Open Access Article
Open Access ArticleBroad-spectrum nanoparticles against bacteriophage infections†
Łukasz
Richter
a,
Karolina
Paszkowska
b,
Urszula
Cendrowska
a,
Francesca
Olgiati
a,
Paulo Jacob
Silva
a,
Matteo
Gasbarri
 a,
Zekiye Pelin
Guven
a,
Jan
Paczesny
b and
Francesco
Stellacci
a,
Zekiye Pelin
Guven
a,
Jan
Paczesny
b and
Francesco
Stellacci
 *ac
*ac
aInstitute of Materials, École Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland. E-mail: francesco.stellacci@epfl.ch
bInstitute of Physical Chemistry of the Polish Academy of Sciences, Kasprzaka 44/52, 01-224 Warsaw, Poland
cInstitute of Bioengineering, École Polytechnique Fédérale de Lausanne, 1015 Lausanne, Switzerland
First published on 26th October 2021
Abstract
Viral infections caused by bacteriophages, i.e., viruses that kill bacteria are one of the most dangerous and common threats for bacteria-based bioreactors. More than 70% of biotechnology companies have admitted to encountering this problem. Despite phage infections being such a dangerous and widespread risk, there are no effective methods to avoid them to date. Herein, we present a novel technology based on nanoparticles that irreversibly deactivates bacteriophages and is safe for bacteria. Our method allows for the unsupervised protection of bacterial processes in the biotechnology industry. Gold nanoparticles coated with a mixture of negatively charged 11-mercapto 1-undecanesulfonic acid (MUS) and hydrophobic 1-octanethiol (OT) ligands are effective at deactivating various types of Escherichia coli-selective phages: T1, T4, and T7. The nanoparticles can lower the titer of phages up to 2 and 5 logs in 6 and 24 h at 50 °C, respectively. A comparative analysis of nanoparticles with different ligand shells illustrates the importance of the combination of negatively charged and hydrophobic ligands that is the key to achieving a good inhibitory concentration (EC50 ≤ 1 μg mL−1) for all tested phages. We show that the nanoparticles are harmless for the commonly used bacteria in industry Escherichia coli and are effective under conditions simulating the environment of bioreactors.
Introduction
Bacteria-based processes are some of the most important in biotechnology and dominate a number of branches of food and agriculture industries. A great number of biotechnology companies exploit the natural metabolic properties of bacteria to produce drugs, vaccines, antibiotics, probiotics, insecticides, dairy products, enzymes, organic acids, fuels, and solvents.1 Events that lead to the closure of bacteria-based factories result in a large economic burden. One of the most dangerous and common factors is infections caused by bacteriophages, i.e., viruses attacking and killing bacteria. In the dairy industry, 1% to 10% of batches of products are lost to phages.1 More than 70% of biotechnology companies have admitted to encountering this problem.1Even a few virions (viral particles) might contaminate a whole industrial production vessel and produce up to 1013 progeny phages per mL within a few hours.2 Hundreds of new virions are released from a single infected bacterium within tens of minutes, thus the fight against phage-based contaminations is extremely difficult. Moreover, some phages can survive high temperatures (e.g., boiling), pasteurization, organic solvents, drying, and even vacuum.3
Despite phage infections being such a dangerous and widespread risk, to date, there are no effective methods to avoid and fight them. Although some approaches have been proposed, they are limited by being only preventive (rotation of bacterial strains4), not very effective (thermal,5 UV radiation4), unsuitable for large volumes (filtration6–8), expensive and overly specific (engineered proteins,9–11 phage-resistant mutants12) or harmful for bacteria (biocides5,13).
Nanoparticles have a constantly growing number of applications in biological systems and they were also used to deactivate phages.14–19 Approaches based on silver,20–24 copper oxide,22,24,25 titanium dioxide26,27 and iron–nickel28 nanoparticles have been proven to be effective in phage deactivation. However, all of them share one of the two possible mechanisms of action: production of toxic silver or copper ions20–22 or generation of reactive oxygen species (ROS).26,27 Both mechanisms are also effective in the deactivation of bacteria29 and therefore cannot be used to selectively protect bacteria from phages in the biotechnology industry. Only nanoparticles effective in phage deactivation and at the same time harmless for bacteria can be used in such applications.
Currently, the most commonly applied solution to a bacteriophage infection is shutting down the reactor and thoroughly disinfecting it, which is an operation that takes days and comes with a significant economic burden. The ideal solution should provide effective eradication of phages before infection of the first bacteria. Such an approach would prevent the most dangerous and difficult scenario: a massive outbreak that leads to large volumes with high concentrations of phages.
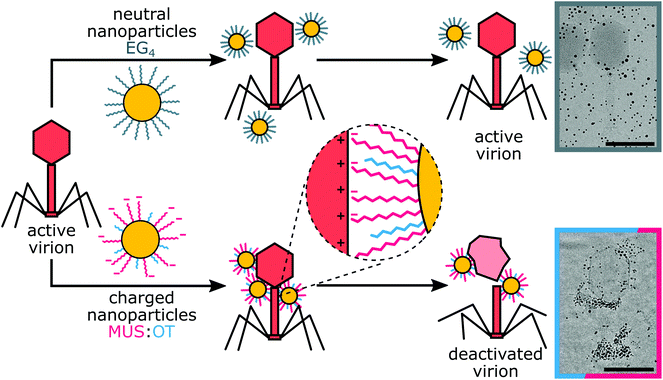
Here we present anti-phage gold nanoparticles that can deactivate bacteriophages and at the same time remain harmless to the bacteria. Negatively charged nanoparticles coated with a mixture of 11-mercapto 1-undecanesulfonic acid (MUS) and hydrophobic 1-octanethiol (OT) first attach to phages through electrostatic forces, and then virions are deactivated by irreversible local distortions caused by hydrophobic interactions (Fig. 1). MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT nanoparticles are effective in conditions similar to the industrial bioreactors and can prevent the infection after just 1 h of the preincubation step.
OT nanoparticles are effective in conditions similar to the industrial bioreactors and can prevent the infection after just 1 h of the preincubation step.
Results and discussion
Our approach to deactivate phages is based on gold nanoparticles coated by a mixture of negatively charged MUS and hydrophobic OT ligands. MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT nanoparticles were already proven to be broad-spectrum antiviral agents against eukaryotic viruses.30 We studied nanoparticles with final ratios of MUS to OT ligands: 70
OT nanoparticles were already proven to be broad-spectrum antiviral agents against eukaryotic viruses.30 We studied nanoparticles with final ratios of MUS to OT ligands: 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 and 85
30 and 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 (Fig. 2a and b). Final ratios of ligands were calculated from NMR, as provided in the ESI.† Nanoparticles were designed to provide deactivation by initial electrostatic attraction followed by hydrophobic interactions causing local irreversible distortions. Long and flexible ligands provide multivalent contact between charges of the nanoparticles and bacteriophage surface. Additionally, we examined positively charged nanoparticles, coated with TMA (Fig. 2c). As a control, non-charged particles with tetra(ethylene glycol) (EG4) ligands were also tested (Fig. 2d). We kept a similar size of particles and length of ligands to directly compare the influence of different nanoparticles on phage virions (Table S1†).
15 (Fig. 2a and b). Final ratios of ligands were calculated from NMR, as provided in the ESI.† Nanoparticles were designed to provide deactivation by initial electrostatic attraction followed by hydrophobic interactions causing local irreversible distortions. Long and flexible ligands provide multivalent contact between charges of the nanoparticles and bacteriophage surface. Additionally, we examined positively charged nanoparticles, coated with TMA (Fig. 2c). As a control, non-charged particles with tetra(ethylene glycol) (EG4) ligands were also tested (Fig. 2d). We kept a similar size of particles and length of ligands to directly compare the influence of different nanoparticles on phage virions (Table S1†).
Influence of nanoparticles on phages and bacteria
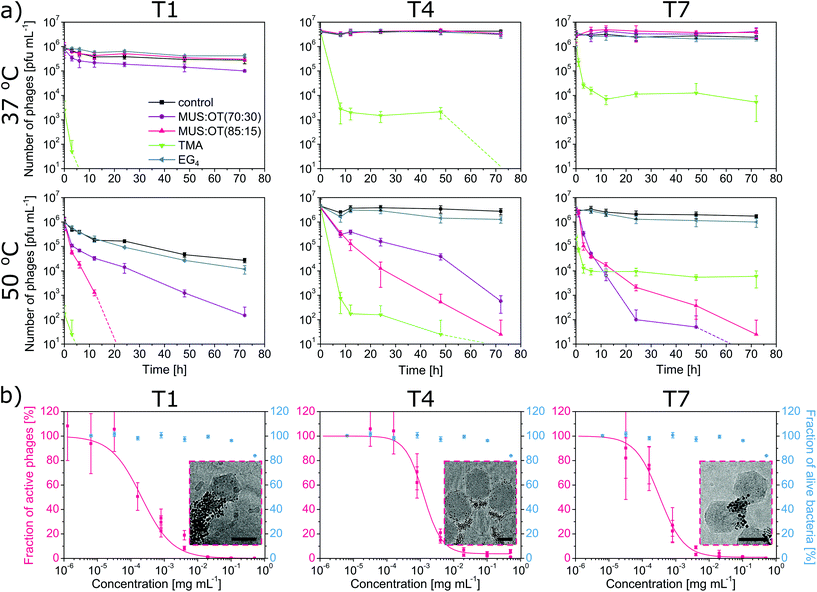
Escherichia coli is not only one of the most commonly used bacteria in the biotechnology industry, but it is also the best-studied model bacterium.31,32 It is the most common bacterial species used for the industrial production of biopharmaceuticals and it is also the second most common system to be used for such a purpose in general.33 Thus, we aimed to effectively deactivate E. coli-specific phages representing three different families: T1 type from Siphoviridae (T1-like phages are causing the majority of E. coli fermentation failures34), T4 type from Myoviridae (one of the best known and well-studied phages35) and T7 type from Podoviridae. All of them share a head-tail structure that is characteristic for more than 95% of all known bacteriophages,36 thus the obtained results can be applied to other industrial bacteria-based systems.As a first test, we incubated bacteriophages with 0.5 mg mL−1 of nanoparticles in TM buffer over three days and analysed the changes in the number of active phages in time (Fig. 3a). We applied two temperatures: 37 °C (optimal for bacteria growth) and 50 °C, a slightly increased temperature that is still accepted in many industrial procedures. Since phage titer measurements include serial dilution steps, all observed deactivation effects are irreversible (i.e., virucidal).
The most efficient deactivation of phages at both temperatures was obtained for TMA nanoparticles. Drops of active phages: 2 logs for T7, 3 logs for T4 and up to 6 logs for T1, were visible already after the first few hours (Fig. 3). Interestingly, an increase in the temperature from 37 °C to 50 °C did not change the magnitude of the observed effects. The strongest deactivation was obtained for T1, where no active virions were visible after 6 hours of incubation. This is in line with the literature since cationic compounds were previously reported to be effective in phage deactivation.37 However, TMA nanoparticles turned out to be unstable in LB medium and toxic against bacteria E. coli (Fig. S3†),38 thus, they were excluded from further tests.
Both studied MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT nanoparticles were effective against all tested types of phages (Fig. 3). At 37 °C no effect was observed, and a higher temperature was required to trigger the deactivation. At 50 °C MUS
OT nanoparticles were effective against all tested types of phages (Fig. 3). At 37 °C no effect was observed, and a higher temperature was required to trigger the deactivation. At 50 °C MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT nanoparticles decreased the titer of phages by up to two logs in 6 h. After 24 h exposure, the decrease was up to 5 logs. To determine the lowest amount of nanoparticles required for effective deactivation, we performed dose–response tests for the best performing nanoparticles (MUS
OT nanoparticles decreased the titer of phages by up to two logs in 6 h. After 24 h exposure, the decrease was up to 5 logs. To determine the lowest amount of nanoparticles required for effective deactivation, we performed dose–response tests for the best performing nanoparticles (MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT, 85
OT, 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
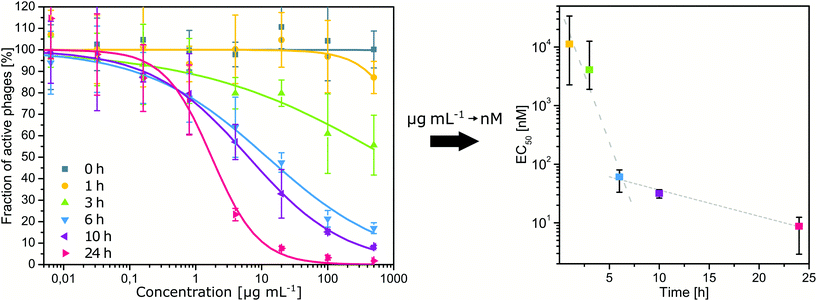
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) against all three types of phages (Fig. 3b). Bacteriophages were incubated with various concentrations of nanoparticles for 24 h, and then the activity of the phages was evaluated. The obtained EC50 values (i.e., effective concentration deactivating 50% of phages) for MUS
15) against all three types of phages (Fig. 3b). Bacteriophages were incubated with various concentrations of nanoparticles for 24 h, and then the activity of the phages was evaluated. The obtained EC50 values (i.e., effective concentration deactivating 50% of phages) for MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT (85
OT (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) nanoparticles were in a similar range of 0.2 to 1.2 μg mL−1 for all types of bacteriophages. Since EC50 depends on the time of the exposition, we also checked the EC50 of MUS
15) nanoparticles were in a similar range of 0.2 to 1.2 μg mL−1 for all types of bacteriophages. Since EC50 depends on the time of the exposition, we also checked the EC50 of MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT (85
OT (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) nanoparticles against phage T4 for various times of incubation (Fig. 4). 6 h of incubation was enough to obtain an EC50 of 12.1 μg mL−1 (i.e., 80 nM) and extending the time to 24 h resulted in an EC50 as low as 1.7 μg mL−1 (i.e., 11 nM). Finally, we should add that despite being effective against phages T1, T4, and T7, MUS
15) nanoparticles against phage T4 for various times of incubation (Fig. 4). 6 h of incubation was enough to obtain an EC50 of 12.1 μg mL−1 (i.e., 80 nM) and extending the time to 24 h resulted in an EC50 as low as 1.7 μg mL−1 (i.e., 11 nM). Finally, we should add that despite being effective against phages T1, T4, and T7, MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT nanoparticles did not interact with other types of phages – spherical phages MS2 that lack the complex head-tail structure (Fig. S4†).
OT nanoparticles did not interact with other types of phages – spherical phages MS2 that lack the complex head-tail structure (Fig. S4†).
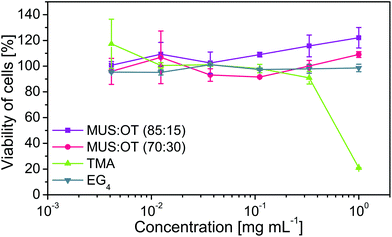
To check the importance of the electrostatic interactions, we used nanoparticles coated with ligands that did not bear a charged group. We could not use non-charged nanoparticles covered only with aliphatic chains since they are not soluble in water. Thus, we used nanoparticles covered with tetra(ethylene glycol) chains, which are also not charged and are soluble in the water environment. Non-charged EG4 nanoparticles showed no effect in all analysed cases, confirming the importance of the electrostatic forces.
To evaluate the significance of the hydrophobic interactions, we tested nanoparticles coated only with MUS ligands (Fig. S5†). Lack of OT ligands decreased the hydrophobicity of MUS nanoparticles. As a result, MUS nanoparticles did not deactivate any of the three types of phages (Fig. S6†) either at 37 °C or 50 °C. These experiments show that the addition of hydrophobic interactions provided by OT ligands is crucial for effective deactivation.
The deactivation test was complemented with toxicity studies against bacteria E. coli BL21. We found that MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT (85
OT (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) nanoparticles were inert for tested bacteria, and no influence on their viability was visible in the tested range (EC50 > 500 mg mL−1), with a small influence observed only at the highest concentration (Fig. 3b and Fig. S7†). Therefore, we showed that MUS
15) nanoparticles were inert for tested bacteria, and no influence on their viability was visible in the tested range (EC50 > 500 mg mL−1), with a small influence observed only at the highest concentration (Fig. 3b and Fig. S7†). Therefore, we showed that MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT nanoparticles have two properties crucial for the protection of the bacteria-based bioreactors: effective at bacteriophage deactivation and at the same time remain harmless for bacteria.
OT nanoparticles have two properties crucial for the protection of the bacteria-based bioreactors: effective at bacteriophage deactivation and at the same time remain harmless for bacteria.
Analysis of the mechanism of action
To better understand interactions between phages and nanoparticles, we focused on the model pair: phage T4 (well-known and studied phage) and best-performing nanoparticles MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT (85
OT (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15). Bacteriophage T4 has a head-tail structure that is a characteristic of more than 95% of all known bacteriophages,36 and consists of a 115 nm-long, 85 nm wide head, and a 100 nm-long, 21-diameter contractile tail terminated with a 46 nm-diameter baseplate and six 145 nm-long fibres attached to the baseplate (Fig. 5a). The mature virus is composed of over forty different types of structural proteins, among which eighteen are exposed to the external environment.35,39 The majority of these structural proteins are negatively charged in pH around 7 (i.e., their pI < 7), with the exception of the ends of fibres, in which pI is higher than 7 (Fig. 5a). Such distribution of charges of phage virions was developed in the process of evolution. Positively charged fibres are electrostatically attracted to bacteria, which have a negatively charged surface. Detailed information about all analysed proteins is provided in Table S2.†
15). Bacteriophage T4 has a head-tail structure that is a characteristic of more than 95% of all known bacteriophages,36 and consists of a 115 nm-long, 85 nm wide head, and a 100 nm-long, 21-diameter contractile tail terminated with a 46 nm-diameter baseplate and six 145 nm-long fibres attached to the baseplate (Fig. 5a). The mature virus is composed of over forty different types of structural proteins, among which eighteen are exposed to the external environment.35,39 The majority of these structural proteins are negatively charged in pH around 7 (i.e., their pI < 7), with the exception of the ends of fibres, in which pI is higher than 7 (Fig. 5a). Such distribution of charges of phage virions was developed in the process of evolution. Positively charged fibres are electrostatically attracted to bacteria, which have a negatively charged surface. Detailed information about all analysed proteins is provided in Table S2.†
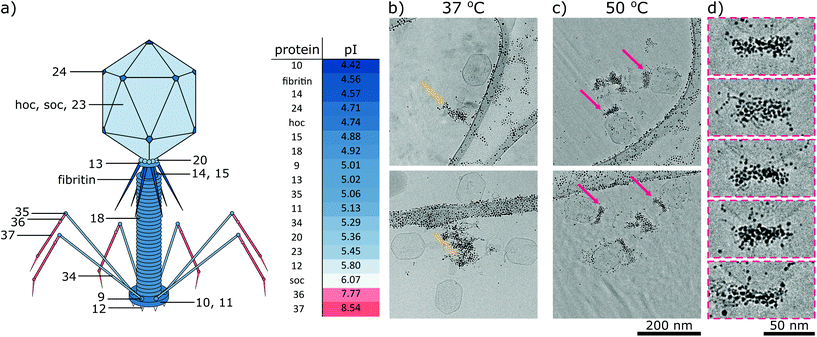 | ||
Fig. 5 (a) Structure of bacteriophage T4. Red or blue colors represent a charge at pH 7.4 that is positive or negative, respectively. The table presents the isoelectric point (pI) of structural proteins. Panels on the right: cryo-TEM analysis of the interaction between T4 phages and MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT (85 OT (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) nanoparticles at various temperatures. T4 bacteriophages were incubated with 0.1 mg mL−1 of nanoparticles at (b) 37 °C and (c) 50 °C for 24 h. Panel (d) presents magnified parts of cryo-TEM images from panel (c). At lower temperatures, negatively charged nanoparticles interacted mainly with positively charged fibers of bacteriophages (yellow arrows). Unexpectedly, at higher temperatures, nanoparticles decorated collars of T4 phages (magenta arrows). As we have shown in Fig. 3, the latter type of interaction caused the deactivation of phages. 15) nanoparticles at various temperatures. T4 bacteriophages were incubated with 0.1 mg mL−1 of nanoparticles at (b) 37 °C and (c) 50 °C for 24 h. Panel (d) presents magnified parts of cryo-TEM images from panel (c). At lower temperatures, negatively charged nanoparticles interacted mainly with positively charged fibers of bacteriophages (yellow arrows). Unexpectedly, at higher temperatures, nanoparticles decorated collars of T4 phages (magenta arrows). As we have shown in Fig. 3, the latter type of interaction caused the deactivation of phages. | ||
We analysed the interactions of phages and nanoparticles using cryo-TEM imaging. At 37 °C MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
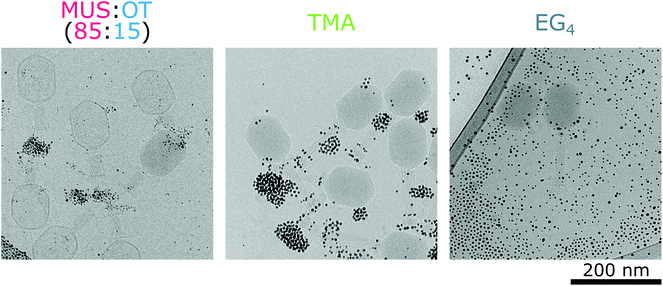
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT nanoparticles adsorbed mostly on the positively charged fibres of the bacteriophages (Fig. 5b). These interactions were already visible after 20 minutes of incubation (Fig. S8†). As shown in Fig. 3a, such adsorption does not cause effective deactivation of bacteriophages. In comparison, positively charged TMA nanoparticles attached under these conditions to negatively charged tails of the T4 phage virions, and neutral EG4 nanoparticles did not interact with T4 phages at all (Fig. 6). Thus, we confirmed that at 37 °C, the interactions between phages and nanoparticles could be predicted based on electrostatic forces between averaged charges of phage proteins (represented by their isoelectric point) and charges of nanoparticles.
OT nanoparticles adsorbed mostly on the positively charged fibres of the bacteriophages (Fig. 5b). These interactions were already visible after 20 minutes of incubation (Fig. S8†). As shown in Fig. 3a, such adsorption does not cause effective deactivation of bacteriophages. In comparison, positively charged TMA nanoparticles attached under these conditions to negatively charged tails of the T4 phage virions, and neutral EG4 nanoparticles did not interact with T4 phages at all (Fig. 6). Thus, we confirmed that at 37 °C, the interactions between phages and nanoparticles could be predicted based on electrostatic forces between averaged charges of phage proteins (represented by their isoelectric point) and charges of nanoparticles.
At a higher temperature of 50 °C we observed that MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT particles bind to the phages in other parts, i.e., collars and heads (Fig. 5c and Fig S9†). Nanoparticles attach mostly to short fibritin fibres, creating a gold “necklace” visible in cryo-TEM images. Attachment of gold nanoparticles to the neck part of bacteriophage T4 seems crucial for effective inhibition and does not match the simple prediction of electrostatic interactions based on overall averaged charges. Almost all proteins are composed of both positively and negatively charged amino acids (Table S2†), and thus have many positive and negative patches or cavities with which nanoparticles can interact.40 We should stress that these studies are qualitative in nature as in TEM one can never be sure that the imaged viruses were infecting before interacting with the nanoparticles.
OT particles bind to the phages in other parts, i.e., collars and heads (Fig. 5c and Fig S9†). Nanoparticles attach mostly to short fibritin fibres, creating a gold “necklace” visible in cryo-TEM images. Attachment of gold nanoparticles to the neck part of bacteriophage T4 seems crucial for effective inhibition and does not match the simple prediction of electrostatic interactions based on overall averaged charges. Almost all proteins are composed of both positively and negatively charged amino acids (Table S2†), and thus have many positive and negative patches or cavities with which nanoparticles can interact.40 We should stress that these studies are qualitative in nature as in TEM one can never be sure that the imaged viruses were infecting before interacting with the nanoparticles.
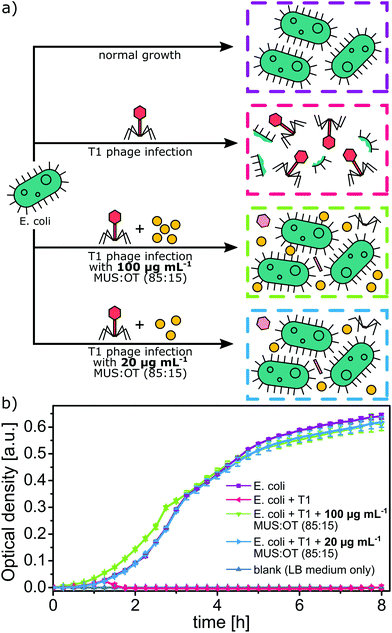
Realistic application test mimicking an infected bioreactor
We tested the best-performing nanoparticles (MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT, 85
OT, 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) in the real-life application of protection of bacteria inside a phage-infected bioreactor. We used phage T1, since T1-like phages are causing the majority of E. coli fermentation failures.34 First, we incubated 200 pfu mL−1 of T1 bacteriophages with MUS
15) in the real-life application of protection of bacteria inside a phage-infected bioreactor. We used phage T1, since T1-like phages are causing the majority of E. coli fermentation failures.34 First, we incubated 200 pfu mL−1 of T1 bacteriophages with MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT (85
OT (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) nanoparticles for a given amount of time at 50 °C to achieve initial deactivation. We tested two cases: (a) incubation with 20 μg mL−1 of nanoparticles for 12 h and (b) incubation with 100 μg mL−1 of nanoparticles for 1 h. Then an inoculum of bacteria E. coli BL21 was added to such a mixture, and bacteria were cultured at 37 °C in LB medium with mixing. As controls, samples with normally growing bacteria and samples contaminated with non-treated phages were prepared (Fig. 7a). In bacterial cultures infected with phages, the growth of bacteria stopped after 1.5 h, and all bacteria were infected and killed. However, in samples protected with anti-phage nanoparticles, no phage infection was visible, and bacterial culture grew as in control experiments (Fig. 7b). The 1 h preincubation step was enough to deactivate all phages and provide a protecting effect of nanoparticles. Thus, in practical application, after the addition of nanoparticles to the bioreactor, the only requirement would be to perform a short preincubation before starting the main bioreactor process.
15) nanoparticles for a given amount of time at 50 °C to achieve initial deactivation. We tested two cases: (a) incubation with 20 μg mL−1 of nanoparticles for 12 h and (b) incubation with 100 μg mL−1 of nanoparticles for 1 h. Then an inoculum of bacteria E. coli BL21 was added to such a mixture, and bacteria were cultured at 37 °C in LB medium with mixing. As controls, samples with normally growing bacteria and samples contaminated with non-treated phages were prepared (Fig. 7a). In bacterial cultures infected with phages, the growth of bacteria stopped after 1.5 h, and all bacteria were infected and killed. However, in samples protected with anti-phage nanoparticles, no phage infection was visible, and bacterial culture grew as in control experiments (Fig. 7b). The 1 h preincubation step was enough to deactivate all phages and provide a protecting effect of nanoparticles. Thus, in practical application, after the addition of nanoparticles to the bioreactor, the only requirement would be to perform a short preincubation before starting the main bioreactor process.
Cytotoxicity and practical applications
Utilized MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT nanoparticles were already proven to be non-toxic for eukaryotic cells in both in vitro and ex vivo tests.30 Nonetheless, to check the cytotoxicity of the nanoparticles used in this study, we performed MTS assay on Vero cells after 24 h of incubation with all four types of nanoparticles (Fig. 8). No cytotoxic effect of both MUS
OT nanoparticles were already proven to be non-toxic for eukaryotic cells in both in vitro and ex vivo tests.30 Nonetheless, to check the cytotoxicity of the nanoparticles used in this study, we performed MTS assay on Vero cells after 24 h of incubation with all four types of nanoparticles (Fig. 8). No cytotoxic effect of both MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT and PEG nanoparticles was visible up to a concentration 1000 μg mL−1. For comparison, EC50 of nanoparticles MUS
OT and PEG nanoparticles was visible up to a concentration 1000 μg mL−1. For comparison, EC50 of nanoparticles MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT (85
OT (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) against bacteriophages was in the range of 1 μg mL−1. Additionally, as expected, positively charged TMA nanoparticles were toxic against tested mammalian cells.
15) against bacteriophages was in the range of 1 μg mL−1. Additionally, as expected, positively charged TMA nanoparticles were toxic against tested mammalian cells.
Another important issue is the removal of the nanoparticles from the final product in downstream processes. Due to the difference in size between nanoparticles (∼3 nm) and bacteria (∼1 μm), nanoparticles can be easily separated by filtration on 0.45 μm filters or centrifugation.
Moreover, downstream processing of bioreactor-based production of biopharmaceuticals involves steps like cell lysis (mechanical or enzymatic), nucleic acid degradation, cell-wall material removal, filtration and final purification via chromatography (e.g., immobilized metal affinity chromatography).41 Such a great number of purification steps provide many possibilities for the effective removal of nanoparticles from the final product.
Finally, the mechanism of deactivation of phages with MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT nanoparticles is based on the interaction with ligands and is independent of the nanoparticle core. Therefore, the developed technology is a first step toward designing better antiphage agents for industrial applications. The idea presented in our study can be in the future transferred to other types of nanoparticles, with cheaper and more biocompatible cores, such as silica or possibly even polymers or biopolymers.
OT nanoparticles is based on the interaction with ligands and is independent of the nanoparticle core. Therefore, the developed technology is a first step toward designing better antiphage agents for industrial applications. The idea presented in our study can be in the future transferred to other types of nanoparticles, with cheaper and more biocompatible cores, such as silica or possibly even polymers or biopolymers.
Conclusions
We believe that the presented approach has the ability to produce safe, non-toxic, and easy-to-apply solutions for the phage problem in the biotechnology industry. To date, nanoparticles designed to deactivate phages were based on the generation of silver/copper ions or ROS – mechanisms which are harmful to all biological species (bacteria, cells, viruses, and phages). Thus, selective protection of bacteria from phages was not possible. The presented solution approaches the problem of phage infection from a completely new perspective. For the first time, technology focusing on the protection of bacteria directly inside the bioreactor is provided. We prepared anti-phage nanoparticles that were effective in the environment of bioreactors and were harmless for bacteria E. coli. The nanoparticles efficiently deactivated all three types of tested phages at reasonable concentrations, including T1, which is the most common cause of failure of E. coli fermentations. The nanoparticles were able to decrease the orders of magnitude of phages within a few hours. We showed that a combination of negatively charged and hydrophobic ligands was the key for effective phage deactivation with good inhibitory concentration (EC50 ≤ 1 μg mL−1). The best-performing nanoparticles MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT (85
OT (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) require just a 1 h preincubation step at 50 °C to fully deactivate T1 phages and protect bacteria E. coli in an environment similar to the inside of the bioreactor.
15) require just a 1 h preincubation step at 50 °C to fully deactivate T1 phages and protect bacteria E. coli in an environment similar to the inside of the bioreactor.
Experimental
Synthesis of MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) OT nanoparticles
OT nanoparticles
The 11-mercapto 1-undecanesulfonic acid (MUS) ligand was synthesized according to the procedure reported by Cendrowska et al. and Guven et al.42,43 Nanoparticles (NPs) were synthesized via the method developed by Zheng et al.44 later modified by Verma et al.45 First, gold salt (HAuCl4·3H2O; 0.9 mmol) and both ligands (0.9 mmol in total) were dissolved in ethanol (220 mL) and mixed for 15 min. The feed ratio between MUS and 1-octanethiol (OT) ligands was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, for MUS
1, for MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT (70
OT (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) and MUS
30) and MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT (85
OT (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) nanoparticles, respectively. Then, sodium borohydride (NaBH4; 50 mmol) dissolved in ethanol (200 mL) was added dropwise to the mixture upon mixing for over 2 h. The reaction was continued for an additional 24 h. Precipitated nanoparticles were then washed by centrifugation using ethanol and finally purified with MilliQ water on Amicon® Ultra-15 centrifugal filter devices (10k or 30k NMWL). The particles were then suspended in a small amount of water (∼2 mL) and freeze-dried. The molecular weight of nanoparticles was calculated as shown in the ESI.†
15) nanoparticles, respectively. Then, sodium borohydride (NaBH4; 50 mmol) dissolved in ethanol (200 mL) was added dropwise to the mixture upon mixing for over 2 h. The reaction was continued for an additional 24 h. Precipitated nanoparticles were then washed by centrifugation using ethanol and finally purified with MilliQ water on Amicon® Ultra-15 centrifugal filter devices (10k or 30k NMWL). The particles were then suspended in a small amount of water (∼2 mL) and freeze-dried. The molecular weight of nanoparticles was calculated as shown in the ESI.†
Synthesis of TMA nanoparticles
N,N,N-Trimethyl(11-mercaptoundecyl)ammonium (TMA) ligands and NPs were prepared according to the procedure reported by Cendrowska et al.42 To prepare nanoparticles, chloro(triphenylphosphine)gold(I) (0.56 mmol) and the TMA ligand (1.2 mmol) were dissolved in ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) toluene mixture (1
toluene mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 230 mL). After 10 min of stirring, borane tert-butylamine complex (1.5 mmol) dissolved in an ethanol
1, 230 mL). After 10 min of stirring, borane tert-butylamine complex (1.5 mmol) dissolved in an ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) toluene mixture (1
toluene mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; 20 mL) was added. The reaction was continued at 125 °C (800 rpm) for 1.5 h. Afterward, the volume was decreased in a rotary evaporator, which caused the precipitation of NPs. Nanoparticles were then washed by centrifugation using diethyl ether and toluene and finally purified with Milli-Q water on Amicon® Ultra-15 centrifugal filter devices (10k or 30k NMWL). The particles were suspended in a small amount of water (∼2 mL) and freeze-dried.
1; 20 mL) was added. The reaction was continued at 125 °C (800 rpm) for 1.5 h. Afterward, the volume was decreased in a rotary evaporator, which caused the precipitation of NPs. Nanoparticles were then washed by centrifugation using diethyl ether and toluene and finally purified with Milli-Q water on Amicon® Ultra-15 centrifugal filter devices (10k or 30k NMWL). The particles were suspended in a small amount of water (∼2 mL) and freeze-dried.
Synthesis of EG4 nanoparticles
Gold salt (HAuCl4·3H2O; 0.2 mmol) and 2-(2-(2-(2-mercaptoethoxy)ethoxy)ethoxy)ethanol (EG4) (0.2 mmol) were dissolved in ethanol (20 mL) and stirred for 15 min. Then, sodium borohydride (NaBH4, 2.5 mmol) dissolved in ethanol (37.5 mL) was added dropwise to the mixture for over 1 minute. The reaction was continued for an additional 24 h. Next, Milli-Q water (50 mL) was added to the mixture, and nanoparticles were washed with Milli-Q water on an Amicon® Ultra-15 centrifugal filter device (30k NMWL). A final volume of 3 mL was evaporated on a rotary evaporator. The remaining nanoparticles were redispersed in an ethanol and diethyl ether mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10; 33 mL). The solvent was removed by centrifugation, and the pellet was dried overnight under vacuum.
10; 33 mL). The solvent was removed by centrifugation, and the pellet was dried overnight under vacuum.
NPs characterization
Size distribution of NPs was calculated based on TEM analysis. A drop of NPs (4 μL; 0.1–0.5 mg mL−1) was deposited onto a 400-mesh carbon-supported copper grid and left to dry. All TEM images were acquired using an FEI TALOS™ electron microscope with an acceleration voltage of 200 kV equipped with a Ceta CCD camera. Images of the NPs were analyzed using Fiji software, and their diameters were calculated using a homemade script compatible with this software.1H-NMR analysis was performed to control the purity of prepared nanoparticles. Assembly of ligands on the nanoparticles causes broadening of their NMR peaks.46–48 This effect was used to evaluate the purity of the prepared nanoparticles. The absence of sharp peaks in the NMR spectrum obtained for the solution of nanoparticles suspended in water indicated a lack of impurities, such as unbound ligands. The ratio between ligands in the case of the mixed ligand nanoparticles was assessed by the 1H-NMR analysis after etching the nanoparticles with iodine. The etching solution was iodine (20 mg) dissolved in methanol-d4 (1 mL). Etching was obtained by suspending NPs (5 mg) in the etching mixture (0.6 mL) for 30 min under sonication. The ligand ratio was calculated according to the integrals of the given peaks (Fig. S1 and S2†).
The zeta potentials of nanoparticles were measured in PBS buffer using a Zetasizer Nano ZS (Malvern). The concentration of the nanoparticles was 0.1 mg mL−1.
Preparation of bacteriophages
Phages were prepared according to a procedure developed by Sambrook et al.49 Briefly, the early logarithmic culture of Escherichia coli was infected with bacteriophages. E. coli BL21 was used for the amplification of phages T1, T4, and T7, and E. coli C3000 was used for phages MS2. After lysis, the phages were precipitated by polyethylene glycol precipitation. Collected phages were washed with TM buffer (10 mM TRIS, 10 mM MgSO4, 5 μM CaCl2, pH 7.4) on Amicon® Ultra-15 centrifugal filter devices (1000k NMWL) and then dialyzed against TM buffer. After completion of the dialysis, DNase I (0.2 μg mL−1) was added to digest the DNA released from the damaged capsids.Virucidal tests
Phages were incubated in TM buffer with 0.5 mg mL−1 of nanoparticles at 37 °C or 50 °C over three days. The number of active phages was measured at different time points. We used the plaque count method for the determination of bacteriophage activity. Plaque forming units (PFU) can be directly correlated with the number of active phages within the sample. In this approach, the bottom LB-agar medium (20 mL) was poured onto a plastic Petri dish and left to solidify. Then, the top LB-agar (i.e., LB medium with 0.5% agar; 4 mL) was mixed with an overnight culture of E. coli (200 μL) and poured onto the plate. E. coli BL21 was used for phages T1, T4, and T7, and E. coli C3000 was used for phages MS2. 10-fold dilutions of phage solution were prepared, and droplets of each dilution (5 μL) were spotted onto the top agar layer. Plaques were counted after the incubation of the plates at 37 °C for 24 h. All experiments were performed in triplicate.Dose–response tests on phages
Phages were incubated in TM buffer with various concentrations of nanoparticles at 50 °C for 24 h. The number of active phages was measured at different time points. As a control, samples with phages without any nanoparticles were used. The number of phages was estimated via the plaque count method described in the previous section. All experiments were performed in duplicate.Dose–response tests on bacteria
Inoculum of bacteria E. coli BL21 was mixed in LB medium with various concentrations of MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT (85
OT (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) nanoparticles. Then, the growth of the bacterial culture was observed by optical density (OD600) measurement. To calculate dose–response, the optical density after 6 h of incubation was compared. All experiments were performed in triplicate.
15) nanoparticles. Then, the growth of the bacterial culture was observed by optical density (OD600) measurement. To calculate dose–response, the optical density after 6 h of incubation was compared. All experiments were performed in triplicate.
Real application test of the infected bacteria-based bioreactor
First, T1 bacteriophages (200 pfu mL−1) were incubated: (a) with MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT (85
OT (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) nanoparticles (100 μg mL−1) at 50 °C for 1 h, and (b) with MUS
15) nanoparticles (100 μg mL−1) at 50 °C for 1 h, and (b) with MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT (85
OT (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) nanoparticles (20 μg mL−1) at 50 °C for 12 h. Then, to such solutions, an inoculum of bacteria E. coli BL21 in LB medium was added. As controls, samples containing only bacteria (normal bacterial growth) and bacteria with T1 bacteriophages were prepared. The growth of bacteria was controlled by measuring the optical density (OD600), which can be directly correlated with the number of bacteria in the solution. All experiments were performed in four repetitions.
15) nanoparticles (20 μg mL−1) at 50 °C for 12 h. Then, to such solutions, an inoculum of bacteria E. coli BL21 in LB medium was added. As controls, samples containing only bacteria (normal bacterial growth) and bacteria with T1 bacteriophages were prepared. The growth of bacteria was controlled by measuring the optical density (OD600), which can be directly correlated with the number of bacteria in the solution. All experiments were performed in four repetitions.
Transmission electron cryo-microscopy
Phages (∼4 × 1010 pfu mL−1) were incubated with nanoparticles (0.1 mg mL−1) in TM buffer at 37 °C or 50 °C for a given amount of time (24 h if not stated otherwise). Then, a droplet of such a solution (3 μL) was deposited onto a lacey carbon film grid (Electron Microscopy Sciences, Germany) and blotted to a thin (100–300 nm) layer of liquid. The grid was then immediately flash-frozen in liquid ethane using an FEI Vitrobot Mark IV. Imaging was performed using a Gatan single tilt cryo holder operated on an FEI Tecnai F20 Cryo 80 kV transmission electron microscope in the low-dose mode to visualize the samples at an average exposure of 1–3 electrons Å−2 on a Ceta camera.Cytotoxicity test
The toxicity of NPs was examined using MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay. Vero cell cultures (African green monkey fibroblastoid kidney cells ATCC CCL-81) seeded in 96-well plates were incubated in DMEM (Gibco-BRL, Gaithersburg, MD) supplemented with 2% FBS with different concentrations of NPs for 24 h. Cell viability was determined using the CellTiter 96 Proliferation Assay Kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. Absorbance was measured using a Microplate Reader (Model 680, BIORAD) at 490 nm. Cells incubated without nanoparticles were used as the negative control, and cells incubated with 30% Triton X-100 were treated as the positive control (very high toxicity, no cells survived). The effect on cell viability at different concentrations of NPs was expressed as a percentage of live cells, by comparing the absorbance of treated cells with those of the cells incubated with the culture medium.Author contributions
Łukasz Richter: investigation, validation, writing – original draft, visualization, funding acquisition. Karolina Paszkowska: investigation. Urszula Cendrowska: resources, investigation. Francesca Olgiati: resources, investigation. Paulo Jacob Silva: resources. Matteo Gasbarri: resources, visualization. Zekiye Pelin Guven: resources. Jan Paczesny: writing – review & editing, supervision, methodology. Francesco Stellacci: writing – review & editing, project administration, funding acquisition, conceptualization.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 754462. The work of Ł.R. was supported by Swiss Government Excellence Scholarship. The work of K.P. and J.P. was supported by the National Science Centre within SONATA BIS grant number 2017/26/E/ST4/00041. M.G. was supported by the National Center of Competence in Research (NCCR) Bio-Inspired Materials. The authors are grateful to Professor Marcin Łoś, from the Phage Consultants Company, for his insight into bacteriophages and phage contaminations in the industry. The authors would also like to acknowledge Professor Robert Hołyst from the Institute of Physical Chemistry Polish Academy of Sciences for fruitful discussions and ideas.References
- G. Bogosian, in The Bacteriophages, ed. R. Calendar, Oxford University Press, 2nd edn, 2006, pp. 667–673 Search PubMed.
- M. Łoś, Eur. Biopharm. Rev., 2010, 51, 78–80 Search PubMed.
- E. Jończyk, M. Kłak, R. Międzybrodzki and A. Górski, Folia Microbiol., 2011, 56, 191–200 CrossRef PubMed.
- Z. Atamer, M. Samtlebe, H. Neve, K. J. Heller and J. Hinrichs, Front. Microbiol., 2013, 4, 1–9 CrossRef PubMed.
- D. M. Guglielmotti, D. J. Mercanti, J. A. Reinheimer and A. del L. Quiberoni, Front. Microbiol., 2012, 2, 1–11 CrossRef PubMed.
- Y. H. Joe, K. Woo and J. Hwang, J. Hazard. Mater., 2014, 280, 356–363 CrossRef CAS PubMed.
- L. Bromberg, D. J. Bromberg, T. A. Hatton, I. Bandín, A. Concheiro and C. Alvarez-Lorenzo, Langmuir, 2012, 28, 4548–4558 CrossRef CAS PubMed.
- T. Matsushita, N. Shirasaki, Y. Matsui and K. Ohno, Chemosphere, 2011, 85, 571–576 CrossRef CAS PubMed.
- A. M. Ledeboer, S. Bezemer, J. J. W. de Haard, I. M. Schaffers, C. T. Verrips, C. van Vliet, E.-M. Düsterhöft, P. Zoon, S. Moineau and L. G. J. Frenken, J. Dairy Sci., 2002, 85, 1376–1382 CrossRef CAS PubMed.
- D. Veesler, B. Dreier, S. Blangy, J. Lichière, D. Tremblay, S. Moineau, S. Spinelli, M. Tegoni, A. Plückthun, V. Campanacci and C. Cambillau, J. Biol. Chem., 2009, 284, 30718–30726 CrossRef CAS PubMed.
- R. D. G. Ferreira, A. R. Azzoni and S. Freitas, Biotechnol. Biofuels, 2018, 11, 81 CrossRef PubMed.
- R. H. Baltz, J. Ind. Microbiol. Biotechnol., 2018, 45, 1003–1006 CrossRef CAS PubMed.
- A. Murata, K. Kitagawa and R. Saruno, Agric. Biol. Chem., 1971, 35, 294–296 CrossRef CAS.
- W. Ma, M. Sun, P. Fu, S. Li, L. Xu, H. Kuang and C. Xu, Adv. Mater., 2017, 29, 1703410 CrossRef PubMed.
- H. Zhang, C. Hao, A. Qu, M. Sun, L. Xu, C. Xu and H. Kuang, Angew. Chem. Int. Ed., 2020, 59, 7131–7138 CrossRef CAS PubMed.
- X. Guo, M. Sun, R. Gao, A. Qu, C. Chen, C. Xu, H. Kuang and L. Xu, Angew. Chem. Int. Ed., 2021, 60, 13073–13080 CrossRef CAS PubMed.
- W. Ma, C. Hao, M. Sun, L. Xu, C. Xu and H. Kuang, Mater. Horiz., 2018, 5, 141–161 RSC.
- M. Sun, L. Xu, A. Qu, P. Zhao, T. Hao, W. Ma, C. Hao, X. Wen, F. M. Colombari, A. F. de Moura, N. A. Kotov, C. Xu and H. Kuang, Nat. Chem., 2018, 10, 821–830 CrossRef CAS PubMed.
- L. Xu, M. Sun, P. Cheng, R. Gao, H. Wang, W. Ma, X. Shi, C. Xu and H. Kuang, Adv. Funct. Mater., 2018, 28, 1707237 CrossRef.
- E. Gilcrease, R. Williams and R. Goel, Water Res., 2020, 181, 115900 CrossRef CAS PubMed.
- K. Gokulan, A. Bekele, K. Drake and S. Khare, Int. J. Nanomed., 2018, 13, 2857–2867 CrossRef CAS PubMed.
- Q. L. Shimabuku, T. Ueda-Nakamura, R. Bergamasco and M. R. Fagundes-Klen, Process Saf. Environ. Prot., 2018, 117, 33–42 CrossRef CAS.
- B. De Gusseme, L. Sintubin, L. Baert, E. Thibo, T. Hennebel, G. Vermeulen, M. Uyttendaele, W. Verstraete and N. Boon, Appl. Environ. Microbiol., 2010, 76, 1082–1087 CrossRef CAS PubMed.
- Q. L. Shimabuku, F. S. Arakawa, M. Fernandes Silva, P. Ferri Coldebella, T. Ueda-Nakamura, M. R. Fagundes-Klen and R. Bergamasco, Environ. Technol., 2017, 38, 2058–2069 CrossRef CAS PubMed.
- J. M. Mazurkow, N. S. Yüzbasi, K. W. Domagala, S. Pfeiffer, D. Kata and T. Graule, Environ. Sci. Technol., 2020, 54, 1214–1222 CrossRef CAS PubMed.
- V. I. Syngouna and C. V. Chrysikopoulos, J. Colloid Interface Sci., 2017, 497, 117–125 CrossRef CAS PubMed.
- M. Cho, E. L. Cates and J.-H. Kim, Water Res., 2011, 45, 2104–2110 CrossRef CAS PubMed.
- R. Cheng, M. Kang, S. Zhuang, S. Wang, X. Zheng, X. Pan, L. Shi and J. Wang, Sci. Total Environ., 2019, 649, 995–1003 CrossRef CAS PubMed.
- E. Sánchez-López, D. Gomes, G. Esteruelas, L. Bonilla, A. L. Lopez-Machado, R. Galindo, A. Cano, M. Espina, M. Ettcheto, A. Camins, A. M. Silva, A. Durazzo, A. Santini, M. L. Garcia and E. B. Souto, Nanomaterials, 2020, 10, 292 CrossRef PubMed.
- V. Cagno, P. Andreozzi, M. D'Alicarnasso, P. J. Silva, M. Mueller, M. Galloux, R. Le Goffic, S. T. Jones, M. Vallino, J. Hodek, J. Weber, S. Sen, E. R. Janecek, A. Bekdemir, B. Sanavio, C. Martinelli, M. Donalisio, M. A. R. Welti, J. F. Eleouet, Y. Han, L. Kaiser, L. Vukovic, C. Tapparel, P. Král, S. Krol, D. Lembo and F. Stellacci, Nat. Mater., 2018, 17, 195–203 CrossRef CAS PubMed.
- M. N. Baeshen, A. M. Al-Hejin, R. S. Bora, M. M. M. Ahmed, H. A. I. Ramadan, K. S. Saini, N. A. Baeshen and E. M. Redwan, J. Microbiol. Biotechnol., 2015, 25, 953–962 CrossRef CAS PubMed.
- C.-J. Huang, H. Lin and X. Yang, J. Ind. Microbiol. Biotechnol., 2012, 39, 383–399 CrossRef CAS PubMed.
- N. A. Baeshen, M. N. Baeshen, A. Sheikh, R. S. Bora, M. M. M. Ahmed, H. A. I. Ramadan, K. S. Saini and E. M. Redwan, Microb. Cell Fact., 2014, 13, 141 CrossRef PubMed.
- M. Los, in Microbial Metabolic Engineering: Methods and Protocols, Springer, New York, NY, 2012, pp. 305–315 Search PubMed.
- M. L. Yap and M. G. Rossmann, Future Microbiol., 2014, 9, 1319–1327 CrossRef CAS PubMed.
- H.-W. Ackermann, D. Tremblay and S. Moineau, World Fed. Cult. Collection Newslett., 2004, 38, 35–40 Search PubMed.
- M. H. Ly-Chatain, S. Moussaoui, A. Vera, V. Rigobello and Y. Demarigny, Front. Microbiol., 2013, 4, 1–6 Search PubMed.
- A. Ortega, S. Farah, P. Tranque, A. V. Ocaña, S. H. Nam-Cha, N. Beyth, C. Gómez-Roldán, R. Pérez-Tanoira, A. J. Domb, F. C. Pérez-Martínez and J. Pérez-Martínez, IET Nanobiotechnol., 2015, 9, 342–348 CrossRef PubMed.
- P. G. Leiman, S. Kanamaru, V. V. Mesyanzhinov, F. Arisaka and M. G. Rossmann, Cell. Mol. Life Sci., 2003, 60, 2356–2370 CrossRef CAS PubMed.
- I. Gitlin, J. D. Carbeck and G. M. Whitesides, Angew. Chem., Int. Ed., 2006, 45, 3022–3060 CrossRef CAS PubMed.
- European Commission EUROSTAT, Nat. Methods, 2008, 5, 135–146 CrossRef PubMed.
- U. Cendrowska, P. J. Silva, N. Ait-Bouziad, M. Müller, Z. P. Guven, S. Vieweg, A. Chiki, L. Radamaker, S. T. Kumar, M. Fändrich, F. Tavanti, M. C. Menziani, A. Alexander-Katz, F. Stellacci and H. A. Lashuel, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 6866–6874 CrossRef CAS PubMed.
- Z. P. Guven, P. H. J. Silva, Z. Luo, U. B. Cendrowska, M. Gasbarri, S. T. Jones and F. Stellacci, J. Visualized Exp., 2019, 2019, 1–11 Search PubMed.
- N. Zheng, J. Fan and G. D. Stucky, J. Am. Chem. Soc., 2006, 128, 6550–6551 CrossRef CAS PubMed.
- A. Verma, O. Uzun, Y. Hu, Y. Hu, H.-S. Han, N. Watson, S. Chen, D. J. Irvine and F. Stellacci, Nat. Mater., 2008, 7, 588–595 CrossRef CAS PubMed.
- A. Badia, W. Gao, S. Singh, L. Demers, L. Cuccia and L. Reven, Langmuir, 1996, 12, 1262–1269 CrossRef CAS.
- L. E. Marbella and J. E. Millstone, Chem. Mater., 2015, 27, 2721–2739 CrossRef CAS.
- M. J. Hostetler, J. E. Wingate, C.-J. Zhong, J. E. Harris, R. W. Vachet, M. R. Clark, J. D. Londono, S. J. Green, J. J. Stokes, G. D. Wignall, G. L. Glish, M. D. Porter, N. D. Evans and R. W. Murray, Langmuir, 1998, 14, 17–30 CrossRef CAS.
- J. Sambrook, E. F. Fritsch and T. Maniatis, Molecular Cloning – A Laboratory Manual, Cold Spring Harbor Press, Cold Spring Harbor, 2nd edn, 1989 Search PubMed.
Footnote |
† Electronic supplementary information (ESI) available: Calculation of the molecular mass of nanoparticles, 1H NMR of MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT nanoparticles, comparison of properties of all studied nanoparticles, the cytotoxic effect of TMA nanoparticles, the effect of nanoparticles against phage MS2, synthesis and analysis of the antiphage effect of MUS nanoparticles, growth of bacteria E. coli in the presence of MUS OT nanoparticles, comparison of properties of all studied nanoparticles, the cytotoxic effect of TMA nanoparticles, the effect of nanoparticles against phage MS2, synthesis and analysis of the antiphage effect of MUS nanoparticles, growth of bacteria E. coli in the presence of MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT nanoparticles, a table describing properties of structural proteins of phage T4, time-dependent cryo-TEM analysis of interactions between T4 phages and MUS OT nanoparticles, a table describing properties of structural proteins of phage T4, time-dependent cryo-TEM analysis of interactions between T4 phages and MUS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) OT nanoparticles. See DOI: 10.1039/d1nr04936d OT nanoparticles. See DOI: 10.1039/d1nr04936d |
| This journal is © The Royal Society of Chemistry 2021 |