Self-assembly of peptide nanofibers for imaging applications
Qiaochu
Jiang†
,
Xiaoyang
Liu†
 ,
Gaolin
Liang
,
Gaolin
Liang
 * and
Xianbao
Sun
*
* and
Xianbao
Sun
*
State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, 2 Sipailou Road, Nanjing 210096, China. E-mail: gliang@seu.edu.cn; xbsun@seu.edu.cn
First published on 1st September 2021
Abstract
Pathological stimuli-responsive self-assembly of peptide nanofibers enables selective accumulation of imaging agent cargos in the stimuli-rich regions of interest. It provides enhanced imaging signals, biocompatibility, and tumor/disease accessibility and retention, thereby promoting smart, precise, and sensitive tumor/disease imaging both in vitro and in vivo. Considering the remarkable significance and recent encouraging breakthroughs of self-assembled peptide nanofibers in tumor/disease diagnosis, this reivew is herein proposed. We emphasize the recent advances particularly in the past three years, and provide an outlook in this field.
1. Introduction
Self-assembly of biocomponents is a crucial biological event for living objects. For example, proteins exhibit physiological functions in their assembled states (tertiary and quaternary structures).1 Of note, abnormal assembly is always closely related to pathological changes, which may help indicate the occurrence of certain diseases (e.g., self-assembly of amyloid-beta is responsible for Alzheimer's disease).2 As a consequence, exploring the self-assembly behaviors of biocomponents can be conducive to understanding certain pathological events at the molecular level.3,4 To this end, biomolecules, particularly peptides with self-assembly ability, are frequently employed as mimics for tumor/disease diagnosis.5–7 Driven by thermodynamics and kinetics,8 these peptides spontaneously form ordered assemblies (e.g., nanoparticles, nanotubes, and nanofibers) through multiple noncovalent interactions between building blocks.9,10Among these peptide assemblies, nanofibers have attracted intensive interest in tumor/disease imaging or therapy. First, in contrast to other assemblies (e.g, nanoparticles, nanomicelles, etc.), nanofibers possess much higher surface-area-to-volume ratios, which provide more functionable and active sites and allow better cooperative interactions with bio-targets, such as cellular mitochondria of tumor,11 thus allowing enhanced tumor accessibility. Second, nanofibers show enhanced retention effects in tumor sites,12 and may exhibit superior tumor accumulation than nanoparticles at the same size,13 therefore facilitating sustained tumor/disease imaging or drug release. Third, intracellularly formed nanofibers may disturb the dynamics of microtubules of glioblastoma cells,14 while pericellular nanofibers can decrease the migration of several cancer cells (e.g., HeLa),15 thus nanofibers hold promise as platforms for fabricating novel theranostic agent. Last and most importantly, nanofibers could further gel water to form supramolecular hydrogels, which are considered as highly promising versatile biomaterials with prominent loading capacity, biocompatibility, and biodegradability for broad applications such as drug delivery, cell culture, and bioimaging.16
To date, self-assembled peptide nanofibers have been recognized as attractive and promising carriers of various imaging agents for smart and precise tumor/disease imaging.17–22 Conjugated with imaging agents,23–25 peptide precursors were rationally designed with responsiveness to tumor/disease-related over-expressed pathological stimuli, such as alkaline phosphatase (ALP),26 matrix metalloproteinase-2 (MMP-2),27 and bacteria surface receptors.28 As such, in situ self-assembly of nanofibers could be smartly activated. This process enabled the smart localization and accumulation of the imaging agents at the target site of interest, thus promoting precise and sensitive imaging.29,30 Moreover, in contrast to the inactivated small-molecule peptide precursors in less-/non-stimuli healthy cells, nanofibers formed in tumor/disease cells could prolong the intracellular retention time of the imaging agents, thus affording enhanced metabolism difference, specificity and sustainability of imaging. Considering that smart self-assembled peptide nanofibers have shown remarkable significance in tumor/disease diagnosis, and notable breakthroughs in this field particularly in the past three years have not been emphatically reviewed, we herein provide this review. We highlighted the recent advance in smart self-assembly of rationally designed nanofibers, which enabled enhanced imaging of tumor/disease with modalities including magnetic resonance imaging (MRI), optical imaging (OI), photoacoustic imaging (PAI), and multi-modality imaging. We anticipate this review will inspire more designs of smart peptide nanofibers for enhanced tumor/disease diagnosis.
2. Self-assembly of nanofibers for imaging applications
2.1 Design principles of smart peptide nanofiber precursors for imaging applications
Smart peptide nanofiber precursors consist of two parts: (1) self-assembly unit, and (2) stimulus–cleavable blocking unit (Scheme 1). Once activated by the stimulus, the self-assembly unit would act as the building block to form nanofibers. | ||
| Scheme 1 Smart peptide nanofiber precursors for imaging applications. S.A. unit, self-assembly unit; S.C.B unit, stimulus–cleavable blocking unit. | ||
| Nanofiber precursor | Peptide self-assembly unit | Stimulus–cleavable blocking unit | Stimulus | Imaging application | Ref. | |
|---|---|---|---|---|---|---|
| Sequence | Driving force | |||||
| NBD-FF-easter-taurine | FF | Hydrogen bonding, π–π stacking | Ester bond | Esterase | Fluorescence imaging of Cellular esterase | 56 |
| Fmoc-K(FITC)FFY(H2PO3) | Fmoc-KFF | Hydrogen bonding, π–π stacking, hydrophobic interactions | Y(H2PO3) | Alkaline phosphatase (ALP) | Fluorescence imaging of tumor | 57 |
| Nap-FFK(DYKDDDDK)-NBD | Nap-FF | Hydrogen bonding, π–π stacking, hydrophobic interactions | DYKDDDDK | Enterokinase (ENTK) | Fluorescence imaging of ENTK activity | 58 |
| Nap-GFFK(Cou)Y(H2PO3)D | Nap-GFF | Hydrogen bonding, π–π stacking, hydrophobic interactions | Y(H2PO3) | Alkaline phosphatase (ALP) | Luminescence imaging of tumor | 59 |
| NBD-FFFGK(succ)G | FFF | Hydrogen bonding, π–π stacking | Succinylated K | SIRT5 | Fluorescence imaging of mitochondria | 60 |
| Nap-FFFYp-EDA-DOTA(Gd) | Nap-FFF | Hydrogen bonding, π–π stacking, hydrophobic interactions | Y(H2PO3) | Alkaline phosphatase (ALP) | Magnetic resonance imaging of tumor | 61 |
| PpiX-PEG8-SSSPLGLAK(DOTA)-PEG6-F4 | FFFF | Hydrogen bonding, π–π stacking | PLGLA | Matrix metalloproteinase-2 (MMP-2) | Magnetic resonance imaging of tumor | 52 |
| GTDTKTGPAKLVFFC(Cyanine)TDTG | KLVFF | Hydrogen bonding, π–π stacking | GPA | FAP-α | Fluorescence imaging of tumor | 62 |
| RGDRDDRDDPLGYLGFFC(Cy) | YLGFFC | Hydrogen bonding, π–π stacking | PLGYLG | Matrix metalloproteinase-2/9 (MMP-2/9) | Fluorescence imaging of renal cell carcinoma | 63 |
| AVPIAQKDEVDKLVFFAEC(Cy)G | KLVFFAECG | Hydrogen bonding, π–π stacking | DEVD | Caspase-3/7 | Fluorescence imaging of tumor | 64 |
| mPEG2000-KLDLKLDLKLDL-p-SCN-deferoxamine-89Zr | KLDLKLDLKLDL | Hydrophobic&electrostatic interactions | N-terminal K | Cathepsin B | Fluorescence/positron emission tomography/computed tomography imaging of tumor | 65 |
| Mannose-YVHDCKK(A-purpurin18) | K(A-purpurin18) | π–π stacking, hydrophobic interactions | YVHDC | Caspase-1 | Photoacoustic imaging of tumor | 66 |
| Purpurin18-PLGVRGRGD | Purpurin18-PLG | π–π stacking, hydrophobic interactions | PLGVRG | Gelatinase | Photoacoustic imaging of tumor | 12 |
2.2 Self-assembly of nanofibers for MRI
Magnetic resonance imaging (MRI) is one of the most prevalent clinical non-invasive diagnostic tools with deep penetration and high spatial resolution.46 In magnetic field, teeny changes in body can significantly influence the relaxation parameters of hydrogen or fluorine nuclei, and further generate MRI signals.47 Nevertheless, MRI still need contrast agents (CAs) as adjuvants to improve its sensitivity in handling clinical issues. According to the ratio between longitudinal relaxation (r1) and transverse relaxation (r2), CAs are mainly divided into T1 and T2 agents. T1 agents produce positive contrast, and are commonly based on stable and inert paramagnetic ions, such as gadolinium (Gd).48 T2 agents produce negative contrasts, and are based on complexes such as superparamagnetic iron oxide (SPIO).25Small molecular Gd-based CAs were proved that they can increase the 1H relaxation rates of nearby water molecules, thereby enhancing MRI contrast signals.24 However, these CAs were subject to fast body clearance after administration, thus showing limited retention ability. To address this issue, Gd-based nanomaterials have been proposed, such as polymers, nanoparticles, micelles, liposomes and nanofibers.47,49–51 For example, Zhang et al. designed a MMP-2-responsive peptide Ppdf-Gd that enabled nanosphere-to-nanofiber transformation in tumor microenvironment (Fig. 1a).52 In brief, the amphiphilic Ppdf-Gd first self-assembled into spherical nanoparticles in physiological conditions to ensure efficient tumor accumulation due to EPR effect. Then the overexpressed MMP-2 in tumor microenvironment recognized and cleaved the peptide backbone, breaking the amphiphilicity and enabling the transformation from nanosphere to nanofiber (Fig. 1b), which further improved the retention time of the CAs. More importantly, this nanosphere-to-nanofiber transformation led to increased relaxation rate of the loaded DOTA-Gd, which afforded amplified tumor MRI signals than the control groups (i.e., free DOTA-Gd, and MMP-2-inert Pdf-Gd) (Fig. 1c). Notably, this strategy utilized in situ morphology transformation (nanoparticle-to-nanofiber) to enhance the retention ability and MR signal of CA, which has shown merits as an attractive methodology for smart delivery of drugs or imaging agents.53 However, for the Phe-Phe-based short peptides, their self-assembly tendency would be significantly impaired after modification with Gd complex.54 To demonstrate this issue, Diaferia et al. replaced the Phe with more aromatic naphthylalanine, and proposed a dinaphthylalanine-Gd-conjugate (DOTA-L6-2Nal2) to form MRI nanofibers.55
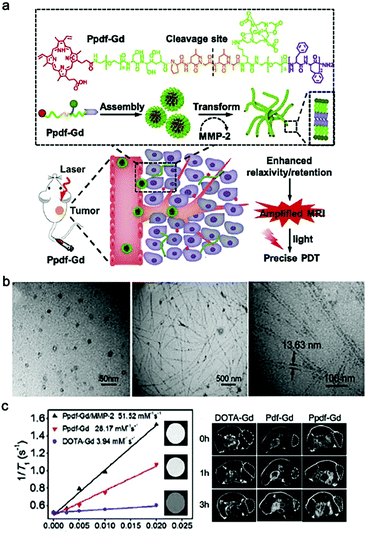 | ||
| Fig. 1 (a) Schematic illustration of MMP-2-triggered transformation of Ppdf-Gd from spherical nanoparticles to nanofibers and the principle of dual-stage-amplified MRI and PDT. Ppdf-Gd can self-assembly to spherical nanoparticles in physiological conditions. When reaching tumor tissue by the EPR effect, the nanoparticles underwent sphere-to-fibers transformation under MMP-2. This transformation can enhance relaxivity and retention of contrast agent in tumor region, which realized amplified MRI and precise PDT. (b) TEM images of Ppdf-Gd solution in the presence or the absence of MMP-2 and amplified TEM image of Ppdf-Gd. (c) Left: longitudinal relaxation rates measurement of Ppdf-Gd, DOTA-Gd, and Ppdf-Gd with enzyme MMP-2. The insets represented the T1 weighted MR images of various groups at same Gd3+concentration; Right: T1-weighted MRI images of mice at 0, 1, and 3 h after intravenous (i.v.) injection (Gd3+dose: 0.05 mmoL kg−1). Modified with permission from ref. 52. Copyright 2018. Elsevier Ltd. | ||
Recently, chemical exchange saturation transfer (CEST)-based MRI has emerged as a highly promising strategy for tumor/disease imaging, owing to its enhanced sensitivity, natural labeling of bioactive molecules, and negligible background interference.67,68 Cui et al. conjugated a peptide precursor with pemetrexed, an FDA-approved anticancer drug, as well as a CEST agent.69 This conjugate could self-assemble into nanofibers in tumors, enabling enhanced tumor CEST MRI. Of note, traditional 1H MRI are commonly limited by inherent noise interference from water molecules.70 By contrast, 19F MRI has shown great advantages due to its high sensitivity, wide chemical shift and negligible background.71 For example, Liang group designed a smart 19F nanofiber that was formed through ALP-instructed self-assembly, enabling sensitive MRI of enzyme activity in vitro and in cell lysates.72
2.3 Self-assembly of nanofibers for OI
Optical imaging (OI) is a non-invasive imaging modality that utilizes light including ultraviolet, visible and infrared regions.73 OI can provide images of tissues and cells with high spatial resolution, but the resolution decreases rapidly with the increment of imaging depth.74 Bioluminescence imaging (BI) and fluorescence imaging (FI) are the two most commonly used techniques. Considering that self-assembled peptide nanofibers for BI have seldom been reported, we thereby only summarize their FI applications.Recently, He et al. designed a branched peptide (D-1![[T with combining macron]](https://www.rsc.org/images/entities/char_0054_0304.gif) FLAG) for targeting and enhanced imaging of mitochondria (Fig. 2a).58 This peptide precursor firstly formed nanoparticles, which could target mitochondria due to their negative charges. Upon the cleavage by enterokinase (ETNK) expressed on the membrane of mitochondria, nanoparticles transfromed into nanofibers by self-assembly on mitochondria membrane (Fig. 2b). As such, the NBD dye, which exhibited aggregation-enhanced emission, efficiently accumulated on mitochondria, and produced enhanced fluorescence signals in tumor cells (Fig. 2c and d). Notably, mitochondria-targeting nanofiber precursor provided significant benefits to the enhancement of fluorescence signals. This is because precursors could be further concentrated after being internalized by mitochondria from cytoplasm, promoting the self-assembly of nanofibers. As a result, subcellular nanofiber precursors that target organelles may be advantageous and deserve to be developed for enhanced bioimaging. Of note, in contrast to conventional fluorescent dyes that are subjected to aggregation-caused quenching effect, NBD shows advantages in helping visualizing in vivo biological events with self-assembled peptide nanofibers. For example, Zhang et al. conjugated NBD on a ALP- and GSH-responsive peptide nanofiber precursor, and achieved fluorescence “on/off” monitoring with the tandem assembly/disassembly of nanofibers in living tumor cells.75 Alternatively, aggregation-induced emission (AIE) dyes also can be employed for the same imaging purpose.76,77
FLAG) for targeting and enhanced imaging of mitochondria (Fig. 2a).58 This peptide precursor firstly formed nanoparticles, which could target mitochondria due to their negative charges. Upon the cleavage by enterokinase (ETNK) expressed on the membrane of mitochondria, nanoparticles transfromed into nanofibers by self-assembly on mitochondria membrane (Fig. 2b). As such, the NBD dye, which exhibited aggregation-enhanced emission, efficiently accumulated on mitochondria, and produced enhanced fluorescence signals in tumor cells (Fig. 2c and d). Notably, mitochondria-targeting nanofiber precursor provided significant benefits to the enhancement of fluorescence signals. This is because precursors could be further concentrated after being internalized by mitochondria from cytoplasm, promoting the self-assembly of nanofibers. As a result, subcellular nanofiber precursors that target organelles may be advantageous and deserve to be developed for enhanced bioimaging. Of note, in contrast to conventional fluorescent dyes that are subjected to aggregation-caused quenching effect, NBD shows advantages in helping visualizing in vivo biological events with self-assembled peptide nanofibers. For example, Zhang et al. conjugated NBD on a ALP- and GSH-responsive peptide nanofiber precursor, and achieved fluorescence “on/off” monitoring with the tandem assembly/disassembly of nanofibers in living tumor cells.75 Alternatively, aggregation-induced emission (AIE) dyes also can be employed for the same imaging purpose.76,77
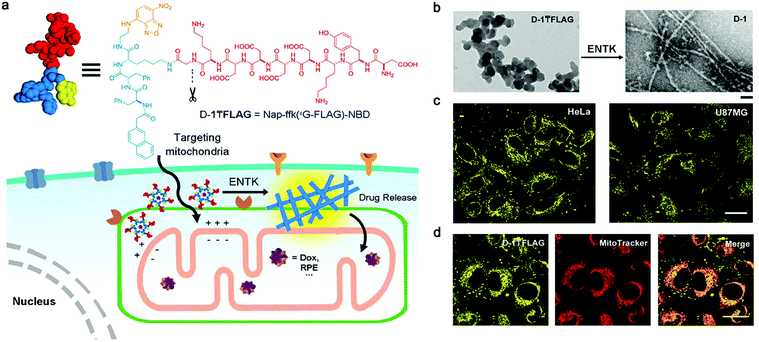 | ||
Fig. 2 (a) Structure of a representative branched peptide and ENTK cleaving the branch to convert micelles to nanofibers on mitochondria. (b) TEM images of D-1![[T with combining macron]](https://www.rsc.org/images/entities/char_0054_0304.gif) FLAG before (left) and after (right) adding ENTK (24 h), scale bar = 100 nm. (c) Fluorescent images of HeLa and U87MG cells incubated with D-1 FLAG before (left) and after (right) adding ENTK (24 h), scale bar = 100 nm. (c) Fluorescent images of HeLa and U87MG cells incubated with D-1![[T with combining macron]](https://www.rsc.org/images/entities/char_0054_0304.gif) FLAG for 2 h. (d) The fluorescent images of D-1TFLAG and mitotracker in HeLa cells. Scale bar = 30 μm in panels c and d. The concentrations of D-1 FLAG for 2 h. (d) The fluorescent images of D-1TFLAG and mitotracker in HeLa cells. Scale bar = 30 μm in panels c and d. The concentrations of D-1![[T with combining macron]](https://www.rsc.org/images/entities/char_0054_0304.gif) FLAG are 200 μM for b–d. Modified with permission from ref. 58. Copyright 2018. American Chemical Society. FLAG are 200 μM for b–d. Modified with permission from ref. 58. Copyright 2018. American Chemical Society. | ||
However, autofluorescence interference and inferior tissue penetration of visible light still largely limit the applications of fluorescent nanofibers in visualizing deep-situated biological events.78 Considering that absorption and scattering ability of biological tissues to photons attenuated rapidly with the increment of light wavelength, NIR light may significantly improve tissue penetration and spatial resolution of imaging.79 In recent years, numerous NIR imaging probes or nanomaterials have been developed. For example, Zhao et al. constructed a smart NIR peptide probe for improved tumor imaging.62 This probe was consisted of two hydrophilic motifs, a tailoring motif, a self-assembly motif, and a NIR cyanine dye. Upon the specific cleavage by fibroblast activation protein-α (FAP-α), the probe would be activated and self-assemble into nanofibers on the surface of tumor-associated fibroblasts (CAFs). Self-assembled nanofibers were confirmed in vitro. Notably, in vivo results demonstrated that self-assembly of nanofibers provided significantly enhanced retention effect and sustained imaging capacity.
2.4 Self-assembly of nanofibers for PAI
Photoacoustic imaging (PAI) is a non-invasive imaging method that combines optical excitation with ultrasonic detection based on photoacoustic effect.80 Compared with traditional OI, PAI owns higher spatial resolution and improved tissue penetration depth (several centimeters deep in biological tissue imaging).81 However, only a few natural light absorbers exists, such as melanin and hemoglobin, while others are not able to send out photoacoustic signals.82 Therefore, various exogenous photoacoustic agents, particularly smart photoacoustic nanomaterials, have been developed to enhance the photoacoustic contrast. For example, Pu's group reported a self-assembled semiconducting polymer amphiphile (SPA).83 As a near-infrared absorbing material, this SPA showed advantages in stability, fluorescence quantum yield, and tumor-targeting ability, enabling precise and sensitive tumor PAI. Remarkably, Liang's group proposed an ALP-responsive NIR probe, which could self-assembled in ALP-rich tumor cells, achieving smart and enhanced in vivo PAI of tumor.84Recently, Wang's group proposed a smart chlorophyll-peptide-based photoacoustic agent (MPC) for imaging intracellular bacterial infection, which was responsible for relevant treatment failure and potential antibiotic resistance.66 This MPC was consisted of a macrophage-targeting motif, a caspase-1-cleavable tailoring motif and a chlorophyll-Cu2+ coordination PA signal motif (Fig. 3a). When infected by bacteria, macrophage immediately expressed caspase-1, which cleaved MPC and activated its self-assembly, further forming nanofibers and quenching the fluorescence (Fig. 3b and c). In the mouse model that was constructed by injecting infected macrophage cells (Fig. 3d), approximately 2.6-fold higher PAI signals were observed upon infection than that of the healthy group (Fig. 3e), and reached the maximum intensity at 8 h (Fig. 3f). The S. aureus infection model verified the chemotaxis-instructed infection of PA detection (Fig. 3g). The infected muscles stained by H&E (Fig. 3h) indicated the inflammation of the S. aureus invasion. This work provided inspirations for PAI of bacterial infection and even the related diseases with smart self-assembled peptide nanofibers. Considering that various peptide nanofibers have been used as advantageous carriers of antimicrobial therapeutic agents,85 it would be promising to combine PAI imaging and therapy agents on smart nanofiber precursors for novel antimicrobial theranostics.
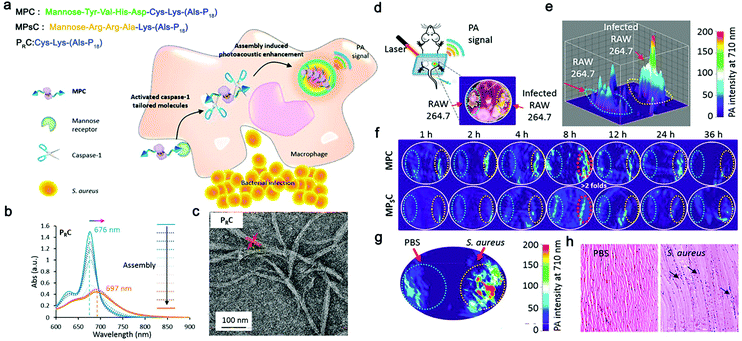 | ||
| Fig. 3 (a) Schematic representation of macrophage chemotaxis-instructed S. aureus infection detection in vivo and the molecular component of the probe (MPC). Structure illustration of MPC, MPSC and PRC. (b) The UV−vis spectra of assembly procedure of molecule PRC in the mixture solution (H2O/DMSO) with different volume ratios (from 0% to 100%). The molecule concentration is 5 × 10−5 M. (c) Transmission electron microscope (TEM) image of PRC fibrous assemblies in the aqueous solution (H2O/DMSO; 95/5; v/v). (d) Chemotaxis-instructed S. aureus infection PA detection in vivo. Schematic illustration of the mice model (intramuscular injection of infected RAW 264.7 cells) and photoacoustic tomography (PAT) detection. The infected RAW 264.7 cells were obtained with the same procedure as before. The mice model was built after intramuscular injection of infected RAW 264.7 (107 cells per injection) for 12 h. (e) PA signal intensity distribution of infected RAW 264.7 cells in vivo after MPC administration with a dose of 35 mg kg−1 though i.v. injection for 8 h. (f) PA images of intracellular infection in vivo between 1 and 36 h after i.v. administration of MPC and MPSC (35 mg kg−1), respectively. The PA intensities per area of MPC and MPSC were calculated based on the red dotted circle area. (g) PA images of muscular infection. The right leg was infected after intramuscular injection of 108 cfu S. aureus cells for 12 h. (h) Representative micrographs of the histology of the muscle sections (H&E staining) of the S. aureus infected and the control (PBS) groups. Black arrows indicate the leukocytes during inflammation of the S. aureus invasion. The number of mice in each group is three. Modified with permission from ref. 66. Copyright 2018. American Chemical Society. | ||
2.5 Self-assembly of nanofibers for multi-modality imaging
In recent years, intensive interest has been raised to develop multi-modality techniques to combine the advantages of various imaging modalities. Smart nanomaterials have been frequently employed as carriers to load multimodal imaging agents. For example, gold nanoparticles were conjugated with the peptide substrate of MMP-2 for smart PET/CT imaging of tumor;86 ALP-responsive Fe3O4-based nanoparticles were proposed for smart MR/Optical Imaging;87 DOTA-Gd and a NIR fluorophore were conjugated on ALP-responsive self-assembling dipeptide for smart and sensitive NIR/MR imaging.88Peptide nanofibers has been explored for multimodal imaging of tumor. For example, Law et al. reported a peptide precursor (NFP) which was capable of self-assembling into nanofibers in tumor regions (Fig. 4a).65 As a carrier, the NFP provided design flexibility for on-demand customization of imaging/therapy agents (Fig. 4b). The optimized GSH-NFP could realize real-time monitoring tumoral delivery by loading Cy5.5 or DFO (Fig. 4c and d). After the cleavage of hydrophilic mPEG by tumor-associated proteases (e.g., cathepsin B), 89Zr-NFP conjugate could self-assemble into large nanofibers (Fig. 4e), providing improved cancer targeting and tissue invasion ability, and enhanced FI/PET/CT multi-modality imaging of tumor (Fig. 4f and g). However, in this work modality agents were not conjugated on one nanofiber precursor, thus a set of precursor analogues were necessitated to realize multi-modality imaging of tumor, which rendered complicated synthesis and less accurate administration of the imaging agents. Unfortunately, smart nanofiber precursors with integrated modality agents still remain to be developed for precise imaging of tumor/disease with the highly promising multi-modality technique.
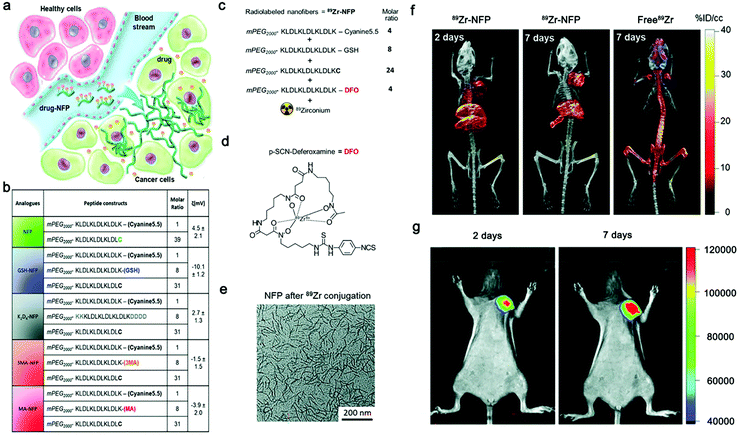 | ||
| Fig. 4 (a) Design of advanced NFP analogues to enhance tumoral uptake, penetration, and local retention. NFP has a high aspect ratio that promotes its uptake by solid tumors. Multiple NFPs can penetrate tumor tissue and subsequently transform into larger interfibril networks via in situ activation by tumor-associated proteases, thus minimizing lymphatic clearance. When used for drug delivery, NFP prolongs the drug–tumor contact time to achieve more effective treatment. (b) A table showing the peptide composition and surface charge (zeta potential) of the NFP analogues. The peptide derivatives were used to coassemble the nanofibers. (c) Synthesis of a dual Cyanine5.5- and 89Zr-labeled GSH-NFP (89Zr-NFP) for studying the biodistribution by fluorescence/PET/CT imaging. The ratio of different peptide constructs used for assembling 89Zr-NFP. Deferoxamine (DFO) served as the chelator of 89Zr. (d) Molecular structure of DFO. (e) TEM analysis of GSH-NFP after DFO-89Zr conjugation. Representative PET/CT whole body images of SCID mice bearing MDA-MB-468 tumors were acquired 2 and 7 days after injection of 100 μCi of 89Zr-NFP or free 89Zr-oxylate as the control (n = 3 per group). Representative fluorescence whole body images of SCID mice bearing MDA-MB-468 tumors (n = 4) 2 and 7 days after IV injection of 89Zr-NFP (100 μCi). Modified with permission from ref. 65. Copyright 2018, Wiley-VCH GmbH. | ||
3. Conclusion and outlook
In this review, we summarized the recent progress on smart self-assembled nanofibers for tumor/disease imaging with various modalities, including magnetic resonance imaging (MRI), optical imaging (OI), photoacoustic imaging (PAI), and multi-modality imaging. These nanofibers were smartly formed through in situ self-assembly of the peptide building blocks, which were activated by tumor/disease-related pathological stimuli of interest (e.g., enzyme). As such, imaging agents loaded on nanofibers were able to accumulate in the stimuli-rich regions with improved tumor/disease accessibility, retention, and metabolism difference, thereby enabling smart, precise, and sensitive tumor/disease imaging both in vitro and in vivo.Nevertheless, some challenges remain to be addressed in this field. First, given that the level and activity of stimuli may differ across types and species ascribed to tumor/disease heterogeneity, therefore highly specific and robust pathological stimuli of tumor/disease of interest still need to be explored for programming more smart nanofibers with reliable responsiveness. Second, nanofibers may transform their morphologies in complicated in vivo conditions due to their dynamic and reversible nature,9,10 thus impairing their properties. As a result, systematical insights into their behaviors in physiological context should be taken to correlate them with their building blocks, which would help establish new design principles of smart nanofiber precursors. More importantly, long-term in vivo pharmacokinetics and biosafety of nanofibers should be strictly evaluated and addressed to promote their clinical translation. Inspiringly, self-assembled peptide nanofibers also show great potential in theranostics89 and therapeutics64,90 of tumor/disease. With this review, we anticipate efforts from multidisciplinary researchers could be devoted to advance the development and clinical translation of smart self-assembled nanofibers for precise and enhanced tumor/disease diagnosis and therapy.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grants 21725505 and 22074016), and the China Postdoctoral Science Foundation funded project (2020M671303).References
- J. Wang, K. Liu, R. Xing and X. Yan, Chem. Soc. Rev., 2016, 45, 5589–5604 RSC.
- Y. J. Chung, K. Kim, B. I. Lee and C. B. Park, Small, 2017, 13, 1700983 CrossRef.
- M. Kaplan, P. Subramanian, D. Ghosal, C. M. Oikonomou, S. Pirbadian, R. Starwalt-Lee, S. K. Mageswaran, D. R. Ortega, J. A. Gralnick, M. Y. El-Naggar and G. J. Jensen, EMBO J., 2019, 38, e100957 CrossRef.
- R. Kubota, S. Liu, H. Shigemitsu, K. Nakamura, W. Tanaka, M. Ikeda and I. Hamachi, Bioconjugate Chem., 2018, 29, 2058–2067 CrossRef CAS PubMed.
- J. Li and K. Pu, Chem. Soc. Rev., 2019, 48, 38–71 RSC.
- Y. Deng, W. Zhan and G. Liang, Adv. Healthcare Mater., 2021, 10, e2001211 CrossRef.
- Q. Miao and K. Pu, Adv. Mater., 2018, 30, e1801778 CrossRef.
- W. Zhang, X. Yu, Y. Li, Z. Su, K. D. Jandt and G. Wei, Prog. Polym. Sci., 2018, 80, 94–124 CrossRef CAS.
- J. Song, R. Xing, T. Jiao, Q. Peng, C. Yuan, H. Mohwald and X. Yan, ACS Appl. Mater. Interfaces, 2018, 10, 2368–2376 CrossRef CAS.
- V. Castelletto, J. Seitsonen, K. M. Tewari, A. Hasan, R. M. Edkins, J. Ruokolainen, L. M. Pandey, I. W. Hamley and K. H. A. Lau, ACS Macro Lett., 2020, 9, 494–499 CrossRef CAS PubMed.
- D. B. Cheng, X. H. Zhang, Y. J. Gao, L. Ji, D. Hou, Z. Wang, W. Xu, Z. Y. Qiao and H. Wang, J. Am. Chem. Soc., 2019, 141, 7235–7239 CrossRef CAS.
- D. Zhang, G. B. Qi, Y. X. Zhao, S. L. Qiao, C. Yang and H. Wang, Adv. Mater., 2015, 27, 6125–6130 CrossRef CAS PubMed.
- A. Wagh, J. Singh, S. Qian and B. Law, Nanomedicine, 2013, 9, 449–457 CrossRef CAS PubMed.
- Y. Kuang and B. Xu, Angew. Chem., Int. Ed., 2013, 52, 6944–6948 CrossRef CAS PubMed.
- Y. Kuang, J. Shi, J. Li, D. Yuan, K. A. Alberti, Q. Xu and B. Xu, Angew. Chem., Int. Ed., 2014, 53, 8104–8107 CrossRef CAS.
- X. Liu, X. Sun and G. Liang, Biomater. Sci., 2021, 9, 315–327 RSC.
- F. Zhang, Y. Ma, Y. Chi, H. Yu, Y. Li, T. Jiang, X. Wei and J. Shi, Sci. Rep., 2018, 8, 8208 CrossRef PubMed.
- B. J. Kim and B. Xu, Bioconjugate Chem., 2020, 31, 492–500 CrossRef CAS PubMed.
- P. Wang, Y. Fan, L. Lu, L. Liu, L. Fan, M. Zhao, Y. Xie, C. Xu and F. Zhang, Nat. Commun., 2018, 9, 2898 CrossRef.
- Y. Li, G. Liu, J. Ma, J. Lin, H. Lin, G. Su, D. Chen, S. Ye, X. Chen, X. Zhu and Z. Hou, J. Controlled Release, 2017, 258, 95–107 CrossRef CAS.
- C. F. Anderson and H. Cui, Ind. Eng. Chem. Res., 2017, 56, 5761–5777 CrossRef CAS.
- X. Hu, F. Li, S. Wang, F. Xia and D. Ling, Adv. Healthcare Mater., 2018, 7, e1800359 CrossRef.
- Z. Zhou, Z. Han and Z. R. Lu, Biomaterials, 2016, 85, 168–179 CrossRef CAS.
- Z. Cai, C. Wu, L. Yang, D. Wang and H. Ai, ACS Biomater. Sci. Eng., 2020, 6, 2533–2542 CrossRef CAS.
- C. Diaferia, E. Gianolio and A. Accardo, J. Pept. Sci., 2019, 25, e3157 CrossRef.
- Z. Hai and G. Liang, Adv. Biosyst., 2018, 2, 1800108 CrossRef.
- N. J. W. Penfold, J. Yeow, C. Boyer and S. P. Armes, ACS Macro Lett., 2019, 8, 1029–1054 CrossRef CAS.
- S. Long, Q. Qiao, L. Miao and Z. Xu, Chin. Chem. Lett., 2019, 30, 573–576 CrossRef CAS.
- L. Wang, C. Gong, X. Yuan and G. Wei, Nanomaterials, 2019, 9, 285 CrossRef CAS PubMed.
- L. J. Chen and H. B. Yang, Acc. Chem. Res., 2018, 51, 2699–2710 CrossRef CAS PubMed.
- S. Zhang, T. Holmes, C. Lockshin and A. Rich, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 3334–3338 CrossRef CAS PubMed.
- T. C. Holmes, S. d. Lacelle, X. Su, G. S. Liu, A. Rich and S. G. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 6728–6733 CrossRef CAS.
- F. Gelain, Z. Luo and S. Zhang, Chem. Rev., 2020, 120, 13434–13460 CrossRef CAS.
- Y. Zhao, W. Yang, C. Chen, J. Wang, L. Zhang and H. Xu, Curr. Opin. Colloid Interface Sci., 2018, 35, 112–123 CrossRef CAS.
- M. Reches and E. Gazit, Science, 2003, 300, 625–627 CrossRef CAS PubMed.
- A. Lampel, R. V. Ulijn and T. Tuttle, Chem. Soc. Rev., 2018, 47, 3737–3758 RSC.
- K. Tao, A. Levin, L. Adler-Abramovich and E. Gazit, Chem. Soc. Rev., 2016, 45, 3935–3953 RSC.
- Y. Zhang, Y. Kuang, Y. Gao and B. Xu, Langmuir, 2011, 27, 529–537 CrossRef CAS PubMed.
- J. Huang, J. Li, Y. Lyu, Q. Miao and K. Pu, Nat. Mater., 2019, 18, 1133–1143 CrossRef CAS PubMed.
- Y. Zhang, S. He, W. Chen, Y. Liu, X. Zhang, Q. Miao and K. Pu, Angew. Chem., Int. Ed., 2021, 60, 5921–5927 CrossRef CAS PubMed.
- S. He, J. Li, Y. Lyu, J. Huang and K. Pu, J. Am. Chem. Soc., 2020, 142, 7075–7082 CrossRef CAS.
- P. Cheng, W. Chen, S. Li, S. He, Q. Miao and K. Pu, Adv. Mater., 2020, 32, e1908530 CrossRef PubMed.
- C. Zhang, Z. Zeng, D. Cui, S. He, Y. Jiang, J. Li, J. Huang and K. Pu, Nat. Commun., 2021, 12, 2934 CrossRef CAS PubMed.
- Y. Xue, H. Bai, B. Peng, B. Fang, J. Baell, L. Li, W. Huang and N. H. Voelcker, Chem. Soc. Rev., 2021, 50, 4872–4931 RSC.
- J. Gao, J. Zhan and Z. Yang, Adv. Mater., 2020, 32, e1805798 CrossRef PubMed.
- P. Padmanabhan, A. Kumar, S. Kumar, R. K. Chaudhary and B. Gulyas, Acta Biomater., 2016, 41, 1–16 CrossRef CAS.
- A. Babic, V. Vorobiev, G. Trefalt, L. A. Crowe, L. Helm, J. P. Vallee and E. Allemann, Chem. Commun., 2019, 55, 945–948 RSC.
- J. Elistratova, B. Akhmadeev, V. Korenev, M. Sokolov, I. Nizameev, A. Gubaidullin, A. Voloshina and A. Mustafina, Soft Matter, 2018, 14, 7916–7925 RSC.
- P. Xie, P. Du, J. Li and P. Liu, Carbohydr. Polym., 2019, 205, 377–384 CrossRef CAS.
- Y. Yuan, J. Zhang, X. Qi, S. Li, G. Liu, S. Siddhanta, I. Barman, X. Song, M. T. McMahon and J. W. M. Bulte, Nat. Mater., 2019, 18, 1376–1383 CrossRef CAS.
- R. An, X. Cheng, S. Wei, Y. Hu, Y. Sun, Z. Huang, H. Y. Chen and D. Ye, Angew. Chem., Int. Ed., 2020, 59, 20636–20644 CrossRef CAS PubMed.
- J. Zhang, Y. L. Mu, Z. Y. Ma, K. Han and H. Y. Han, Biomaterials, 2018, 182, 269–278 CrossRef CAS PubMed.
- L. Zhang, D. Jing, N. Jiang, T. Rojalin, C. M. Baehr, D. Zhang, W. Xiao, Y. Wu, Z. Cong, J. J. Li, Y. Li, L. Wang and K. S. Lam, Nat. Nanotechnol., 2020, 15, 145–153 CrossRef CAS PubMed.
- C. Diaferia, E. Gianolio, P. Palladino, F. Arena, C. Boffa, G. Morelli and A. Accardo, Adv. Funct. Mater., 2015, 25, 7003–7016 CrossRef CAS.
- C. Diaferia, E. Gianolio, T. Sibillano, F. A. Mercurio, M. Leone, C. Giannini, N. Balasco, L. Vitagliano, G. Morelli and A. Accardo, Sci. Rep., 2017, 7, 307 CrossRef PubMed.
- J. Zhou, X. Du, J. Li, N. Yamagata and B. Xu, J. Am. Chem. Soc., 2015, 137, 10040–10043 CrossRef CAS PubMed.
- L. Dong, Q. Miao, Z. Hai, Y. Yuan and G. Liang, Anal. Chem., 2015, 87, 6475–6478 CrossRef CAS PubMed.
- H. He, J. Wang, H. Wang, N. Zhou, D. Yang, D. R. Green and B. Xu, J. Am. Chem. Soc., 2018, 140, 1215–1218 CrossRef CAS PubMed.
- Y. Zhong, J. Zhan, G. Xu, Y. Chen, Q. Qin, X. Liao, S. Ma, Z. Yang and Y. Cai, Angew. Chem., Int. Ed., 2021, 60, 8121–8129 CrossRef CAS PubMed.
- L. Yang, R. Peltier, M. Zhang, D. Song, H. Huang, G. Chen, Y. Chen, F. Zhou, Q. Hao, L. Bian, M. L. He, Z. Wang, Y. Hu and H. Sun, J. Am. Chem. Soc., 2020, 142, 18150–18159 CrossRef CAS PubMed.
- L. Dong, J. Qian, Z. Hai, J. Xu, W. Du, K. Zhong and G. Liang, Anal. Chem., 2017, 89, 6922–6925 CrossRef CAS PubMed.
- X. X. Zhao, L. L. Li, Y. Zhao, H. W. An, Q. Cai, J. Y. Lang, X. X. Han, B. Peng, Y. Fei, H. Liu, H. Qin, G. Nie and H. Wang, Angew. Chem., Int. Ed., 2019, 58, 15287–15294 CrossRef CAS PubMed.
- H. W. An, D. Hou, R. Zheng, M. D. Wang, X. Z. Zeng, W. Y. Xiao, T. D. Yan, J. Q. Wang, C. H. Zhao, L. M. Cheng, J. M. Zhang, L. Wang, Z. Q. Wang, H. Wang and W. Xu, ACS Nano, 2020, 14, 927–936 CrossRef CAS.
- R. Zheng, J. Yang, M. Mamuti, D. Y. Hou, H. W. An, Y. Zhao and H. Wang, Angew. Chem., Int. Ed., 2021, 60, 7809–7819 CrossRef CAS.
- V. Bellat, R. Ting, T. L. Southard, L. Vahdat, H. Molina, J. Fernandez, O. Aras, T. Stokol and B. Law, Adv. Funct. Mater., 2018, 28, 1803969 CrossRef.
- Q. Cai, Y. Fei, L. Hu, Z. Huang, L. L. Li and H. Wang, Nano Lett., 2018, 18, 6229–6236 CrossRef CAS PubMed.
- K. M. Ward, A. H. Aletras and R. S. Balaban, J. Magn. Reson., 2000, 143, 79–87 CrossRef CAS PubMed.
- G. Liu, X. Song, K. W. Chan and M. T. McMahon, NMR Biomed., 2013, 26, 810–828 CrossRef CAS.
- L. L. Lock, Y. Li, X. Mao, H. Chen, V. Staedtke, R. Bai, W. Ma, R. Lin, Y. Li, G. Liu and H. Cui, ACS Nano, 2017, 11, 797–805 CrossRef CAS.
- I. Tirotta, V. Dichiarante, C. Pigliacelli, G. Cavallo, G. Terraneo, F. B. Bombelli, P. Metrangolo and G. Resnati, Chem. Rev., 2015, 115, 1106–1129 CrossRef CAS PubMed.
- X. Tang, X. Gong, A. Li, H. Lin, C. Peng, X. Zhang, X. Chen and J. Gao, Nano Lett., 2020, 20, 363–371 CrossRef CAS PubMed.
- Z. Zheng, H. Sun, C. Hu, G. Li, X. Liu, P. Chen, Y. Cui, J. Liu, J. Wang and G. Liang, Anal. Chem., 2016, 88, 3363–3368 CrossRef CAS PubMed.
- J. Song, X. Yang, Z. Yang, L. Lin, Y. Liu, Z. Zhou, Z. Shen, G. Yu, Y. Dai, O. Jacobson, J. Munasinghe, B. Yung, G. J. Teng and X. Chen, ACS Nano, 2017, 11, 6102–6113 CrossRef CAS PubMed.
- G. D. Luker and K. E. Luker, J. Nucl. Med., 2008, 49, 1–4 CrossRef.
- M. Zhang, C. Wang, C. Yang, H. Wu, H. Xu and G. Liang, Anal. Chem., 2021, 93, 5665–5669 CrossRef CAS.
- Y. Qian, W. Wang, Z. Wang, X. Jia, Q. Han, I. Rostami, Y. Wang and Z. Hu, ACS Appl. Mater. Interfaces, 2018, 10, 7871–7881 CrossRef CAS.
- A. Han, H. Wang, R. T. Kwok, S. Ji, J. Li, D. Kong, B. Z. Tang, B. Liu, Z. Yang and D. Ding, Anal. Chem., 2016, 88, 3872–3878 CrossRef CAS PubMed.
- Kenry, Y. Duan and B. Liu, Adv. Mater., 2018, 30, e1802394 CrossRef CAS PubMed.
- Y. Hu, Y. Wang, X. Wen, Y. Pan, X. Cheng, R. An, G. Gao, H. Y. Chen and D. Ye, Research, 2020, 2020, 4087069 CrossRef CAS PubMed.
- P. Beard, Interface Focus, 2011, 1, 602–631 CrossRef PubMed.
- S. R. Kothapalli, T. J. Ma, S. Vaithilingam, O. Oralkan, B. T. Khuri-Yakub and S. S. Gambhir, IEEE Trans. Biomed. Eng., 2012, 59, 1199–1204 Search PubMed.
- A. Dragulescu-Andrasi, S. R. Kothapalli, G. A. Tikhomirov, J. Rao and S. S. Gambhir, J. Am. Chem. Soc., 2013, 135, 11015–11022 CrossRef CAS PubMed.
- C. Xie, X. Zhen, Q. Lei, R. Ni and K. Pu, Adv. Funct. Mater., 2017, 27, 1605397 CrossRef.
- C. Wu, R. Zhang, W. Du, L. Cheng and G. Liang, Nano Lett., 2018, 18, 7749–7754 CrossRef CAS PubMed.
- B. Hu, C. Owh, P. L. Chee, W. R. Leow, X. Liu, Y. L. Wu, P. Guo, X. J. Loh and X. Chen, Chem. Soc. Rev., 2018, 47, 6917–6929 RSC.
- W. Mao, H. S. Kim, Y. J. Son, S. R. Kim and H. S. Yoo, J. Controlled Release, 2018, 269, 52–62 CrossRef CAS PubMed.
- X. R. Song, S. H. Li, J. Dai, L. Song, G. Huang, R. Lin, J. Li, G. Liu and H. H. Yang, Small, 2017, 13, 1603997 CrossRef PubMed.
- R. Yan, Y. Hu, F. Liu, S. Wei, D. Fang, A. J. Shuhendler, H. Liu, H. Y. Chen and D. Ye, J. Am. Chem. Soc., 2019, 141, 10331–10341 CrossRef CAS PubMed.
- N. Liu, L. Zhu, Z. Li, W. Liu, M. Sun and Z. Zhou, Biomater. Sci., 2021, 9, 5427–5436 RSC.
- E. Arslan, I. C. Garip, G. Gulseren, A. B. Tekinay and M. O. Guler, Adv. Healthcare Mater., 2014, 3, 1357–1376 CrossRef CAS.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |
