolution over time scales of several microseconds.
Liposome–liposome interface geometries explained by different NP embedding
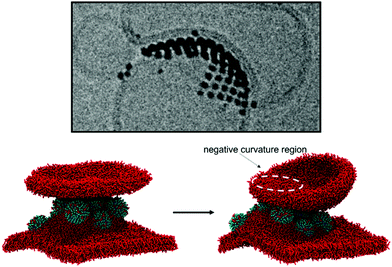
We have shown that isolated and aggregated NPs can significantly affect membrane curvature, and that different degrees of NP embedding lead to the stabilization of different curvatures. We can thus propose an interpretation for the heterogeneous liposome–liposome interfaces shown in Fig. 1c. At the L1–L2 interface, the NP aggregate is simply adsorbed on both liposomes. The symmetric, flat L1–L2 interface maximizes the favorable bilayer-NP contacts, overcoming the energetic penalty associated to bilayer flattening. At the L2–L3 interface, NPs are only adsorbed on the L2 liposome and semi-snorkeled in the L3 bilayer. The symmetry is broken: the curvature of the L3 bilayer is extremely stable due to the presence of the semi-snorkeled NP aggregate. The enthalpic advantage resulting from the adhesion of the L2 bilayer to the L3-embedded NPs is large enough to compensate for the elastic deformation of the L2 bilayer, which adapts to the curvature imposed by L3. As a final test, to demonstrate the validity of our interpretation, we designed an in silico experiment. We built a simulation box containing a membrane with a semi-snorkeled NP aggregate, and a bicelle (See Fig. 4). If isolated, the bicelle would close and form a small liposome. However, after a few tens of nanoseconds, the bicelle lays on top of the underlying NP aggregate, developing a local negative curvature.Conclusions
In conclusion, we have shown that during the different stages of Au NP–liposome interaction the NP is able to impose different curvatures to the bilayer. NPs which are simply adsorbed at the membrane surface induce a slight negative curvature, with a range of deformation that does not exceed one or two NP diameters. The most pronounced effect on membrane curvature is obtained when the NPs are in the a-symmetric semi-snorkeled configuration, causing the bilayer to develop a positive curvature over length scales of several NP diameters. The length of the anionic NP ligands, which anchor to the distal leaflet and heavily deform it, is a key parameter to obtain such a significant curvature.The slow kinetics of NP embedding plays a major role. As the characteristic time for the complete embedding of a single NP is longer than that characterizing NP diffusion, NPs can form aggregates and impose different curvatures to the underlying membrane on large spatial scales. Aggregates of adsorbed NPs flatten the liposome membrane, aggregates of semi-snorkeled NPs well adapt to the positive curvature of the liposome and further stabilize it. The molecular mechanisms we have observed via molecular simulations offer an interpretation to the puzzling experimental observation of both flat and curved liposome–liposome interfaces. These results reveal that the kinetics of NP–membrane interaction should be regarded as a key physical descriptor guiding the design of nanoparticles with controlled interaction with plasma membranes.
Overall, our results contribute to the fundamental understanding of synthetic NP–membrane interactions, and highlight the importance of the physical properties of the NP ligand shell. This understanding is fundamental to correctly interpret the outcomes of experimental assays on model lipid membranes, and to guide the design of monolayer-protected nanoparticles with controlled interactions with cell membranes.
Author contributions
Conceptualization: GR, DB and FS. MD simulations: EL with supervision of GR and DB. NP synthesis and cryo-EM experiments: ZPG with supervision of FS. Data curation: FO. Writing – original draft: EL and DB. Writing – review&editing: GR, DB and FS.Conflicts of interest
There are no conflicts to declare.Acknowledgements
G.R. acknowledges funding from the ERC Starting Grant BioMNP-677513 and from the Curiosity Driven grant 2019-2021 funded by the University of Genoa. G. R., D. B. and E. L. acknowledge MIUR funding (DIFI – Dipartimento di Eccellenza 2018-2022) for computational resources.Notes and references
- A. Hossein and M. Deserno, Biophys. J., 2020, 118, 624–642 CrossRef CAS PubMed.
- M. M. Kozlov, F. Campelo, N. Liska, L. V. Chernomordik, S. J. Marrink and H. T. McMahon, Curr. Opin. Cell Biol., 2014, 29, 53–60 CrossRef CAS PubMed.
- B. J. Peter, H. M. Kent, I. G. Mills, Y. Vallis, P. J. G. Butler, P. R. Evans and H. T. McMahon, Science, 2004, 303, 495–499 CrossRef CAS PubMed.
- M. Simunovic, G. A. Voth, A. Callan-Jones and P. Bassereau, Trends Cell Biol., 2015, 25, 780–792 CrossRef CAS PubMed.
- P. D. Blood and G. A. Voth, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15068–15072 CrossRef CAS PubMed.
- T. Baumgart, B. R. Capraro, C. Zhu and S. L. Das, Annu. Rev. Phys. Chem., 2011, 62, 483–506 CrossRef CAS PubMed.
- J. G. Joseph, C. Osorio, V. Yee, A. Agrawal and A. P. Liu, Commun. Biol., 2020, 3, 1–16 CrossRef PubMed.
- L. Johannes, W. Pezeshkian, J. H. Ipsen and J. C. Shillcock, Trends Cell Biol., 2018, 28, 405–415 CrossRef CAS PubMed.
- W. Pezeshkian, A. G. Hansen, L. Johannes, H. Khandelia, J. C. Shillcock, P. B. S. Kumar and J. H. Ipsen, Soft Matter, 2016, 12, 5164–5171 RSC.
- B. J. Reynwar, G. Illya, V. A. Harmandaris, M. M. Müller, K. Kremer and M. Deserno, Nature, 2007, 447, 461–464 CrossRef CAS PubMed.
- J. Hsin, J. Gumbart, L. G. Trabuco, E. Villa, P. Qian, C. N. Hunter and K. Schulten, Biophys. J., 2009, 97, 321–329 CrossRef CAS PubMed.
- H. G. Franquelim, A. Khmelinskaia, J. P. Sobczak, H. Dietz and P. Schwille, Nat. Commun., 2018, 9, 1–10 CrossRef CAS PubMed.
- E. Lavagna, J. Barnoud, G. Rossi and L. Monticelli, Nanoscale, 2020, 12, 9452–9461 RSC.
- A. H. Bahrami and T. R. Weikl, Nano Lett., 2018, 18, 1259–1263 CrossRef CAS PubMed.
- S. Dasgupta, T. Auth and G. Gompper, J. Phys.: Condens. Matter, 2017, 29, 373003 CrossRef CAS PubMed.
- S. Rana, A. Bajaj, R. Mout and V. M. Rotello, Adv. Drug Delivery Rev., 2012, 64, 200–216 CrossRef CAS PubMed.
- P. Pengo, M. Şologan, L. Pasquato, F. Guida, S. Pacor, A. Tossi, F. Stellacci, D. Marson, S. Boccardo, S. Pricl and P. Posocco, Eur. Biophys. J., 2017, 46, 749–771 CrossRef CAS PubMed.
- Y. Jiang, S. Huo, T. Mizuhara, R. Das, Y. Lee, S. Hou, D. F. Moyano, B. Duncan, X. Liang and V. M. Rotello, ACS Nano, 2015, 9, 9986–9993 CrossRef CAS PubMed.
- R. C. Van Lehn, P. U. Atukorale, R. P. Carney, Y.-S. Yang, F. Stellacci, D. J. Irvine and A. Alexander-Katz, Nano Lett., 2013, 13, 4060–4067 CrossRef CAS PubMed.
- F. Lolicato, L. Joly, H. Martinez-Seara, G. Fragneto, E. Scoppola, F. Baldelli Bombelli, I. Vattulainen, J. Akola and M. Maccarini, Small, 2019, 15, 1805046 CrossRef PubMed.
- P. U. Atukorale, Z. P. Guven, A. Bekdemir, R. P. Carney, R. C. Van Lehn, D. S. Yun, P. H. Jacob Silva, D. Demurtas, Y. S. Yang, A. Alexander-Katz, F. Stellacci and D. J. Irvine, Bioconjugate Chem., 2018, 29, 1131–1140 CrossRef CAS PubMed.
- T. Pfeiffer, A. De Nicola, C. Montis, F. Carlà, N. F. A. Van Der Vegt, D. Berti and G. Milano, J. Phys. Chem. Lett., 2019, 10, 129–137 CrossRef CAS PubMed.
- E. Canepa, S. Salassi, A. L. de Marco, C. Lambruschini, D. Odino, D. Bochicchio, F. Canepa, C. Canale, S. Dante, R. Brescia, F. Stellacci, G. Rossi and A. RELINI, Nanoscale, 2020, 12, 19746–19759 RSC.
- M. A. Tahir, Z. P. Guven, L. R. Arriaga, B. Tinao, Y. S. S. Yang, A. Bekdemir, J. T. Martin, A. N. Bhanji, D. Irvine, F. Stellacci and A. Alexander-Katz, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 18470–18746 CrossRef CAS PubMed.
- F. Simonelli, D. Bochicchio, R. Ferrando and G. Rossi, J. Phys. Chem. Lett., 2015, 6, 3175–3179 CrossRef CAS.
- R. C. Van Lehn, M. Ricci, P. H. J. Silva, P. Andreozzi, J. Reguera, K. Voïtchovsky, F. Stellacci and A. Alexander-Katz, Nat. Commun., 2014, 5, 4482 CrossRef CAS PubMed.
- R. C. Van Lehn and A. Alexander-Katz, Soft Matter, 2015, 11, 3165–3175 RSC.
- P. Gkeka, P. Angelikopoulos, L. Sarkisov and Z. Cournia, PLoS Comput. Biol., 2014, 10, e1003917 CrossRef PubMed.
- S. Salassi, F. Simonelli, D. Bochicchio, R. Ferrando and G. Rossi, J. Phys. Chem. C, 2017, 121, 10927–10935 CrossRef CAS.
- R. C. Van Lehn and A. Alexander-Katz, PLoS One, 2019, 14, 1–19 CrossRef PubMed.
- Z. P. Guven, P. H. Silva, Z. Luo, U. B. Cendrowska, M. Gasbarri, S. T. Jones and F. Stellacci, J. Visualized Exp., 2019, 149, e58872 Search PubMed.
- Z. P. Guven, PhD thesis, École Polytechnique Fédérale de Lausanne, 2014, DOI:10.5075/epfl-thesis-8963.
- M. Hu, P. Diggins IV and M. Deserno, J. Chem. Phys., 2013, 138, 1–13 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1nr05067b |
| This journal is © The Royal Society of Chemistry 2021 |