Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Culturing human iPSC-derived neural progenitor cells on nanowire arrays: mapping the impact of nanowire length and array pitch on proliferation, viability, and membrane deformation†
Jann
Harberts
 *a,
Katja
Bours
a,
Malte
Siegmund
a,
Carina
Hedrich
a,
Michael
Glatza
b,
Hans R.
Schöler
b,
Undine
Haferkamp
*a,
Katja
Bours
a,
Malte
Siegmund
a,
Carina
Hedrich
a,
Michael
Glatza
b,
Hans R.
Schöler
b,
Undine
Haferkamp
 c,
Ole
Pless
c,
Ole
Pless
 c,
Robert
Zierold
c,
Robert
Zierold
 *a and
Robert H.
Blick
*a and
Robert H.
Blick
 ad
ad
aCenter for Hybrid Nanostructures (CHyN), Universität Hamburg, Luruper Chaussee 149, 22761 Hamburg, Germany. E-mail: jann.harberts@chyn.uni-hamburg.de; robert.zierold@chyn.uni-hamburg.de
bDepartment of Cell and Developmental Biology, Max Planck Institute for Molecular Biomedicine, Röntgenstraße 20, 48149 Münster, Germany
cFraunhofer Institute for Translational Medicine and Pharmacology (ITMP), ScreeningPort, Schnackenburgallee 114, 22525 Hamburg, Germany
dMaterial Science and Engineering, College of Engineering, University of Wisconsin-Madison, Madison, Wisconsin 53706, USA
First published on 29th November 2021
Abstract
Nanowire arrays used as cell culture substrates build a potent tool for advanced biological applications such as cargo delivery and biosensing. The unique topography of nanowire arrays, however, renders them a challenging growth environment for cells and explains why only basic cell lines have been employed in existing studies. Here, we present the culturing of human induced pluripotent stem cell-derived neural progenitor cells on rectangularly arranged nanowire arrays: In detail, we mapped the impact on proliferation, viability, and topography-induced membrane deformation across a multitude of array pitches (1, 3, 5, 10 μm) and nanowire lengths (1.5, 3, 5 μm). Against the intuitive expectation, a reduced proliferation was found on the arrays with the smallest array pitch of 1 μm and long NWs. Typically, cells settle in a fakir-like state on such densely-spaced nanowires and thus experience no substantial stress caused by nanowires indenting the cell membrane. However, imaging of F-actin showed a distinct reorganization of the cytoskeleton along the nanowire tips in the case of small array pitches interfering with regular proliferation. For larger pitches, the cell numbers depend on the NW lengths but proliferation generally continued although heavy deformations of the cell membrane were observed caused by the encapsulation of the nanowires. Moreover, we noticed a strong interaction of the nanowires with the nucleus in terms of squeezing and indenting. Remarkably, the cell viability is maintained at about 85% despite the massive deformation of the cells. Considering the enormous potential of human induced stem cells to study neurodegenerative diseases and the high cellular viability combined with a strong interaction with nanowire arrays, we believe that our results pave the way to apply nanowire arrays to human stem cells for future applications in stem cell research and regenerative medicine.
Introduction
Human induced pluripotent stem cells (iPSCs) have the potential to maximize the throughput and efficacy of clinical trials and applications.1 Many ethical and political controversies are avoided by relinquishing embryonic stem cells and the limited availability of primary human cells is overcome.2 Moreover, iPSCs are patient-specific since the initial cell sample is commonly obtained by an ordinary skin biopsy.3 The direct use of human cells also reduces the high failure rate of clinical translation created by differences of disease-associated pathways between human and animal cells.4–6 Access to a multitude of human iPSC-derived cell types such as dopaminergic or glutamatergic neurons might allow for novel treatment modalities with improved pre-clinical efficacy and safety assessment.7 Thus, human pathophysiological conditions including neurodegenerative conditions such as Alzheimer's disease or Parkinson's disease can be explored more efficiently and potentially even be ameliorated.8,9 Neuroscience research and clinical translation not only benefit from the feasibility to derive all major neuronal cell types but also from the progress to create more advanced cell cultures such as brain organoids or blood–brain barrier models.10–12 Hence, iPSC technologies have profoundly changed the operation routines of basic, pre-clinical, and clinical research in neuroscience since their initial discovery in 2006.13The cell culturing of these complex systems including the administration of therapeutic agents is commonly performed in regular Petri dishes or multi-well plates. However, replacing the passive surface of these dishes with an active substrate would possibly render another dimension to test or to manipulate the cells. Here, the continuous progress in micro- and nanofabrication offers novel strategies to create such functionalized biocompatible (semi-conductor) materials to be employed as cell culture substrates.14,15
Tailor-made substrates were published in numerous studies demonstrating their applicability but up until now primarily standard human cell lines (e.g., HEK293 or HeLa cells) or primary rodent cells were employed.16–19 The influence of the substrate in terms of chemical and topological properties has been tested, for example, for adhesion, proliferation, viability, migration, and guidance of seeded cells.20–26 In this context, a particular subset of substrates featuring upright arranged high aspect ratio nanostructures—so-called nanowire (NW) arrays—play an increasingly important role.27–33
Nanowire arrays can affect biological parameters such as cellular growth, viability, morphology, and mechanotransduction machinery,34–39 while electrophysiological parameters are maintained.40–43 Furthermore, NW arrays have been used to measure mechanical properties,44–46 to interact with the cell's nucleus,47 to constrain movement and spreading,48–52 or to direct cell polarization such as outgrowth of neurites.53–57 In addition to the aforementioned passive applications, functionalized NW arrays were employed to incorporate an executing role, such as drug delivery,29,58–63 cell transfection,64–67 electrical stimulation/sensing,68–72 or biosensing,73 to name a few. To address specific applications, the interaction of the cell and the NWs can be tuned by adjusting the length and diameter of the NWs and the pitch of the array.52,74–76 Here, the settling regimes of the cells are ranging from a fakir-like state on a bed-of-nails to a complete encapsulation of the NWs. A model to estimate the regimes was developed by Buch-Månson et al.77 Even though these NW substrates offer many advantages over conventional cell culture dishes, the application of human iPSCs on NW arrays is less prevalent. This is potentially due to the fact that the NW arrays also create a challenging topography that must be tolerated by the cells during culture. For example, material cues have been discussed to be a regulator for epigenetics and stem cell function,78 and NW forests have been used to influence intracellular signaling, gene regulation, or basic cell differentiation.79–83 To make use of different NW arrays for human iPSCs it is hence crucial to ensure that, e.g., proliferation and viability of the cells are maintained.
In this work, we present the cultivation of human iPSC-derived small molecule neural progenitor cells (smNPCs) on silicon (Si) NW arrays with varying lengths of 1.5, 3, and 5 μm (L1.5, L3, L5) and array pitches of 1, 3, 5, and 10 μm (P1, P3, P5, P10). Such array parameters cover multiple interaction regimes between cells and NWs. The proliferation and cell viability were monitored for 4 days and compared to control substrates (glass and planarly etched silicon). On substrates with P ≥ 3, where the cells encapsulate the NWs, the cell numbers varied with the degree of topological stress, namely, the number of NWs per cell and the NW length. Remarkably, proliferation was also significantly reduced on P1 NW arrays (i.e., densely-packed NW arrays) at which the cells settle in a fakir-like state on a ‘bed-of-nails’. In general, such a settling state is considered non-invasive compared to encapsulating settling states on NW substrates with P ≥ 3. Imaging of F-actin showed that the cells cultured on P1 arrays underwent a reorganization of the cytoskeleton along the NW tips, which influenced normal cell division. However, reduced cell numbers on P1 arrays were only observed for L3 and L5 but not for L1.5. This was in agreement with further analyses that showed an intermediate settling regime for P1 L1.5 NW arrays, thus diminishing the impact on normal proliferation. In any case, on L1.5 NW arrays, the cell numbers after 4 days in vitro (DIV) were not only independent from the pitch and but also equal to the control. The cell viability after 4 DIV was excellent on the majority of the NW arrays with about 85% viable cells. Only on P1 L3, P1 L5, and P3 L5 substrates where the cells were either constrained to align the cytoskeleton along the NWs or deeply indented at multiple sites, viability was slightly reduced (at worst to about 75%). Three-dimensional imaging of the cells on the NW arrays showed that the cells strongly interact with NWs. Moreover, the level of NW encapsulation was defined by the NW length and array pitch. In severe appearing cases with long wires and medium array pitch, not only the cell membrane was deformed massively but also the nucleus was substantially indented or squeezed. Considering the strong interaction with the NWs and the outstanding cell viability, we believe that our results open new pathways to apply NW arrays in human stem cell biology and regenerative medicine technologies.
Results
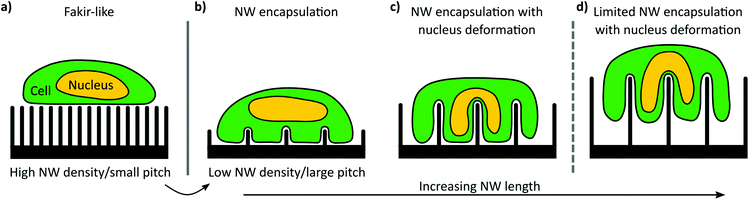
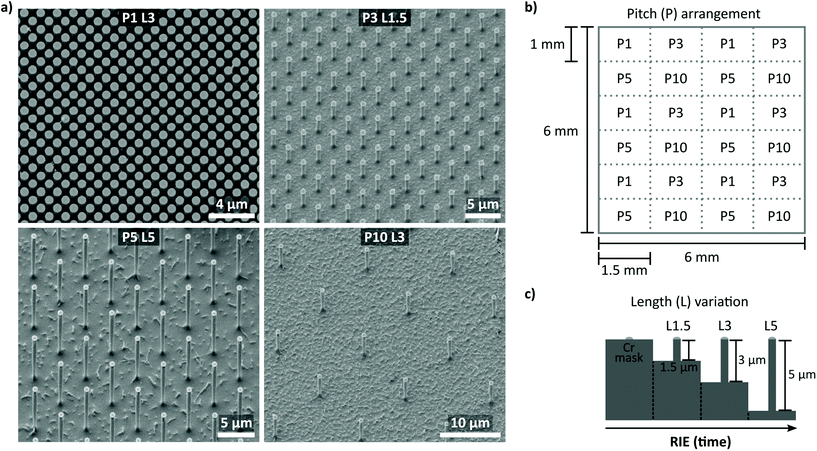
The settling regime of cells cultured on NW arrays strongly depends on the interplay of NW length and array pitch. Specifically, the cell settling can be tuned from a fakir-like state (Fig. 1a) to complete encapsulation of the NWs (Fig. 1b–d): In the fakir-like state, the cells rest on a ‘bed-of-nails’ being only in contact with the very NW tips (panel a). This state is predominantly caused by high array densities, i.e., small pitches. With increasing pitch, the cells start to encapsulate the NWs (panel b). Depending on the NW length, not only the cell membrane is deformed but also the nucleus is affected when the NW length is in the order of the cell's height (panel c). In the case of even longer NWs, the cells might lose contact with the bottom of the substrate creating an intermediate settling state between a fakir-like state and full encapsulation (panel d).The design and fabrication method of the rectangularly arranged Si NW arrays is summarized in Fig. 2. To map the effect of different array pitches and NW lengths on the culturing of smNPCs, we prepared NW arrays with array pitches of 1, 3, 5, and 10 μm (P1, P3, P5, P10) combined with NW lengths of 1.5, 3, and 5 μm (L1.5, L3, L5) exemplarily shown in Fig. 2a. The utilized pitches result in array densities of 100, 11.1, 4, and 1 NWs per 100 μm2, respectively. The NWs were produced in a top-down process via reactive ion etching (RIE) using chromium dots with a diameter of 700 nm as a hard mask. The hard mask for the array arrangement was defined by electron beam lithography on a 6 × 6 mm2 area with altering pitches (Fig. 2b). The NW length was controlled by the time used for RIE per chip containing the 6 × 6 mm2 patterned area (Fig. 2c). For control, areas of planarly etched Si right next to the region covered with NWs were used. Of note, cells grown here showed similar proliferation, viability, and spreading compared to controls grown on conventional glass coverslips (Fig. S1a–g†). Thus, smNPCs grown on NW arrays were compared in the following only to the Si control.
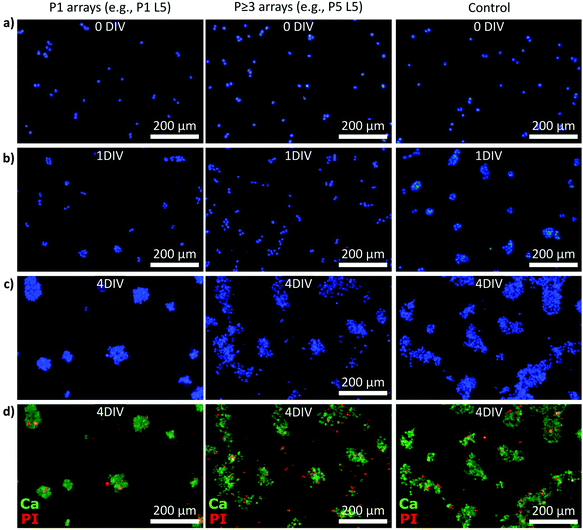
Data collection to determine the cell numbers and viability was performed on five consecutive days (0–4 DIV) using Hoechst, calcein, and propidium iodide (PI) stainings (Fig. 3). Specifically, we imaged the samples after initial cell attachment (≥1 h, referred to as 0 DIV) and the following four 24 h intervals. Fig. 3a–c show examples of Hoechst-stained smNPCs cultured on NW arrays with different densities but the same NW lengths (here, e.g., P1 L5 and P5 L5) and control samples after 0, 1, and 4 DIV. Right after initial cell adhesion, the cells were found randomly distributed on the substrate as the cells sediment and allocate from the cell suspension in a random manner (panel a). After one day, the cells started to form cell clusters (Fig. 3b) and larger colonies were formed after 4 DIV on both NW arrays and control substrate (panel c). Note, solely by visual inspection, the area covered with cells appeared to be reduced using a P1 NW array sample. Cell viability was assayed using calcein to identify viable cells and PI to stain for dead cells (examples shown in Fig. 3d). In a next step, we quantified the images taken and determined cell numbers, viability, and spreading of the cells on all types of NW arrays from 0 to 4 days of cultivation.
The quantification of the cell numbers, viability, and spreading using an automized software-assisted image analysis are summarized in Fig. 4. Specifically, Fig. 4a displays the cell numbers for all types of NW substrates from 0–4 DIV compared to the control. Initially, about 250 cells per captured image were determined. For unaffected cells on the control samples, the cell numbers reached about 1500 cells per image on day 4. On the NW samples, the cell numbers also increased in general over time but the final number of cells after 4 DIV depended on the degree of topological stress caused by the specific type of NW array. The overall cell numbers were reduced for encapsulating regimes (P ≥ 3) mainly with increasing number of NWs per cell (P3 NW arrays: ≈−40%, P5 NW arrays: ≈−25%, P10 NW arrays: ≈−10%) where L5 NW arrays decreased the cell numbers slightly more than L3 NW arrays. Note, L1.5 NW arrays showed no change in the cell numbers. Remarkably, the cell numbers for P1 L3 and P1 L5 NW arrays also showed significantly reduced values although such densely-spaced NW arrays are expected to produce a fakir-like settling state that is considered non-invasive. However, P1 L1.5 NW arrays showed like all L1.5 no change in cell numbers. The cell viabilities were quantified from the calcein and PI stainings and the final viabilities after 4 DIV are presented (Fig. 4b). For most of the NW substrates, a uniform and high cell viability of more than 85% similar to the control was found. For P1 L3, P1 L5, and P3 L5 NW arrays, however, the cell viability was reduced (at worst to about 75%) which correlates with the most prominent reduction in cell numbers on these substrates. Thus, not only the proliferation but also the viability suggests a strong interaction with the substrate. The intermediate viabilities for 0–3 DIV are shown in the ESI (Fig. S2a†). The cell clustering was analysed using a Density-Based Spatial Clustering of Applications with Noise (DBSCAN) algorithm applied to the viable cells and the bar charts of clustering and cluster density are shown in Fig. 4c and d. The proportions of cells that have been classified as part of a cell cluster with more than three cells are plotted for 0–2 DIV in Fig. 4c (3–4 DIV: Fig. S2b†). Immediately after seeding, only 30% of the cells were located in clusters. After one day, already about 80% of the cells formed clusters until almost all cells (approx. 95%) contributed to the colonies (4 DIV). In principle, no underlying dependence of the proportion of cells in clusters could be extracted with respect to the NW characteristics. However, in the early stages between 0 and 1 DIV, the clusters must be mainly produced by cell migration instead of cell division because the cell numbers within that time hardly increased. For that reason, the spatial distance of cells in the clusters (relating to the centres of the nuclei) was quantified as shown in Fig. 4d for 1 and 2 DIV to indicate cell movement (Fig. S2c for 3–4 DIV†). After one day, the mean distance between viable cells cultured on P3, P5, and P10 NW arrays was slightly yet significantly increased from 8.9 μm on the control to 9.5 μm indicating that the NWs prevented the cells from moving closer together. This inhibited movement was also visible in the Hoechst-stained images in Fig. 3b for the P5 L5 NW sample at 1 DIV where cells in clusters maintain larger gaps between the cells. This phenomenon was even more pronounced when only viable cells were imaged (Fig. S3†). With time, the difference vanished since proliferation overcame movement as the driving force in cluster formation. For a pitch of 1 μm where a fakir-like settling state is likely, the movement was not impaired compared to the control.
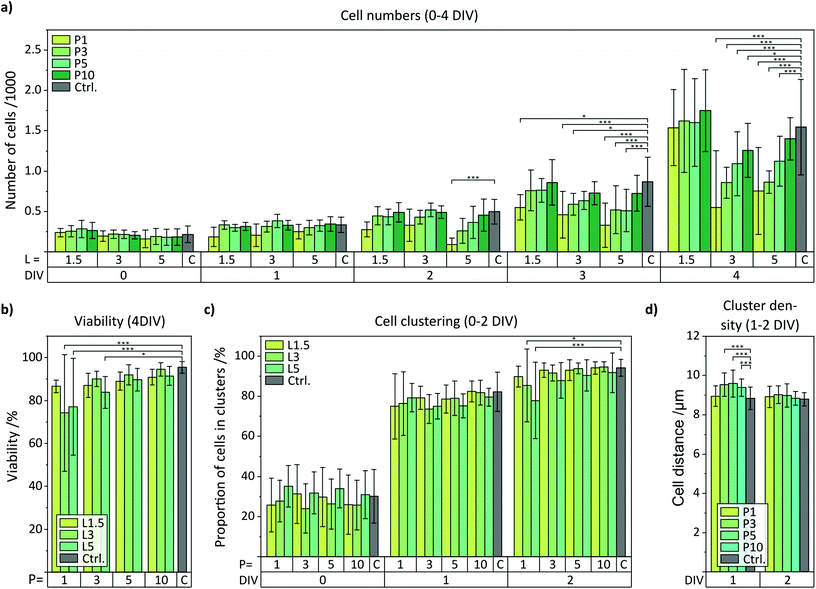 | ||
| Fig. 4 Quantification of the fluorescent microscopy images in terms of proliferation, viability, and spreading of cells cultured on samples with NW arrays featuring array pitches of 1, 3, 5, and 10 μm (P1, P3, P5, P10) and NW lengths of 1.5, 3, and 5 μm (L1.5, L3, L5) in comparison to control (C). (a) Number of viable cells cultured on NW arrays from 0 DIV (right after seeding) up to 4 DIV. (b) Viability after 4 DIV for all NW densities and NW lengths. (c) Proportion of viable cells in clusters from 0–2 DIV for all NW densities and NW lengths. (d) Distance between viable cells in clusters for 1–2 DIV for all array pitches and combined NW lengths. Data not shown here is compiled in Fig. S2 in the ESI.† Error bars are standard deviation (SD). Indicated significances: *α = 0.05, **α = 0.01, ***α = 0.001. | ||
Scanning electron microscopy (SEM) was used to study the interaction between the cells and the NW substrates (Fig. 5). In particular, Fig. 5a–c display a collection of top-view SEM images of cells cultured on P1 NW arrays with the lengths L1.5, L3, and L5. Repeatedly, smNPCs at the edge of a cluster showed distinct extensions along the x- and y-direction of the array arrangement. Remarkably, these extensions appeared on all NW substrates independent of the NW length. Hence, this observation obtained by SEM imaging did not fully correlate with the reduced proliferation on P1 L3 and P1 L5 NW arrays but normal proliferation on P1 L1.5 NW arrays. Nevertheless, on NW arrays with P ≥ 3 ordered extensions along the array axes could not be observed as presented in the tilted SEM images in Fig. 5d–f (exemplarily for P3 L3, P5 L1.5, and P10 L5 NW arrays; images of all remaining substrates in Fig. S4†). Even though no influence of the NW array's geometry on the cell's extensions was apparent for P ≥ 3 in the SEM images, one still was able to obtain a first impression of how the cells interact with the NWs. For example, in the case of the P3 L3 NW arrays, the cells in the outer region interacted with the NW tips but also grew between the NWs (detail in Fig. 5d). A similar interplay is seen for P5 L1.5 in the detail section of Fig. 5e. For P10 L5, the smNPCs also interacted with the shaft of the NWs (detail image in Fig. 5f). Note, the SEM technique visualizes merely the outer face of the cells and the substrate. To gain insight into the interior of the cells and to image the deformation of the membrane and the nucleus by the NWs, a complementary fluorescent imaging technique was utilized and the results are presented later (Fig. 7 and 8).
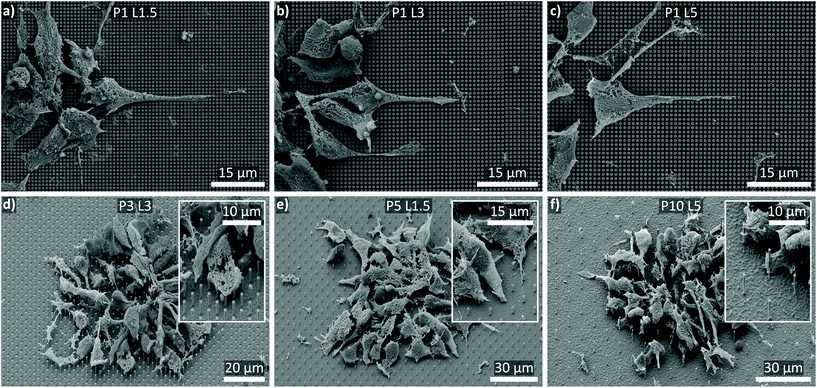 | ||
| Fig. 5 SEM images of smNPCs cultured on NW arrays. (a–c) Exemplary top-views of cells cultured on P1 NW arrays with increasing NW lengths (L1.5, L3, L5). Repeatedly, the cells showed distinct elongations along the NW tips independent of the NW length. (d–f) Exemplary tilted views of cells cultured on P ≥ 3 NW arrays with different lengths (L3, L1.5, L5). The cells did strongly interact with the NWs (insets) but a fundamental alignment alongside the NWs was not observed. Tilt is 45° with tilt correction. Corresponding SEM images of cells cultured on the remaining NW samples and on control substrates are shown in Fig. S4 in the ESI.† | ||
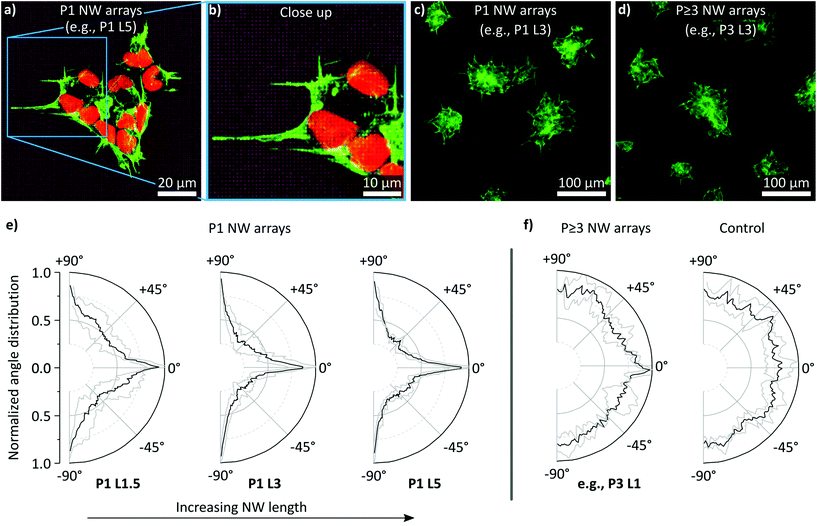 | ||
| Fig. 6 Confocal laser scanning microscopy (CLSM) and epifluorescent imaging of cells cultured on NW arrays (NW tips' reflections in purple) stained with phalloidin (green) for labeling the cytoskeleton (F-actin) and quantification of angle distribution of the cellular extensions. The nuclei were stained with DRAQ5 (red). (a) Exemplary down projection of multiple x–y slices in z-direction prepared by high-resolution CLSM of a cell cluster cultured on P1 NW arrays. Cells at the border of the cluster showed multiple alignments of the cytoskeleton along the x- and y-axis of the NW array (a close-up is shown in panel b). (c and d) Exemplary epifluorescent overview images of cells cultured on P1 and P ≥ 3 NW arrays. For P1 NW arrays, an alignment of the cytoskeleton along the array axes was observable. A corresponding image of cells cultured on a control substrate is shown in Fig. S5 in the ESI.† (e) Angle distribution of the cellular extensions for P1 NW arrays and varying lengths (L1.5, L3, L5). The grey line indicates the standard deviation. (f) Angle distribution of the cellular extensions for P ≥ 3 NW arrays (representative plot, e.g., P3 L1) and control substrates. The grey line indicates the standard deviation. n (cluster) > 50. | ||
The cytoskeleton of the smNPCs cultured on the NW arrays was imaged using confocal laser scanning microscopy (CLSM) and epifluorescent microscopy exemplarily shown in Fig. 6a–d. Here, we focused on the analysis of the cell extensions along the array axes that were previously observed by SEM imaging. The cytoskeleton was labelled with phalloidin to indicate F-actin. Specifically, Fig. 6a displays an exemplary CLSM image of a cell cluster on a P1 array sample (shown: P1 L5). The corresponding close-up shown in Fig. 6b illustrates that parts of the cytoskeleton were elongated in both x- and y-direction within a small area. Subsequently, widefield microscopy images were prepared of F-actin stained cells being cultured on all types of substrates (Fig. 6c and d, representative images of P1 and P ≥ 3 NW arrays, e.g., P1 L3, P3 L3. Control: Fig. S5†). We determined the angles of the protrusions emerging from the cell clusters and plotted the normalized angle distribution of the occurring orientation from −90° to +90° as shown for P1 NW arrays and increasing NW lengths (L1.5, L3, L5) in Fig. 6e. All the distributions revealed peaks with amplitudes of >0.8 at angles of ±90° and 0° independent of the NW length. These angles represent the rectangular axes of the NW arrays, hence, the majority of the extensions were aligned with the orientation of the array. However, the values of the minima between the peaks at ±90° and 0° changed depending on the NW lengths. Specifically, for L3 and L5 NWs, distinct minima of about 0.25 were observed at angles of about ±45° which would belong to the diagonal axes of the NW array. In the case of the L1.5 NWs, the minima around ±45° were less pronounced and the normalized amplitudes added up to a higher value of approx. 0.5 compared to only 0.25 using L3 and L5 NWs. Taken together, cells cultured on L3 and L5 NWs combined with a P1 array pitch showed reduced proliferation and systematically arranged elongations of the cytoskeleton. In the case of L1.5 NW arrays, where proliferation was normal at a P1 array pitch, the effect of angle sorting was less distinct. P ≥ 3 NW array and control substrates showed no systematic orientation of the extensions (Fig. 6f).
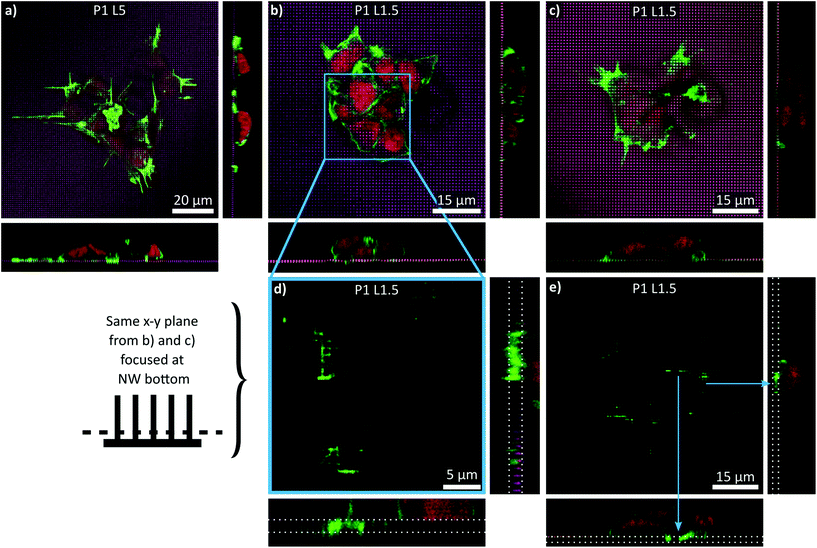
The difference between P1 L1.5 and P1 L ≥ 3 NW arrays related to the interaction of the cells with the NW arrays was investigated in more detail by three-dimensional (3D) CLSM imaging. 3D reconstructions were prepared from images recorded from varying z-planes (z-stacks) and then used to visualize different x–y planes and cross sections along the z-axis (Fig. 7). In particular, the main panel in Fig. 7a displays the example of a cell colony on a P1 L ≥ 3 NW array (e.g., P1 L5) that was already shown as a down projection beforehand, but now as a single image from the x–y plane focused on the NW tips. The cross-sections (narrow panels, right/bottom) demonstrated as expected that the cells are resting in a fakir-like state on the NWs as already seen in the SEM images. A similar settling regime was observed in the case of cells cultured on P1 L1.5 NW arrays (Fig. 7b and c). The cross-sections precisely positioned along the NWs showed no detrimental difference compared to the previously shown micrographs as the cells appeared to be lying on a bed-of-nails as well. However, differences were observed when the focus was adjusted to the bottom of the NWs (Fig. 7d and e). Specifically, panel d displays a close-up of the cell cluster shown in Fig. 7b. With the focus at the base of the NWs, CLSM imaging revealed that part of the cytoskeleton was also in contact with the substrate's bottom. Cross-sections placed now between the NWs then illustrated that F-actin is found below the level of the NW tips. A similar behaviour is depicted in Fig. 7e which moreover exemplifies that the cell's extensions also aligned with the NW axes at the bottom of the substrate. To briefly conclude, for P1 L1.5 NW arrays, the cells were found to be in an intermediate state where the cells did not only rest on the NW tips but also interacted with the bottom of the substrate. Since the interaction regime differed from cells cultured on P1 L3 and P1 L5 NW arrays, the development of ordered extension was less prominent and more importantly, proliferation was apparently no longer hindered.
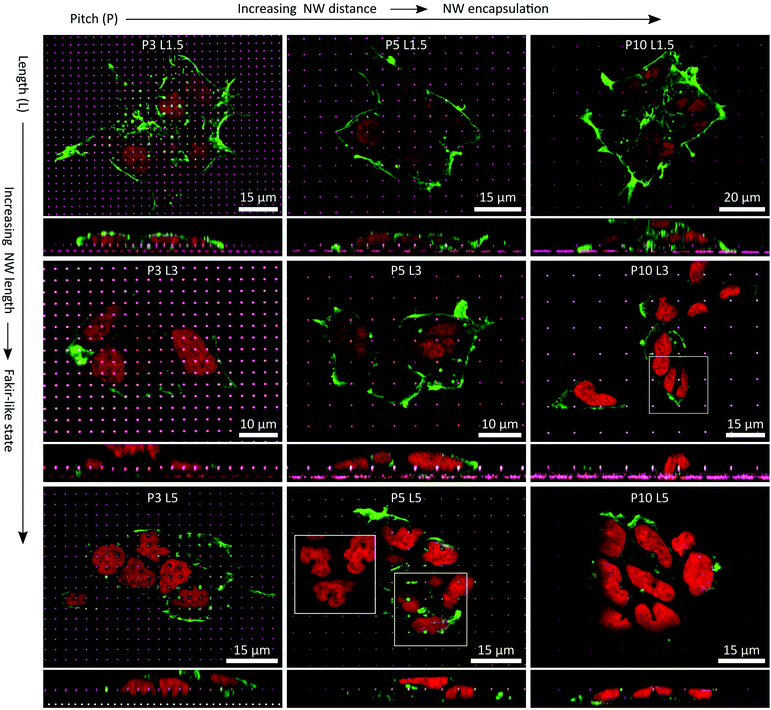
P ≥ 3 NW arrays were finally imaged as well using CLSM (Fig. 8). Two fundamental statements regarding the settling regimes of the cells on the NW arrays could be made: first, with increasing NW pitch, cells were more inclined to encapsulate the NWs. Secondly, with increasing NW length a fakir-like resting state was favoured. Nonetheless, for some fixed pitches or fixed lengths, the settling regimes of the cells on the NWs were maintained while changing the other parameter. In particular, for L1.5 and L3 NW arrays, the NWs were very likely to be encapsulated. Of course, the impact of the shortest NWs on the cells and especially the nuclei was rather small since the NW indent the cell by at most 1.5 μm which was negligible in comparison to a cell height of a few micrometres. The deformations induced by the 3 μm long wires were more distinct and thus, the interactions with the nuclei were more present. Yet, the cells did not seem to use all available means to completely avoid deformations of the nucleus. For instance, at the P10 L3 NW array, one nucleus was exactly positioned on a NW (white square) although there would have been enough free space next to the NW to avoid the very nanostructure. Instead, the nucleus was stretching down to the flat area of the substrate causing a u-shaped deformation seen in the cross-sectional panel below the main panel. For the P3 L5 NW arrays, the cells settled in an intermediate state on the NWs where only the top part was engulfed by the cell. Remarkably, this situation went along with heavy deformations of the nuclei where each nucleus was indented by several NWs by about 2.5 μm. Also, such strong reshaping of the nucleus occurred frequently with larger pitches where the cells encapsulated the L5 NWs completely. Here, the nuclei were also either indented by NWs, or strong curvatures were generated at nuclei that were squeezed by NWs or were curled around an individual NW (e.g., in the white square of the P5 L5 NW array). Especially for P10 NW arrays, cells could have avoided these heavy interactions of the nucleus with the NWs by resting between the NWs, but noteworthy this was not necessarily the case.
Discussion
The fabrication of Si nanostructures in a top-down approach via RIE is a well-established routine in the semiconductor community.84 In addition, e-beam lithography is a versatile nanofabrication technique to fine-tune characteristics such as the NW spacing of highly ordered arrays and can also be operated at wafer-scale to enable mass production.85 Reliable fabrication of highly ordered Si NW arrays using a single step RIE process has been reported before but typically shorter NWs of only about one micron are achieved.86 Hence, we produced comparatively long nanowires of 5 μm which are usually achieved in a multi-step deep RIE (DRIE) process.87 In general, other (semiconductor) materials, such as optically active III–V semiconductors, could be processed by these techniques further broadening the opportunities in potential future applications, e.g., optical stimulation or read-out.88 However, choosing Si as the essential material for the substrate not only benefits from years of experience in the microchip industry but more importantly, Si was chosen as it is considered a promising material for prospective applications such as subcellular bio-interfaces.89The interactions of cells with NW arrays were mapped to a certain extent before, but usually fewer array parameters were altered and only basic cell lines were used. For instance, the culturing of fibroblast cells was studied using varying array densities of randomly arranged NWs but with a fixed NW length of 2 μm.52 In the present work, we used highly ordered NW arrays with varying pitches of 1, 3, 5, and 10 μm resulting in densities of 100, 11.1, 4, and 1 NWs per 100 μm2 and varying lengths of 1.5, 3, and 5 μm to allow for a comprehensive study. Both pitch and length cover the parameter space frequently used in the field.33 Moreover, these characteristics encase sufficiently diverse parameter sets to expect changing settling regimes from a fakir-like state to NW encapsulation modeled by Buch-Månson et al.77 In contrast to practically all of these studies which used basic cell lines or, e.g., primary rodents cells,27 we employed human induced stem cell-derived neural progenitor cells in our studies. Utilizing human iPSCs as the basis for the experiments greatly improves the pertinence for neurodegenerative disease studies such as Alzheimer's disease or Parkinson's disease by enabling the use of patient-specific cells.3,90 Feasibility to culture these patient-derived cells reliably on NW arrays would possibly render many applications such as cell transfection, biomolecule delivery, or biosensing as demonstrated on other cells.29,62,67,73,91–93
Human induced stem cell-derived smNPCs were now cultured successfully on a multitude of different NW arrays. For most of the different arrays used, we can report excellent cell viability of more than 85% which was stable over a culturing period of 4 days and similar to viability in control cells. Neglectable impact on the viability of cells cultured on NW arrays is overall in accordance with the literature.31,34,43,94,95 However, universal conclusions are difficult to draw since many factors such as cell type, culturing time, NW length, and array pitch occasionally play important roles. For example, HEK293 cells showed lower viability when cultured on nanowire arrays with a pitch of 2 μm compared to larger pitches and control.96 Note, the HEK293 cells were in a NW encapsulating regime with partly to fully enwrapping the NWs. Interestingly, reduced viability of the smNPCs of about 75% after 4 DIV was found for the highest NW density combined with long NWs (P1 L ≥ 3). Particularly here, unchanged viability was expected since for high NW densities, the cells rest in a fakir-like state on the NWs without any deformation by the NWs.40,97 In addition, we observed reduced cell numbers on these P1 NW arrays. SEM imaging of these particular samples then showed that the cells form elongations along the array axes.
Imaging of F-actin revealed that the extensions along the array axes were part of the cytoskeleton. In literature, it is reported that well-ordered NW arrays can generate guiding forces to polarize cells or to direct neurite growth.41,54–57,75,98 Many array parameters were modified in these studies, but in the end, primarily the spacing of the nanostructures was contributing decisively, and guiding only occurs within a certain range of array pitches. Nonetheless, a direct comparison can only be made with Bucaro et al. because they used comparable array pitches from 0.8 to 5 μm (but only at a fixed NW length of 5 μm) to fine-tune the polarization of human mesenchymal and rodent stem cells.53 In contrast to our results showing polarization along the array axis with a pitch of 1 μm, polarization appeared in their experiments only with array pitches larger than 1.25 μm and smaller than 3.5 μm. Below 1.25 μm, the surface appears homogeneous to the cells; above 3.5 μm, the cells were not able to reach the next NW. Furthermore, Bucaro et al. explicitly mention that further studies are needed to elaborate whether the morphological changes correlate with proliferation since such analysis was not conducted in their work.
The proliferation as a function of time was investigated here by determining the cell numbers on five consecutive days. As already mentioned in the discussion, we observed fewer cells on the P1 NW arrays with L ≥ 3 NWs. In general, reduced viability and proliferation of cells cultured on densely-spaced NW arrays (fakir-like state) are in contrast to literature. For example, Yan et al. demonstrated that proliferation and viability of murine neural stem cells can be improved using high-density but randomly distributed NW arrays.99 Hence, we assume in the case of the ordered arrays that particularly the reorganization of F-actin at the elongations along the defined array axes interfered with normal proliferation by changing the dynamics of the cytoskeleton.100,101 For the shorter wires, proliferation was normal, since the settling regime changed to an intermediate state and thus the stress on the cytoskeleton was generated to an endurable extent. On samples with larger array pitches (P ≥ 3) and longer NWs (L ≥ 3), namely, encapsulating states, we also found fewer cells. Reduced proliferation on such NW arrays with encapsulation of individual NWs is reasonable due to the challenging topology and is in accordance with the literature.37,50,102 Other studies moreover described actin reorganization at encapsulated nanostructures using osteosarcoma cells which we yet did not spot in the vicinity of the NWs.103 For the shortest NWs (L1.5), we identified normal proliferation independent of the pitch. Although it has been demonstrated that even short NWs can have a negative impact on proliferation,50 usually nanostructures of about 1 μm show no detrimental influence.67,93 The reduced mobility of cells that encapsulate NWs is in good accordance with literature30,37,81,104 whereas mobility is not influenced in a fakir-like regime.52,105
The interaction of the cells with NWs was imaged for all pitches and lengths. The fundamental behavior that shorter nanowires/larger array pitches favor NW encapsulation and longer wires/smaller array pitches encourage a fakir-like regime is in accordance with the literature.52,75,77 Deformation of the nucleus by the nanostructures was also reported when using, for example, fibroblasts or mesenchymal stem cells.50,74,106 Considering the severe deformations of not only the cell but also the nucleus, it is remarkable that the cells maintained their viability even in the case of the longest NWs. The close interaction between the NW and the nucleus might offer the opportunity to use the NW substrates for gene transduction or biomolecule delivery.59,66,107 We believe that the employment of human induced stem cell-derived neural progenitors not only allows for advanced patient-specific neurodegenerative disease studies or pharmacological drug screenings,108 but also that our findings are an excellent starting point to establish a more advanced cell type for next-generation biological metamaterials featuring high aspect ratio nanostructures.109
Summary and conclusions
The cultivation of human induced stem cells and their derivatives on NW arrays to investigate cellular characteristics is still underrepresented in the field. One potential reason is the challenging topography of such substrates in combination with the demanding nature of stem cell culture. In this work, we show that human iPSC-derived neural progenitor cells can be successfully cultured on ordered Si NW arrays featuring a wide range of NW lengths (1.5 to 5 μm) and array pitches (1 to 10 μm). In general, the cells maintained the ability to proliferate on the NW arrays albeit the overall cell numbers varied in correlation with the topological difficulty, e.g., long NWs. A special case was found for high-density NW arrays, where we observed a reorganization of the cytoskeleton along the array axes which interfered with normal proliferation. We mapped the interaction of the human smNPCs with the NWs and found that in some configurations the cells encapsulated the NWs completely. Moreover, in these cases, the NWs strongly interacted with the nuclei. Remarkably, despite the heavy deformations of the cells, the viability was essentially unimpaired. Thus, we are convinced that our results are a promising starting point to apply human neural progenitor cells to nanowire arrays for future applications in stem cell research and regenerative medicine.Materials and methods
Nanowire fabrication
The NWs were fabricated in a top-down process with reactive ion etching (RIE) using a chromium hard mask. The mask was predefined via electron-beam lithography (Raith Voyager e-beam lithography system) in a 90 nm thick layer of positive photoresists (PMMA, AllResist, AR-P 671.02). Circles with 700 nm diameter were defined in a rectangular arrangement with pitches of 1, 3, 5, and 10 μm. After developing (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 MIBK
3 MIBK![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IPA, 2.7% H2O for 30 s and IPA for 30 s), a 65 nm thick layer of chromium was deposited in a physical vapor deposition (PVD) process. The samples were etched with an inductively coupled plasma (ICP)-RIE (Sentech SI500) using 200 W ICP power, 50 W RF power, 2 Pa pressure, 25 sccm SF6, 50 sccm C4F8 at 0 °C electrode temperature. The etch rate was about 300 nm min−1. To remove residues from the RIE procedure, the samples were processed with a KOH dip for a few seconds at 40 °C. To ease handling, the substrates were glued with polydimethylsiloxane (PDMS, SYLGARD® 184 Elastomer Kit, Dow Corning) to 12 × 12 mm2 glass coverslips. For cell culture, the samples were placed in a well of a 12-well plate, sterilized in 70% ethanol for 5 min, and coated with Matrigel® (1 ml per well, cf. section ‘cell culture’). After cell culture and data collection, samples were cleaned with Terg-a-zyme® (10 mg ml−1) for 30 min at 37 °C for reuse purposes.
IPA, 2.7% H2O for 30 s and IPA for 30 s), a 65 nm thick layer of chromium was deposited in a physical vapor deposition (PVD) process. The samples were etched with an inductively coupled plasma (ICP)-RIE (Sentech SI500) using 200 W ICP power, 50 W RF power, 2 Pa pressure, 25 sccm SF6, 50 sccm C4F8 at 0 °C electrode temperature. The etch rate was about 300 nm min−1. To remove residues from the RIE procedure, the samples were processed with a KOH dip for a few seconds at 40 °C. To ease handling, the substrates were glued with polydimethylsiloxane (PDMS, SYLGARD® 184 Elastomer Kit, Dow Corning) to 12 × 12 mm2 glass coverslips. For cell culture, the samples were placed in a well of a 12-well plate, sterilized in 70% ethanol for 5 min, and coated with Matrigel® (1 ml per well, cf. section ‘cell culture’). After cell culture and data collection, samples were cleaned with Terg-a-zyme® (10 mg ml−1) for 30 min at 37 °C for reuse purposes.
Cell culture
The smNPCs used in this study were derived from human iPSCs as previously described by Reinhardt et al.110 All experiments were conducted in accordance with the ethical statement in Reinhardt et al.110 The generated smNPCs were then cultivated in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of DMEM/F12 and Neurobasal medium supplemented with 1% penicillin/streptomycin/glutamine (100×), 1% B27 supplement without vitamin A (50×), 0.5% N2 supplement (100×) (Life Technologies, Carlsbad, CA, USA), 100 μM ascorbic acid (Sigma-Aldrich, St Louis, MO, USA), 0.5 μM smoothened agonist (Biomol, Hamburg, Germany), 3 μM CHIR 99021 (Axon MedChem, Groningen, Netherlands) in a humidified atmosphere at 37 °C and 5% CO2. The medium was exchanged every 2–3 days. Cells were kept in Matrigel-coated 6-well plates and split in a ratio of 1
1 mixture of DMEM/F12 and Neurobasal medium supplemented with 1% penicillin/streptomycin/glutamine (100×), 1% B27 supplement without vitamin A (50×), 0.5% N2 supplement (100×) (Life Technologies, Carlsbad, CA, USA), 100 μM ascorbic acid (Sigma-Aldrich, St Louis, MO, USA), 0.5 μM smoothened agonist (Biomol, Hamburg, Germany), 3 μM CHIR 99021 (Axon MedChem, Groningen, Netherlands) in a humidified atmosphere at 37 °C and 5% CO2. The medium was exchanged every 2–3 days. Cells were kept in Matrigel-coated 6-well plates and split in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 to 1
10 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 every 4–5 days close to confluency using Accutase® (Sigma-Aldrich). Coating with Matrigel (Corning 354263, stored in a 1
20 every 4–5 days close to confluency using Accutase® (Sigma-Aldrich). Coating with Matrigel (Corning 354263, stored in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 dilution at −20 °C and additionally diluted 1
5 dilution at −20 °C and additionally diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 for application, diluted in KnockOut DMEM, Life Technologies) was performed with 1.5 mL per well overnight at room temperature or for 1 h at 37 °C. Per well of the 12-well plate which contained the samples, 125k cells were seeded. The preservation of the neural progenitor cell (NPC) state after 4 days of culturing on the NW arrays was verified using NPC markers (SOX2 and NES (nestin), protocol in the ESI†) and the recordings for NW and control samples are shown in Fig. S6 (SOX2) and Fig. S7 (NES) in the ESI.†
30 for application, diluted in KnockOut DMEM, Life Technologies) was performed with 1.5 mL per well overnight at room temperature or for 1 h at 37 °C. Per well of the 12-well plate which contained the samples, 125k cells were seeded. The preservation of the neural progenitor cell (NPC) state after 4 days of culturing on the NW arrays was verified using NPC markers (SOX2 and NES (nestin), protocol in the ESI†) and the recordings for NW and control samples are shown in Fig. S6 (SOX2) and Fig. S7 (NES) in the ESI.†
Cell viability
The cells were stained with calcein (0.5 μM, acetoxymethyl esters (calcein-AM), ex/em: 495/515 nm, Thermo Fisher Scientific, Waltham, MA, USA) to identify viable cells and with propidium iodide (PI, 10 μM, Sigma-Aldrich, ex/em: 535/617 nm) to indicate dead cells. Counterstain was Hoechst 33342 (5 mg mL−1 in H2O, ex/em 361/497 nm). Samples were washed with Dulbecco's phosphate-buffered saline (PBS) and staining was performed with calcein-AM, PI, and Hoechst diluted in PBS for 15 min at 37 °C and 5% CO2 in the incubator. Subsequently, the samples were rinsed three times prior to imaging (Nikon Eclipse FN1 microscope with a 10× objective and DSRi2 camera). Data were collected in three independent experiments with each 5–6 images per combination of NW length and array pitch. Control was collected from planarly etched silicon wafer without NWs next to the NW arrays. Note, that proliferation, viability, and spreading on the etched Si were similar compared to control on conventional glass coverslips as shown in the ESI (Fig. S1†). Image size was 1282 × 853 μm2.Confocal laser scanning microscopy
A Leica TCS SP8 microscope in upright configuration equipped with 488 and 638 nm wavelength laser sources was used for confocal scanning laser microscopy. Cells were stained with phalloidin (ActinGreen™ 488 ReadyProbes™ Reagent, ex/em: 495/518 nm Thermo Fisher Scientific, Massachusetts, USA) and DRAQ5 (DRAQ5 fluorescent probe, ex/em: 647/681 nm, Thermo Fisher Scientific). Samples were rinsed with PBS, fixed with 4% formaldehyde in PBS for 10 min at room temperature (RT), and rinsed three times with PBS. Cells were permeabilized and blocked with 3% bovine serum albumin (BSA), 0.1% Tween 20, and 0.1% Triton X-100 in PBS for 45 min at RT. Staining with phalloidin (2 drops per mL) and DRAQ5 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) in PBS with 0.1% BSA was performed in the dark for 1 h at RT. Samples were washed three times with 0.05% Tween 20 in PBS in the dark for 5 min at RT. For imaging, the samples were transferred from the 12-well plate onto a microscope slide and covered with a precision glass cover slide to use the water immersion objectives. Z-stacks were recorded with a slicing step size of 172 nm and analysed using Leica LAS X Core software.
1000) in PBS with 0.1% BSA was performed in the dark for 1 h at RT. Samples were washed three times with 0.05% Tween 20 in PBS in the dark for 5 min at RT. For imaging, the samples were transferred from the 12-well plate onto a microscope slide and covered with a precision glass cover slide to use the water immersion objectives. Z-stacks were recorded with a slicing step size of 172 nm and analysed using Leica LAS X Core software.
Scanning electron microscopy
SEM images were prepared with a Crossbeam 550 from Zeiss. For this purpose, the cells were rinsed with PBS and fixed with 4% formaldehyde in PBS for 10 min at RT. The solution was exchanged with deionized water followed by dehydration in a step-wise ethanol exchange. Subsequently, the cells were critical point dried (Tousimis Autosamdri-815) and sputter-coated with a 20 nm gold layer to avoid charging effects.Image and data analysis
Cells were identified in the Hoechst counterstain channel and cross-correlated to viable and dead cells in the calcein and PI channel, respectively, with CellProfiler 4.1.3.111 Images of identified viable cells were exported in order to analyse clustering with ImageJ/Fiji112 using the ‘SSIDC Cluster Indicator’ in the BioVoxxel toolbox.113 Distances of cells in clusters were determined with the ‘nearest neighbour distances’ plugin for ImageJ/Fiji. Orientations of the extensions were quantified with OrientationJ in ImageJ/Fiji. Data were processed and plots were prepared with Origin (v.2021). Statistical analysis was done by an ANOVA analysis with a post-hoc Tukey's test using the ‘paired comparison plot’ application. Differences were considered significant for α = 0.05, α = 0.01, and α = 0.001. For publication, the images were optimized in contrast and brightness.Author contributions
JH, KB, MS conducted cell culture. JH, KB, MS conducted epifluorescence microscopy imaging. UH, OP helped with smNPC characterization. JH conducted confocal microscopy imaging. JH, CH prepared the NW substrates. JH, KB analysed the data. JH wrote the manuscript. JH, RZ conceptualized the study. RZ, RHB supervised the study. JH, KB, MS, RZ, MG, HRS revised the manuscript. RZ, RHB acquired funding. All authors conducted proofreading and approved the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by the Deutsche Forschungs-gemeinschaft (DFG) via the Priority Program ‘ESSENCE’ (Schwerpunktprogramm SPP-1857), via the Excellence Cluster ‘Advanced Imaging of Matter, AIM’ (EXC-2056), via the Collaborative Research Initiative SFB 986 ‘Tailor-Made Multi-Scale Materials Systems’ (project number 192346071), and the Joachim-Herz Foundation via the Research Group, ‘Infecto-Physics’, via the Focused Research Group, ‘Bio-Pict’. We also acknowledge the Federal Ministry for Research (BMBF) for support within the ‘Forschungslabor Mikroelektronik Deutschland, ForLab’ via the HELIOS-group. The study was further funded by the BMBF project NeuRIMS (FKZ 16GW0309). Michael Glatza was sponsored by a fellowship from the Hans and Ilse Breuer Stiftung.References
- J. Deinsberger, D. Reisinger and B. Weber, NPJ Regen. Med., 2020, 5, 15 CrossRef PubMed.
- B. Lo and L. Parham, Endocr. Rev., 2009, 30, 204–213 CrossRef PubMed.
- Y. Li, H. V. Nguyen and S. H. Tsang, Methods Mol. Biol., 2015, 1353, 77–88 CrossRef PubMed.
- M. Dragunow, Nat. Rev. Drug Discovery, 2008, 7, 659–666 CrossRef CAS PubMed.
- J. Seok, H. S. Warren, A. G. Cuenca, M. N. Mindrinos, H. V. Baker, W. Xu, D. R. Richards, G. P. McDonald-Smith, H. Gao, L. Hennessy, C. C. Finnerty, C. M. López, S. Honari, E. E. Moore, J. P. Minei, J. Cuschieri, P. E. Bankey, J. L. Johnson, J. Sperry, A. B. Nathens, T. R. Billiar, M. A. West, M. G. Jeschke, M. B. Klein, R. L. Gamelli, N. S. Gibran, B. H. Brownstein, C. Miller-Graziano, S. E. Calvano, P. H. Mason, J. P. Cobb, L. G. Rahme, S. F. Lowry, R. V. Maier, L. L. Moldawer, D. N. Herndon, R. W. Davis, W. Xiao and R. G. Tompkins, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 3507–3512 CrossRef CAS PubMed.
- X. Xu, E. I. Stoyanova, A. E. Lemiesz, J. Xing, D. C. Mash and N. Heintz, eLife, 2018, 7, e37551 CrossRef PubMed.
- H. C. Ko and B. D. Gelb, Stem Cells Transl. Med., 2014, 3, 500–509 CrossRef CAS PubMed.
- J. A. Garcia-Leon, L. Caceres-Palomo, E. Sanchez-Mejias, M. Mejias-Ortega, C. Nuñez-Diaz, J. J. Fernandez-Valenzuela, R. Sanchez-Varo, J. C. Davila, J. Vitorica and A. Gutierrez, Int. J. Mol. Sci., 2020, 21, 6867 CrossRef CAS PubMed.
- B. Xiao, H. H. Ng, R. Takahashi and E.-K. Tan, J. Neurol. Neurosurg. Psychiatry, 2016, 87, 697–702 CrossRef PubMed.
- L. M. Smits, L. Reinhardt, P. Reinhardt, M. Glatza, A. S. Monzel, N. Stanslowsky, M. D. Rosato-Siri, A. Zanon, P. M. Antony, J. Bellmann, S. M. Nicklas, K. Hemmer, X. Qing, E. Berger, N. Kalmbach, M. Ehrlich, S. Bolognin, A. A. Hicks, F. Wegner, J. L. Sterneckert and J. C. Schwamborn, npj Parkinson's Dis., 2019, 5, 5 CrossRef PubMed.
- A. Appelt-Menzel, S. Oerter, S. Mathew, U. Haferkamp, C. Hartmann, M. Jung, W. Neuhaus and O. Pless, Curr. Protoc. Stem Cell Biol., 2020, 55, e122 CAS.
- M. A. Lancaster, M. Renner, C.-A. Martin, D. Wenzel, L. S. Bicknell, M. E. Hurles, T. Homfray, J. M. Penninger, A. P. Jackson and J. A. Knoblich, Nature, 2013, 501, 373–379 CrossRef CAS PubMed.
- K. Takahashi and S. Yamanaka, Cell, 2006, 126, 663–676 CrossRef CAS PubMed.
- M. W. Tibbitt, C. B. Rodell, J. A. Burdick and K. S. Anseth, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 14444–14451 CrossRef CAS PubMed.
- I. Armentano, L. Tarpani, F. Morena, S. Martino, L. Latterini and L. Torre, Curr. Org. Chem., 2018, 22, 1193–1204 CrossRef CAS.
- C. Leclech and C. Villard, Front. Bioeng. Biotechnol., 2020, 8, 1198 Search PubMed.
- C. Simitzi, A. Ranella and E. Stratakis, Acta Biomater., 2017, 51, 21–52 CrossRef CAS PubMed.
- M. Marcus, K. Baranes, M. Park, I. S. Choi, K. Kang and O. Shefi, Adv. Healthcare Mater., 2017, 6, 1700267 CrossRef PubMed.
- W. Zhang, Y. Yang and B. Cui, Curr. Opin. Solid State Mater. Sci., 2021, 25, 100873 CrossRef CAS PubMed.
- M. Lampin, R. Warocquier-Clérout, C. Legris, M. Degrange and M. F. Sigot-Luizard, J. Biomed. Mater. Res., 1997, 36, 99–108 CrossRef CAS PubMed.
- Y. H. Kim, N. S. Baek, Y. H. Han, M. A. Chung and S. D. Jung, J. Neurosci. Methods, 2011, 202, 38–44 CrossRef CAS PubMed.
- C. Fendler, J. Harberts, L. Rafeldt, G. Loers, R. Zierold and R. H. Blick, Nanoscale Adv., 2020, 2, 5192–5200 RSC.
- A. Koitmäe, J. Harberts, G. Loers, M. Müller, C. S. Bausch, D. Sonnenberg, C. Heyn, R. Zierold, W. Hansen and R. H. Blick, Adv. Mater. Interfaces, 2016, 3, 1600746 CrossRef.
- A. Koitmäe, M. Müller, C. S. Bausch, J. Harberts, W. Hansen, G. Loers and R. H. Blick, Langmuir, 2018, 34, 1528–1534 CrossRef PubMed.
- C. Leclech and A. I. Barakat, Cytoskeleton, 2021, 78, 284–292 CrossRef PubMed.
- J. Harberts, C. Fendler, J. Teuber, M. Siegmund, A. Silva, N. Rieck, M. Wolpert, R. Zierold and R. H. Blick, ACS Nano, 2020, 14, 13091–13102 CrossRef CAS PubMed.
- G. He, N. Hu, A. M. Xu, X. Li, Y. Zhao and X. Xie, Adv. Funct. Mater., 2020, 30, 1909890 CrossRef CAS.
- M. L. Khraiche and R. El Hassan, J. Sci. Adv. Mater. Devices, 2020, 5, 279–294 CrossRef.
- A. Tay and N. Melosh, Acc. Chem. Res., 2019, 52, 2462–2471 CrossRef CAS PubMed.
- A. F. McGuire, F. Santoro and B. Cui, Annu. Rev. Anal. Chem., 2018, 11, 101–126 CrossRef PubMed.
- T. Berthing, S. Bonde, C. B. Sørensen, P. Utko, J. Nygård and K. L. Martinez, Small, 2011, 7, 640–647 CrossRef CAS PubMed.
- R. Elnathan, M. Kwiat, F. Patolsky and N. H. Voelcker, Nano Today, 2014, 9, 172–196 CrossRef CAS.
- S. Bonde, N. Buch-Månson, K. R. Rostgaard, T. K. Andersen, T. Berthing and K. L. Martinez, Nanotechnology, 2014, 25, 362001 CrossRef PubMed.
- W. Hällström, T. Mårtensson, C. Prinz, P. Gustavsson, L. Montelius, L. Samuelson and M. Kanje, Nano Lett., 2007, 7, 2960–2965 CrossRef PubMed.
- Z. Li, R. Yang, M. Yu, F. Bai, C. Li and Z. L. Wang, J. Phys. Chem. C, 2008, 112, 20114–20117 CrossRef CAS.
- G. Piret, M.-T. Perez and C. N. Prinz, Biomaterials, 2013, 34, 875–887 CrossRef CAS PubMed.
- Z. Li, S. Kamlund, T. Ryser, M. Lard, S. Oredsson and C. N. Prinz, J. Mater. Chem. B, 2018, 6, 7042–7049 RSC.
- Z. Li, H. Persson, K. Adolfsson, S. Oredsson and C. N. Prinz, Sci. China: Life Sci., 2018, 61, 427–435 Search PubMed.
- C. S. Hansel, S. W. Crowder, S. Cooper, S. Gopal, M. João Pardelha da Cruz, L. de Oliveira Martins, D. Keller, S. Rothery, M. Becce, A. E. G. Cass, C. Bakal, C. Chiappini and M. M. Stevens, ACS Nano, 2019, 13, 2913–2926 CrossRef CAS PubMed.
- J. Harberts, R. Zierold, C. Fendler, A. Koitmäe, P. Bayat, I. Fernandez-Cuesta, G. Loers, B.-P. Diercks, R. Fliegert, A. H. Guse, C. Ronning, G. Otnes, M. Borgström and R. H. Blick, RSC Adv., 2019, 9, 11194–11201 RSC.
- J. Harberts, U. Haferkamp, S. Haugg, C. Fendler, D. Lam, R. Zierold, O. Pless and R. H. Blick, Biomater. Sci., 2020, 8, 2434–2446 RSC.
- J. Harberts, M. Siegmund, M. Schnelle, T. Zhang, Y. Lei, L. Yu, R. Zierold and R. H. Blick, Sci. Rep., 2021, 11, 18819 CrossRef CAS PubMed.
- G. Tullii, F. Giona, F. Lodola, S. Bonfadini, C. Bossio, S. Varo, A. Desii, L. Criante, C. Sala, M. Pasini, C. Verpelli, F. Galeotti and M. R. Antognazza, ACS Appl. Mater. Interfaces, 2019, 11, 28125–28137 CrossRef CAS PubMed.
- Z. Li, J. Song, G. Mantini, M.-Y. Lu, H. Fang, C. Falconi, L.-J. Chen and Z. L. Wang, Nano Lett., 2009, 9, 3575–3580 CrossRef CAS PubMed.
- W. Hällström, M. Lexholm, D. B. Suyatin, G. Hammarin, D. Hessman, L. Samuelson, L. Montelius, M. Kanje and C. N. Prinz, Nano Lett., 2010, 10, 782–787 CrossRef PubMed.
- Z. Li, H. Persson, K. Adolfsson, L. Abariute, M. T. Borgström, D. Hessman, K. Åström, S. Oredsson and C. N. Prinz, Nanoscale, 2017, 9, 19039–19044 RSC.
- L. Hanson, W. Zhao, H.-Y. Lou, Z. C. Lin, S. W. Lee, P. Chowdary, Y. Cui and B. Cui, Nat. Nanotechnol., 2015, 10, 554–562 CrossRef CAS PubMed.
- K. S. Beckwith, S. Ullmann, J. Vinje and P. Sikorski, Small, 2019, 15, 1902514 CrossRef CAS PubMed.
- S. Qi, C. Yi, S. Ji, C.-C. Fong and M. Yang, ACS Appl. Mater. Interfaces, 2009, 1, 30–34 CrossRef CAS PubMed.
- H. Persson, C. Købler, K. Mølhave, L. Samuelson, J. O. Tegenfeldt, S. Oredsson and C. N. Prinz, Small, 2013, 9, 4006–4016 CrossRef CAS PubMed.
- H. Persson, Z. Li, J. O. Tegenfeldt, S. Oredsson and C. N. Prinz, Sci. Rep., 2015, 5, 18535 CrossRef CAS PubMed.
- N. Buch-Månson, D.-H. Kang, D. Kim, K. E. Lee, M.-H. Yoon and K. L. Martinez, Nanoscale, 2017, 9, 5517–5527 RSC.
- M. A. Bucaro, Y. Vasquez, B. D. Hatton and J. Aizenberg, ACS Nano, 2012, 6, 6222–6230 CrossRef CAS PubMed.
- F. Milos, A. Belu, D. Mayer, V. Maybeck and A. Offenhäusser, Adv. Biol., 2021, 5, 2000248 CrossRef CAS.
- V. Gautam, S. Naureen, N. Shahid, Q. Gao, Y. Wang, D. Nisbet, C. Jagadish and V. R. Daria, Nano Lett., 2017, 17, 3369–3375 CrossRef CAS PubMed.
- H. Amin, M. Dipalo, F. De Angelis and L. Berdondini, ACS Appl. Mater. Interfaces, 2018, 10, 15207–15215 CrossRef CAS PubMed.
- M. Park, E. Oh, J. Seo, M.-H. Kim, H. Cho, J. Y. Choi, H. Lee and I. S. Choi, Small, 2016, 12, 1148–1152 CrossRef CAS PubMed.
- J. J. VanDersarl, A. M. Xu and N. A. Melosh, Nano Lett., 2012, 12, 3881–3886 CrossRef CAS PubMed.
- Y. Wang, Y. Yang, L. Yan, S. Y. Kwok, W. Li, Z. Wang, X. Zhu, G. Zhu, W. Zhang, X. Chen and P. Shi, Nat. Commun., 2014, 5, 4466 CrossRef CAS PubMed.
- X. Xie, A. Aalipour, S. V. Gupta and N. A. Melosh, ACS Nano, 2015, 9, 11667–11677 CrossRef CAS PubMed.
- C. Chiappini, J. O. Martinez, E. De Rosa, C. S. Almeida, E. Tasciotti and M. M. Stevens, ACS Nano, 2015, 9, 5500–5509 CrossRef CAS PubMed.
- Y. Cao, H. Chen, R. Qiu, M. Hanna, E. Ma, M. Hjort, A. Zhang, R. S. Lewis, J. C. Wu and N. A. Melosh, Sci. Adv., 2018, 4, eaat8131 CrossRef CAS PubMed.
- S. Gopal, C. Chiappini, J. Penders, V. Leonardo, H. Seong, S. Rothery, Y. Korchev, A. Shevchuk and M. M. Stevens, Adv. Mater., 2019, 31, 1806788 CrossRef PubMed.
- C. Chiappini, E. De Rosa, J. O. Martinez, X. Liu, J. Steele, M. M. Stevens and E. Tasciotti, Nat. Mater., 2015, 14, 532–539 CrossRef CAS PubMed.
- W. Kim, J. K. Ng, M. E. Kunitake, B. R. Conklin and P. Yang, J. Am. Chem. Soc., 2007, 129, 7228–7229 CrossRef CAS PubMed.
- R. Elnathan, B. Delalat, D. Brodoceanu, H. Alhmoud, F. J. Harding, K. Buehler, A. Nelson, L. Isa, T. Kraus and N. H. Voelcker, Adv. Funct. Mater., 2015, 25, 7215–7225 CrossRef CAS.
- Y. Chen, S. Aslanoglou, G. Gervinskas, H. Abdelmaksoud, N. H. Voelcker and R. Elnathan, Small, 2019, 15, 1904819 CrossRef CAS PubMed.
- C. Xie, Z. Lin, L. Hanson, Y. Cui and B. Cui, Nat. Nanotechnol., 2012, 7, 185–190 CrossRef CAS PubMed.
- J. T. Robinson, M. Jorgolli, A. K. Shalek, M.-H. Yoon, R. S. Gertner and H. Park, Nat. Nanotechnol., 2012, 7, 180–184 CrossRef CAS PubMed.
- K.-Y. Lee, I. Kim, S.-E. Kim, D.-W. Jeong, J.-J. Kim, H. Rhim, J.-P. Ahn, S.-H. Park and H.-J. Choi, Nanoscale Res. Lett., 2014, 9, 56 CrossRef PubMed.
- Z. C. Lin, C. Xie, Y. Osakada, Y. Cui and B. Cui, Nat. Commun., 2014, 5, 3206 CrossRef PubMed.
- R. Liu, R. Chen, A. T. Elthakeb, S. H. Lee, S. Hinckley, M. L. Khraiche, J. Scott, D. Pre, Y. Hwang, A. Tanaka, Y. G. Ro, A. K. Matsushita, X. Dai, C. Soci, S. Biesmans, A. James, J. Nogan, K. L. Jungjohann, D. V. Pete, D. B. Webb, Y. Zou, A. G. Bang and S. A. Dayeh, Nano Lett., 2017, 17, 2757–2764 CrossRef CAS PubMed.
- X. Li, J. Mo, J. Fang, D. Xu, C. Yang, M. Zhang, H. Li, X. Xie, N. Hu and F. Liu, J. Mater. Chem. B, 2020, 8, 7609–7632 RSC.
- L. Hanson, Z. C. Lin, C. Xie, Y. Cui and B. Cui, Nano Lett., 2012, 12, 5815–5820 CrossRef CAS PubMed.
- N. Buch-Månson, A. Spangenberg, L. P. C. Gomez, J.-P. Malval, O. Soppera and K. L. Martinez, Sci. Rep., 2017, 7, 9247 CrossRef PubMed.
- F. Santoro, W. Zhao, L.-M. Joubert, L. Duan, J. Schnitker, Y. van de Burgt, H.-Y. Lou, B. Liu, A. Salleo, L. Cui, Y. Cui and B. Cui, ACS Nano, 2017, 11, 8320–8328 CrossRef CAS PubMed.
- N. Buch-Månson, S. Bonde, J. Bolinsson, T. Berthing, J. Nygård and K. L. Martinez, Adv. Funct. Mater., 2015, 25, 3246–3255 CrossRef.
- S. W. Crowder, V. Leonardo, T. Whittaker, P. Papathanasiou and M. M. Stevens, Cell Stem Cell, 2016, 18, 39–52 CrossRef CAS PubMed.
- H.-Y. Lou, W. Zhao, Y. Zeng and B. Cui, Acc. Chem. Res., 2018, 51, 1046–1053 CrossRef CAS PubMed.
- R. Liu and J. Ding, ACS Appl. Mater. Interfaces, 2020, 12, 35799–35812 CrossRef CAS PubMed.
- H. Kim, I. Kim, H.-J. Choi, S. Y. Kim and E. G. Yang, Nanoscale, 2015, 7, 17131–17138 RSC.
- C. H. Rasmussen, P. M. Reynolds, D. R. Petersen, M. Hansson, R. M. McMeeking, M. Dufva and N. Gadegaard, Adv. Funct. Mater., 2016, 26, 815–823 CrossRef CAS.
- H. S. Kim and H. S. Yoo, RSC Adv., 2015, 5, 49508–49512 RSC.
- H. Jansen, H. Gardeniers, M. de Boer, M. Elwenspoek and J. Fluitman, J. Micromech. Microeng., 1996, 6, 14–28 CrossRef CAS.
- Y. Chen, Microelectron. Eng., 2015, 135, 57–72 CrossRef CAS.
- Y.-J. Hung, S.-L. Lee, B. J. Thibeault and L. A. Coldren, IEEE J. Sel. Top. Quantum Electron., 2011, 17, 869–877 CAS.
- Y. Q. Fu, A. Colli, A. Fasoli, J. K. Luo, A. J. Flewitt, A. C. Ferrari and W. I. Milne, J. Vac. Sci. Technol., B: Microelectron. Nanometer Struct.–Process., Meas., Phenom., 2009, 27, 1520 CrossRef CAS.
- F. Karouta, J. Phys. D: Appl. Phys., 2014, 47, 233501 CrossRef.
- H. Acaron Ledesma and B. Tian, J. Mater. Chem. B, 2017, 5, 4276–4289 RSC.
- A. P. Reddy, J. Ravichandran and N. Carkaci-Salli, Biochim. Biophys. Acta, Mol. Basis Dis., 2020, 1866, 165506 CrossRef CAS PubMed.
- X. Xu, S. Hou, N. Wattanatorn, F. Wang, Q. Yang, C. Zhao, X. Yu, H.-R. Tseng, S. J. Jonas and P. S. Weiss, ACS Nano, 2018, 12, 4503–4511 CrossRef CAS PubMed.
- M. Lard, H. Linke and C. N. Prinz, Nanotechnology, 2019, 30, 214003 CrossRef CAS PubMed.
- L. Schmiderer, A. Subramaniam, K. Žemaitis, A. Bäckström, D. Yudovich, S. Soboleva, R. Galeev, C. N. Prinz, J. Larsson and M. Hjort, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 21267–21273 CrossRef CAS PubMed.
- A. K. Shalek, J. T. Gaublomme, L. Wang, N. Yosef, N. Chevrier, M. S. Andersen, J. T. Robinson, N. Pochet, D. Neuberg, R. S. Gertner, I. Amit, J. R. Brown, N. Hacohen, A. Regev, C. J. Wu and H. Park, Nano Lett., 2012, 12, 6498–6504 CrossRef CAS PubMed.
- C. N. Prinz, J. Phys.: Condens. Matter, 2015, 27, 233103 CrossRef PubMed.
- S. Bonde, T. Berthing, M. H. Madsen, T. K. Andersen, N. Buch-Månson, L. Guo, X. Li, F. Badique, K. Anselme, J. Nygård and K. L. Martinez, ACS Appl. Mater. Interfaces, 2013, 5, 10510–10519 CrossRef CAS PubMed.
- S.-M. Kim, S. Lee, D. Kim, D.-H. Kang, K. Yang, S.-W. Cho, J. S. Lee, I. S. Choi, K. Kang and M.-H. Yoon, Nano Res., 2018, 11, 2532–2543 CrossRef CAS.
- A. Kundu, L. Micholt, S. Friedrich, D. R. Rand, C. Bartic, D. Braeken and A. Levchenko, Lab Chip, 2013, 13, 3070 RSC.
- Q. Yan, L. Fang, J. Wei, G. Xiao, M. Lv, Q. Ma, C. Liu and W. Wang, J. Biomater. Sci., Polym. Ed., 2017, 28, 1394–1407 CrossRef CAS PubMed.
- P. P. Provenzano and P. J. Keely, J. Cell Sci., 2011, 124, 1195–1205 CrossRef CAS PubMed.
- N. Bendris, B. Lemmers and J. M. Blanchard, Cell Cycle, 2015, 14, 1786–1798 CrossRef CAS PubMed.
- D. Ning, B. Duong, G. Thomas, Y. Qiao, L. Ma, Q. Wen and M. Su, Langmuir, 2016, 32, 2718–2723 CrossRef CAS PubMed.
- H.-Y. Lou, W. Zhao, X. Li, L. Duan, A. Powers, M. Akamatsu, F. Santoro, A. F. McGuire, Y. Cui, D. G. Drubin and B. Cui, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 23143–23151 CrossRef CAS PubMed.
- C. Xie, L. Hanson, W. Xie, Z. Lin, B. Cui and Y. Cui, Nano Lett., 2010, 10, 4020–4024 CrossRef CAS PubMed.
- F. Viela, D. Granados, A. Ayuso-Sacido and I. Rodríguez, Adv. Funct. Mater., 2016, 26, 5599–5609 CrossRef CAS.
- J. Carthew, H. H. Abdelmaksoud, M. Hodgson-Garms, S. Aslanoglou, S. Ghavamian, R. Elnathan, J. P. Spatz, J. Brugger, H. Thissen, N. H. Voelcker, V. J. Cadarso and J. E. Frith, Adv. Sci., 2021, 8, 2003186 CrossRef CAS PubMed.
- J.-A. Huang, V. Caprettini, Y. Zhao, G. Melle, N. Maccaferri, L. Deleye, X. Zambrana-Puyalto, M. Ardini, F. Tantussi, M. Dipalo and F. De Angelis, Nano Lett., 2019, 19, 722–731 CrossRef CAS PubMed.
- A. Farkhondeh, R. Li, K. Gorshkov, K. G. Chen, M. Might, S. Rodems, D. C. Lo and W. Zheng, Drug Discovery Today, 2019, 24, 992–999 CrossRef CAS PubMed.
- S. G. Higgins, M. Becce, A. Belessiotis-Richards, H. Seong, J. E. Sero and M. M. Stevens, Adv. Mater., 2020, 32, 1903862 CrossRef CAS PubMed.
- P. Reinhardt, M. Glatza, K. Hemmer, Y. Tsytsyura, C. S. Thiel, S. Höing, S. Moritz, J. A. Parga, L. Wagner, J. M. Bruder, G. Wu, B. Schmid, A. Röpke, J. Klingauf, J. C. Schwamborn, T. Gasser, H. R. Schöler and J. Sterneckert, PLoS One, 2013, 8, e59252 CrossRef CAS PubMed.
- C. McQuin, A. Goodman, V. Chernyshev, L. Kamentsky, B. A. Cimini, K. W. Karhohs, M. Doan, L. Ding, S. M. Rafelski, D. Thirstrup, W. Wiegraebe, S. Singh, T. Becker, J. C. Caicedo and A. E. Carpenter, PLoS Biol., 2018, 16, e2005970 CrossRef PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- J. Brocher, The BioVoxxel Image Processing and Analysis Toolbox, EuBIAS-Conference, 2015, Jan 5 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1: Comparison of cells cultured on glass control and planarly etched Si substrate. Fig. S2: Data from quantification that is not shown in the main manuscript. Fig. S3: Viable cells 1 DIV after plating. Fig. S4: SEM images of smNPCs cultured on the remaining NW substrates and on a control substrate. Fig. S5: F-actin/cytoskeleton on control substrate. Fig. S6: Exemplary anti-SOX2 stainings on all types of NW substrates and glass and Si control after 4 DIV. Fig. S7: Exemplary images of anti-NES stainings on all types of NW substrates and glass and Si control after 4 DIV. See DOI: 10.1039/d1nr04352h |
| This journal is © The Royal Society of Chemistry 2021 |